Abstract
Wastewater1 surveillance of SARS-CoV-2 may be a useful supplement to clinical surveillance as it is shed in feces, there are many asymptomatic cases, and diagnostic testing can have capacity limitations and extended time to results. Although numerous studies have utilized wastewater surveillance for SARS-CoV-2, the methods used were developed and/or standardized for other pathogens. This study evaluates multiple methods for concentration and recovery of SARS-CoV-2 and seeded human coronavirus OC43 from municipal primary wastewater and/or sludge from the Greater Seattle Area (March–July 2020). Methods evaluated include the bag-mediated filtration system (BMFS), with and without Vertrel™ extraction, skimmed milk flocculation, with and without Vertrel™ extraction, polyethylene glycol (PEG) precipitation, ultrafiltration, and sludge extraction. Total RNA was extracted from wastewater concentrates and analyzed for SARS-CoV-2 and OC43 with RT-qPCR. Skimmed milk flocculation without Vertrel™ extraction performed consistently over time and between treatment plants in Seattle-area wastewater with the lowest average OC43 Cq value and smallest variability (24.3; 95% CI: 23.8–24.9), most frequent SARS-CoV-2 detection (48.8% of sampling events), and highest average OC43 percent recovery (9.1%; 95% CI: 6.2–11.9%). Skimmed milk flocculation is also beneficial because it is feasible in low-resource settings. While the BMFS had the highest average volume assayed of 11.9 mL (95% CI: 10.7–13.1 mL), the average OC43 percent recovery was low (0.7%; 95% CI: 0.4–1.0%). Ultrafiltration and PEG precipitation had low average OC43 percent recoveries of 1.0% (95% CI: 0.5–1.6%) and 3.2% (95% CI: 1.3–5.1%), respectively. The slopes and efficiency for the SARS-CoV-2 standard curves were not consistent over time, confirming the need to include a standard curve each run rather than using a single curve for multiple plates. Results suggest that the concentration and detection methods used must be validated for the specific water matrix using a recovery control to assess performance over time.
Keywords: SARS-CoV-2, Wastewater, Environmental surveillance
Graphical abstract
1. Introduction
In December 2019, an outbreak of pneumonia of unknown etiology associated with the live animal market in the Hubei Province of China was first reported to the World Health Organization (WHO) (World Health Organization, n.d.). This pneumonia would later be classified as COVID-19, the disease caused by the novel coronavirus SARS-CoV-2. SARS-CoV-2 is a novel coronavirus like those that caused severe acute respiratory syndrome (SARS) in 2003 and Middle East respiratory syndrome (MERS) in 2012 (Fani et al., 2020). SARS-CoV-2 spread quickly throughout the world and was declared a pandemic by the WHO on March 11, 2020 (https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020, 2020).
Symptoms of COVID-19 may include fever, dry cough, tiredness, sore throat, body aches, and diarrhea (Zhang et al., 2020). Because viral shedding can occur before an individual becomes symptomatic, the rapid worldwide spread can partially be attributed to transmission by pre-symptomatic and asymptomatic individuals (He et al., 2020). Additionally, individuals infected with SARS-CoV-2 shed the virus in their stool (Wang et al., 2020). Together, this suggests that wastewater can be easily collected to conduct disease surveillance in low prevalence areas or before clinical cases are identified (Randazzo et al., 2020; Mallapaty, 2020).
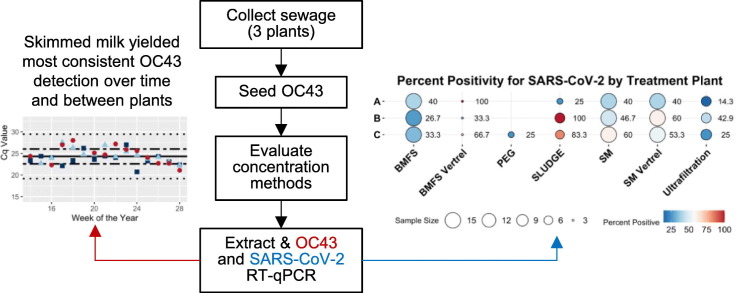
Public Health departments and research groups around the world are conducting wastewater surveillance for SARS-CoV-2 using various methods, including ultrafiltration, polyethylene glycol (PEG) precipitation, direct sludge extraction, and skimmed milk flocculation (Ahmed et al., 2020a; Peccia et al., 2020; Medema et al., 2020). However, many of the methods being used have been optimized for non-enveloped enteric viruses. Because SARS-CoV-2 is an enveloped virus, the existing methods are not optimized for its surveillance. Additionally, wastewater profiles can vary greatly due to geography, population, treatment processes at the plant, and where the sample is collected. A comparison of concentration methods is necessary to develop effective environmental wastewater surveillance and to standardize the methods being used across communities. To compare and optimize methods currently being used around the world, the following methods were carried out on primary wastewater from three wastewater treatment plants (WWTPs) in the Seattle area: the Bag-Mediated Filtration System (BMFS) (Zhou et al., 2019), modified skimmed milk flocculation from Calgua et al. 2008 (Calgua et al., 2008; Falman et al., 2019), polyethylene glycol precipitation (PEG) (Falman et al., 2019; Lewis and Metcalf, 1988), and ultrafiltration via Millipore filtration concentration (Ahmed et al., 2020a). Direct sludge extraction was also carried out on primary sludge samples from the same WWTPs. This methods comparison evolved as the pandemic evolved and was not designed to result in statistical comparisons, but rather to describe how the methods perform using a recovery control. This is the first SARS-CoV-2 wastewater concentration methods comparison performed in the United States to date and provides useful guidance for monitoring wastewater for SARS-CoV-2.
2. Methods
2.1. Generation of OC43 stock
Human coronavirus OC43 (OC43; ATCC VR-1558) stocks were prepared by infecting confluent HCT-8 cells (ATCC CCL-244) in flasks at 33 °C in RPMI-1640, 2% fetal bovine serum, and vancomycin/gentamycin. Six days post infection, virus was harvested by freeze/thawing. Lysates were clarified by centrifugation 2500 ×g for 20 min at 4 °C. Supernatant was collected and stored at −80 °C or was further concentrated by PEG precipitation. Further PEG precipitation included adding 9% PEG-8000 (Sigma-Aldrich, St. Louis, MO, USA) and 0.5 M NaCl (ThermoFisher Scientific, Waltham, MA, USA) to supernatant and shaking overnight at 5 °C. PEG slurry was centrifuged at 5500 ×g for 30 min at 4 °C. Virus pellets were resuspended for a 10-fold concentration in PBS and stored at −80 °C.
Stocks of OC43 were quantified using a modified version of a previously described immunoperoxidase assay by fixing cells 4 dpi using 2% paraformaldehyde (Lambert et al., 2008). Fixed cells were permeabilized using PBS with 0.5% TritonX-100 and 20 mM Glycine. Cells were labeled and stained using Anti-Coronavirus OC43 nucleoprotein monoclonal mouse antibody at 1:900 (MilliporeSigma, MAB9013), IgG (H + L) Cross-absorbed Goat anti-Mouse HRP at 1:1000 (Invitrogen, G21040) and Thermo-Scientific Pierce DAB substrate kit. Infected wells were counted, and TCID50/ml was calculated using the Spearman & Kärber algorithm.
2.2. Wastewater concentration
Primary wastewater was grab sampled weekly from three Seattle area wastewater treatment plants from late-March to July 2020 (Supplementary material Table A1). All wastewater was stored at 4 °C and used within one week of collection. For 1.0 L grab samples collected over the course of a single day, they were composited and mixed prior to seeding with OC43. OC43 was seeded at a concentration of 3.3 × 104 TCID50/L of wastewater.
Seeded wastewater samples were aliquoted and concentrated using four methods: the BMFS (with and without Vertrel™ extraction), skimmed-milk flocculation (with and without Vertrel™ extraction), PEG precipitation, or ultrafiltration (Table 1 ) (Appendix B). The BMFS concentrated an average of 2.63 L (Table 1) of primary wastewater using previously published methods, including filtration, elution, and secondary concentration with a two-hour shake at 200 RPM and pellet resuspension in 4 mL of sterile PBS (pH 7.4) (Zhou et al., 2019; Falman et al., 2019; Fagnant et al., 2018). Skimmed milk flocculation was also carried out on primary wastewater samples in volumes of 0.1 L, 0.5 L, and 1.0 L following the protocol used during BMFS processing. Sample volumes varied to try to optimize the effective volume assayed and processing time, 0.5 L was used most often because it best balanced these two priorities. Skimmed milk flocculation pellets were resuspended in either 4 mL or 6 mL of sterile PBS (pH = 7.4). Initial volumes of 0.1 L were resuspended in 4 mL, and initial volumes of 0.5 L and 1.0 L were resuspended in 6 mL. Resuspensions of both BMFS and skimmed milk samples were then divided into two volumes: one for RNA extraction and the other to separate the viruses from the solids with Vertrel XF™ (Miller-Stephenson, Inc., Danbury, CT, USA) (Falman et al., 2019). PEG precipitation was carried out on 0.5 L of primary wastewater by shaking for 4 h (4 °C at 200RPM) and resuspended in 10 mL of sterile PBS (pH = 7.4) following previously published methods (Falman et al., 2019). There were a limited number of replicates of this method due to the time required for processing and restrictions on people and laboratory space. Ultrafiltration with Centricon Plus-70 centrifugal filter devices (MilliporeSigma, Burlington, MA, USA) were used to concentrate 0.1 L of composite wastewater following manufacturer instructions. The number of ultrafiltration replicates was limited by supply chain constraints. Prior to ultrafiltration, 0.1 L of wastewater was centrifuged at 6800 ×g and 4 °C for 30 min to pellet out the solids. The supernatant was applied to the filter device, discarded after filtration (repeated once to allow full volume to pass through), and the retentate retained for detection (Ahmed et al., 2020a). Resuspension volumes for all methods are reported in Supplemental material Fig. A1. All concentrates were stored at −80 °C for RNA extraction.
Table 1.
Number of total samples for each concentration method and average volume sampled across all weeks.
| Plant A |
Plant B |
Plant C |
|||||
|---|---|---|---|---|---|---|---|
| Number | Average volume (L) | Number | Average volume (L) | Number | Average volume (L) | ||
| BMFS | Vertrel | 15 | 1.23 | 15 | 1.23 | 15 | 2.63 |
| No vertrel | 3 | 3 | 3 | ||||
| Skimmed milk | Vertrel | 0.25 L = 15 | 0.25 | 0.25 L = 15 | 0.25 | 0.25 L = 15 | 0.25 |
| 0.5 L = 1 | 0.5 L = 1 | 0.5 L = 1 | |||||
| 0.05 L = 1 | 0.05 L = 1 | 0.05 L = 1 | |||||
| No vertrel | 0.25 L = 15 | 0.25 | 0.25 L = 15 | 0.25 | 0.25 L = 15 | 0.25 | |
| 0.5 L = 1 | 0.5 L = 1 | 0.5 L = 1 | |||||
| 0.05 L = 1 | 0.05 L = 1 | 0.05 L = 1 | |||||
| Polyethylene glycol precipitation | 0 | – | 0 | – | 4 | 0.50 | |
| Ultrafiltration | 7 | 0.1 | 7 | 0.1 | 8 | 0.1 | |
| Sludge extraction | 4 | 0.0025 | 6 | 0.0025 | 6 | 0.0025 | |
2.3. RNA extraction
RNA extraction was carried out on all concentrated wastewater in duplicate using the QIAamp Viral RNA Mini Kit (QIAGEN, Germantown, MD, USA). For liquid samples, the input volume was 280uL with the exception of ultrafiltration, which had an input volume of 140uL due to the small retentate volume generated. The ultrafiltration pellet from 50 mL of sewage entered the kit via resuspension in Buffer AVL. Each sample was eluted in 60uL and duplicates were combined before being re-aliquoted into 60uL aliquots and frozen at −20 °C.
2.4. Sludge extraction
Primary composite sludge was collected on the same day as primary wastewater and was not seeded with OC43 prior to extraction following published methods by Peccia et al. (Peccia et al., 2020) Briefly, viral RNA was extracted using the QIAGEN RNeasy PowerSoil Total RNA kit by adding 2.5 mL of well mixed sludge directly to the commercial kit with an elution volume of 100 μL.
2.5. RT-qPCR
Reverse-transcription qPCR (RT-qPCR) for OC43 and SARS-CoV-2 was carried out on all RNA extracts using the iTaq Universal Probes One-Step Kit (Bio-Rad Laboratories, Hercules, CA, USA) with a total reaction volume of 20uL. All samples were run with both undiluted RNA extracts and 10−1 dilutions of the RNA extracts. OC43 was detected using a previously published protocol with 0.3uM of primers and 0.2uM of the FAM probe targeting the M protein, the membrane glycoprotein (Vijgen et al., 2005). The US Centers for Disease Control and Prevention SARS-CoV-2 research-use only detection kit provided by IDT (Integrated DNA Technologies, Inc., Coralville, IA, USA) was used targeting three regions of the N gene (US Centers for Disease Control and Prevention, n.d.). RT-qPCR cycling conditions for both assays were 50 °C for 10 min, 95 °C for 3 min, and 45 cycles of 95 °C for 15 s and 60 °C for 30 s. Positive control standard curves for both viruses were done in duplicate using serial 10-fold dilutions in nuclease free water. Positive control standard curves for the N1, N2, and N3 assays were carried out using a plasmid control containing the target genes for SARS-CoV-2 (Integrated DNA Technologies, Inc., Coralville, IA, USA). Positive control standard curves for OC43 were generated from extractions of serially diluted human coronavirus OC43 (OC43; ATCC VR-1558) enumerated by TCID50 on HCT-8 cells (ATCC CCL-244). Negative controls were nuclease free water. No negative controls had detections for either OC43 or SARS-CoV-2. An off-target control was used for each assay, with SARS-CoV-2 serving as this control for OC43 and OC43 serving as this control for the SARS-CoV-2 targets. There was no cross-reactivity in the assays. Limits of detection (LOD) and quantification (LOQ) were determined in nuclease free water and in RNA extracted skimmed milk flocculation concentrated wastewater using standard curves with ten replicates at each concentration. The LOD was determined as the concentration below which less than 90% of replicates are detected as positive (Burd, 2010). The LOQ is the lowest concentration with a coefficient of variation below 35% (Klymus et al., 2020).
2.6. Data analysis
Bio-Rad CFX Maestro for Mac (Bio-Rad Laboratories, Hercules, CA, USA) was used to analyze all RT-qPCR tests, and data were collated and managed using Microsoft Excel (Microsoft Corp., Redmond, WA, USA). Cycle threshold levels were manually set at the point where the positive controls start exponentially multiplying. Samples with non-exponential multiplication were considered false positives, and samples with reduced fluorescence as evident in the qPCR curves were considered inhibited. All samples with a Cq larger than 40 in both the undiluted and the 10−1 dilution were considered negative. All figures were generated using RStudio (RStudio, PBC, Boston, MA, USA).
3. Results and discussion
3.1. Method comparison using OC43 as a surrogate coronavirus
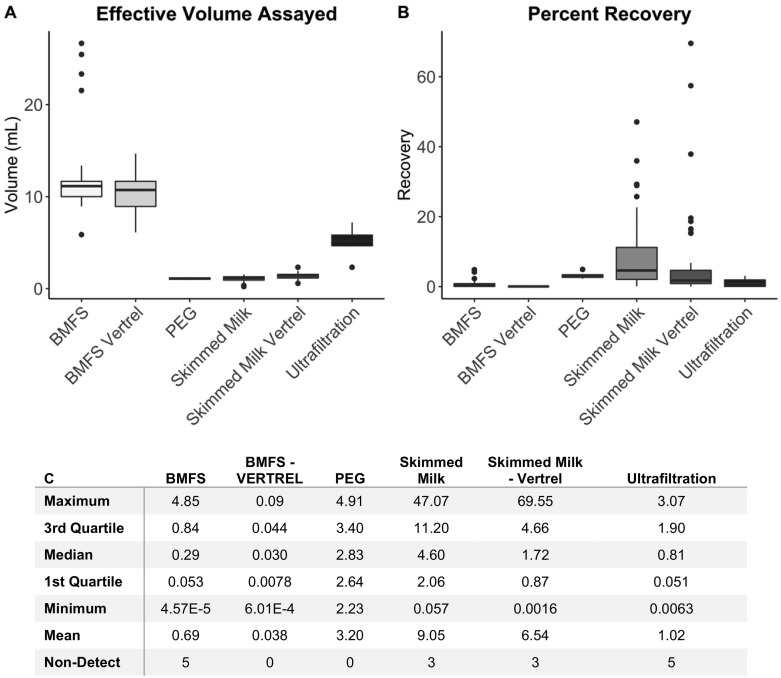
Multiple factors were considered when using OC43 to compare concentration methods including the effective volume assayed, OC43 percent recovery, detection frequency, detection consistency, and concentration. The total volume assayed for each method was assessed by calculating the proportion of the initial volume of concentrated wastewater that was assayed by RT-qPCR. The BMFS method, with and without Vertrel™ extraction, had the highest average effective volume assayed (11.90 and 10.41 mL, respectively) when compared to the other four methods (Fig. 1A, SI Table A2). PEG precipitation and skimmed milk flocculation, with and without Vertrel™ extraction, had the three lowest average effective volume assayed of the methods of 1.09 mL, 1.10 mL, and 1.37 mL, respectively (Fig. 1A, SI Table A2). Ultrafiltration had an average effective volume assayed of 5.31 mL (Fig. 1A, SI Table A2).
Fig. 1.
Effective volume assayed for and percent recovery for each method. A) The effective volume assayed is the proportion of the original wastewater sample assayed by RT-qPCR. The BMFS, with and without Vertrel, have the largest average effective volume assayed per reaction. PEG precipitation and skimmed milk flocculation, with and without Vertrel, have the smallest average effective volume assayed per reaction. B) The percent recovery is calculated using the standard curves for the RT-qPCR assay generated with each experimental run and the Cq value for each undiluted sample. Skimmed milk flocculation has the highest average percent recovery, followed by skimmed milk flocculation with Vertrel extraction. Both BMFS methods had the two lowest average percent recoveries. All methods tested had some non-detections in the undiluted sample except PEG precipitation and BMFS with Vertrel extraction. C) Percent recovery descriptive statistics by method.
The percent recovery for the spiked OC43 recovery control was calculated for each method using the standard curves generated for each RT-qPCR assay and the estimated spiked concentration (Eqs. (1), (2), (3)).
| (1) |
where: C q is the cycle quotient as determined using standard curves generated from OC43 stock solutions, C OC43 is log concentration in
| (2) |
| (3) |
where: V sample is volume of sample adjusted for amount entering qPCR, V inoc is fraction of volume assayed relative to volume processed.
The highest average percent recoveries across all methods were obtained for skimmed milk flocculation with Vertrel™ extraction (9%) and without (6%) (Fig. 1B, C). Ultrafiltration and the BMFS, with and without Vertrel™ extraction, had the three lowest average percent recoveries across all methods compared (1.0%, 0.04%, and 0.7%, respectively) (Fig. 1C). The only two methods that did not have non-detections in the undiluted sample were PEG precipitation and BMFS with Vertrel™ extraction (Fig. 1C). However, these two methods had fewer total replicates compared to the other methods (Table 1) and are therefore not directly comparable.
Because the BMFS concentrates and assays the largest volumes of sewage of the methods tested, it is likely that the percent recovery is reduced due to inhibitors concentrated with the wastewater as larger volumes of concentrated water exhibit reduced detection (Loge et al., 2002). Additionally, because all of the 10−1 diluted samples detected OC43 (data not presented), it is probable that inhibitors present in the un-diluted samples reduced or blocked detection. Our recovery values are similar to those reported for Bovine-Coronavirus (BCoV) (Jafferali et al., 2020), but substantially lower than recoveries reported for murine hepatitis virus (MHV) (Ahmed et al., 2020b). MHV is an enteric virus and, therefore, potentially persists longer in sewage than OC43 or BCoV. These results emphasize the importance of using seeded recovery controls to correct for matrix effects and inhibition when selecting a method for SARS-CoV-2 wastewater surveillance.
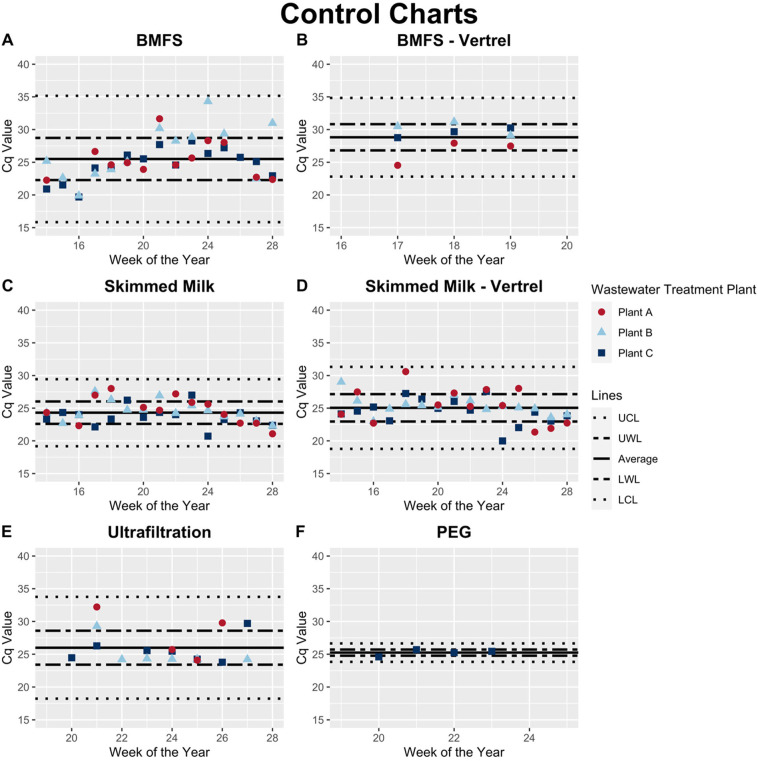
To compare the efficiency of the methods across treatment plants and time, control charts were generated for each method (Fig. 2 ). Upper and lower warning limits (UWL and LWL) were set to one standard deviation from the mean Cq value for that concentration method. Upper and lower control limits (UCL and LCL) were set to three standard deviations from the mean Cq value for that concentration method.
Fig. 2.
OC43 RT-qPCR control charts. Average Cq values were calculated for each method by averaging across treatment plants and time. The UWL and LWL, or upper and lower warning limits, for each method were calculated by adding or subtracting, respectively, the standard deviation from the average Cq. The UCL and LCL, or the upper and lower control limits, for each method were calculated by adding or subtracting, respectively, three times the standard deviation from the average Cq. Anything detected at or above a Cq of 40 was considered a non-detection. All samples that had non-detections by RT-qPCR in the undiluted samples reported here had detection in the 10−1 dilution. BMFS without Vertrel extraction (A) has a lower average Cq compared to BMFS with Vertrel extraction (B), but has a substantially larger range of data. Skimmed milk flocculation without Vertrel extraction (C) and with Vertrel extraction (D) have similar average Cq's, control limits, and warning limits. Ultrafiltration (E) had a similar average Cq to both skimmed milk methods, but had a larger variability in the data and fewer detections in the undiluted samples. PEG precipitation (F) had a low average Cq and variability around the mean, but only one treatment plant was tested with this method and it is therefore not directly comparable.
The BMFS without Vertrel™ extraction has a lower mean Cq (25.6) compared to BMFS with Vertrel™ extraction (28.8) (Fig. 2A & B, Table 2 ). Both skimmed milk with and without Vertrel™ extraction have similar mean Cq values of 24.3 and 25.1, respectively (Fig. 2C & D, Table 2). However, skimmed milk with Vertrel™ extraction has a higher standard deviation and variance compared to skimmed milk without Vertrel™ extraction (Table 2). Ultrafiltration has a comparable mean Cq value (26.0) to the skimmed milk methods, but it has a much higher standard deviation and variance around the mean compared to both skimmed milk methods (Table 2).
Table 2.
Descriptive statistics of OC43 Cq values by RT-qPCR for each method.
| Average | Standard deviation | Variance | Max | Min | Non-detection | number | |
|---|---|---|---|---|---|---|---|
| BMFS | 25.5 | 3.2 | 10.4 | 34.3 | 19.7 | 5 | 15 |
| BMFS - vertrel | 28.8 | 2.0 | 4.0 | 31.2 | 24.5 | 0 | 9 |
| Skimmed milk | 24.3 | 1.7 | 2.9 | 28.0 | 20.7 | 2 | 15 |
| Skimmed milk - vertrel | 25.1 | 2.1 | 4.4 | 30.6 | 20.0 | 2 | 15 |
| Ultrafiltration | 26.0 | 2.6 | 6.7 | 32.2 | 23.8 | 7 | 22 |
| PEG | 25.2 | 0.5 | 0.2 | 25.7 | 24.6 | 0 | 4 |
The only two methods that detected OC43 from all undiluted samples were the BMFS with Vertrel™ extraction and PEG precipitation. However, these have fewer samples and are therefore not directly comparable. BMFS with Vertrel™ extraction and PEG precipitation were dropped early in the methods comparison because they were time consuming and did not add any additional detection power for SARS-CoV-2. Both skimmed milk flocculation methods had three non-detected samples by RT-qPCR in the undiluted reaction (all detected in 10−1 diluted reaction), and ultrafiltration had five non-detected samples in the undiluted reaction (all detected in 10−1 diluted reaction). The BMFS had the highest number of non-detected samples with six samples being non-detected in the undiluted RT-qPCR reaction (Table 2). Therefore, all concentration methods were susceptible to inhibition.
Taken together, these results suggest that, of the evaluated methods, skimmed milk without Vertrel™ extraction performs well for detection of OC43. Skimmed milk flocculation without Vertrel™ extraction had the highest mean percent recovery (Fig. 1B), consistent performance over time and across treatment plants (Fig. 2C), lowest mean Cq for OC43 detection (Table 2), and lowest variability around the mean (Table 2). Although Vertrel™ extraction on the skimmed milk resuspensions performed similarly to the non- Vertrel™ extracted samples, Vertrel™ extraction is not an optimal step for human coronavirus detection as it was developed for non-enveloped viruses and added substantial processing time (Mendez et al., 2000). Because human coronaviruses are enveloped, Vertrel™ extraction is not an ideal method to separate residual solids from the virions after pellet resuspension.
3.2. RT-qPCR efficiency
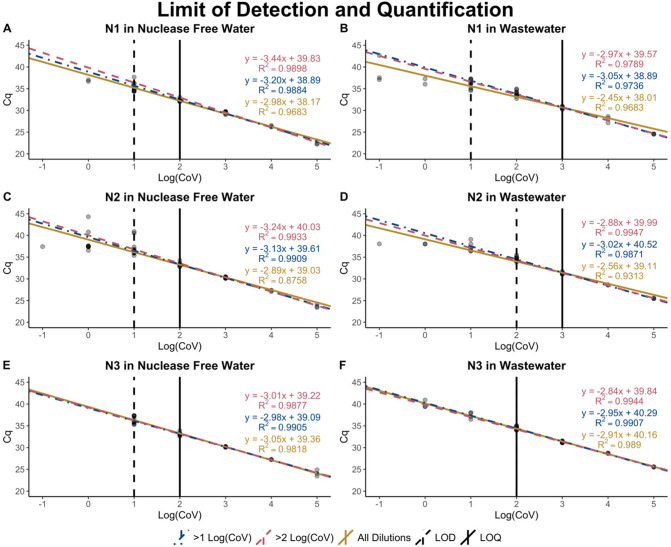
By graphing all of the standard curves in nuclease free water for a single assay (OC43, N1, N2, or N3) on the same plot (SI Fig. A2, SI Fig. A3) and extracting the slopes and intercepts for the standards when including all dilutions or the 10−1 through 10−3 dilutions, substantial variation in the standard curves for the three SARS-CoV2 assays is observed. This is evidenced by variable slopes and intercepts for each assay. OC43 standard curves in nuclease free water were much more consistent as the slopes and intercepts were less variable between assays. Variability in SARS-CoV-2 standard curves makes sample quantification difficult because of its poor-performance assay to assay.
The LOD for all three SARS-CoV-2 assays in nuclease free water is 10 gene copies (Fig. 3 ), while the LOQ in nuclease free water is 100 gene copies. While the LOD for N1 in Seattle wastewater is the same as in nuclease free water (10 gene copies), the LOD is increased for N2 and N3 to 100 gene copies, likely due to inhibitors present in the water matrix (Loge et al., 2002). Additionally, all three assays have reduced efficiency in wastewater compared to nuclease free water. The LOQ for N1 and N2 in Seattle wastewater is 1000 gene copies, and the LOQ for N3 in Seattle wastewater is 100 gene copies. This highlights the importance of validating assays in different water matrices and using recovery controls to quantify inhibition. The LOD and LOQ were not determined for the OC43 assay because all samples were detected within the ranges of standard curve and there was little variability in the standard curves over time.
Fig. 3.
Limits of detection and quantification for SARS-CoV-2 assays. The limit of detection and limit of quantification for each SARS-CoV-2 assay is dependent on the water matrix. Greater inhibition of the assays were seen when standard curves were prepared using skimmed milk wastewater extracts as the diluent (B, D, F) than when standard curves were prepared using nuclease free water as the diluent (A, C, E), as seen by decreased efficiency and higher limits of detection. In panels D and F, the single vertical line indicates the LOD and LOQ are on top of each other.
3.3. SARS-CoV-2 results
Samples positive for SARS-CoV-2 were identified using the following criteria:
-
•
Detection for at least one of the three SARS-CoV-2 assays (N1, N2, or N3) AND
-
•
Amplification below Cq = 40 in non-diluted reaction OR
-
•
Amplification below Cq = 40 in 10−1 diluted reaction.
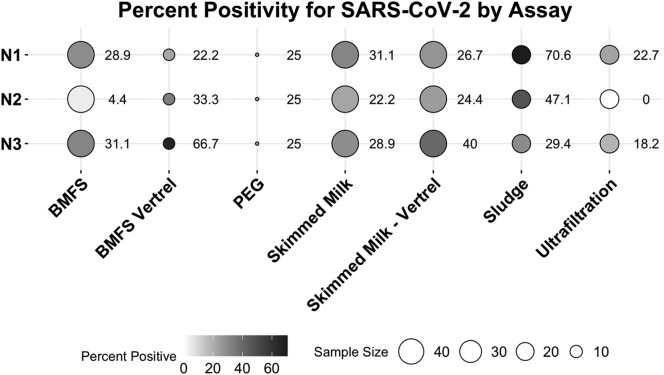
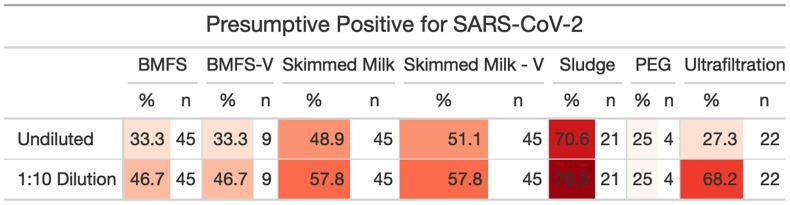
Many of the samples across methods amplified at ranges that were unquantifiable at a genome copy level but were considered positive if they amplified below Cq of 40. The performance of each assay varied by method (Fig. 4 ), with N1 and N2 having the highest detection in direct sludge extraction, but N3 having the highest detection in skimmed milk – Vertrel™. Across all methods looking at wastewater, skimmed milk flocculation provided more positive samples in undiluted assays (48.9%, n = 45) (Fig. 5 ) with lower Cq values than other methods surveying wastewater, despite the lower effective volume assayed (Fig. 1A). Surveying sludge/biosolids yielded the lowest Cq values for SARS-CoV-2 (SI Fig. A4), demonstrating its ability to concentrate viral particles. However, it is not as comparable to water-based samples and methods because treatment plants handle biosolids differently. Each WWTP has a different number of settling tanks, the holding time in these tanks is dependent on the flow and what flocculants used, if any, and the sample is not a composite from all clarifiers. Therefore, a sludge sample does not reflect an influent sample and is variable and difficult to interpret.
Fig. 4.
Percent positivity for SARS-CoV-2: samples were considered positive if the Cq ≤ 40. Direct sludge extraction had highest percent positive for N1 and N2 but had fewer samples. N3 had the highest percent positive in skimmed milk flocculation with Vertrel™ extraction. All three assays had roughly 30% positive with skimmed milk flocculation.
Fig. 5.
Presumptive positive for SARS-CoV-2 by method. A sample was considered “presumptive positive” if at least one of the SARS-CoV-2 assays had a Cq ≤ 40 in either the non-diluted or 10−1 dilution reaction.
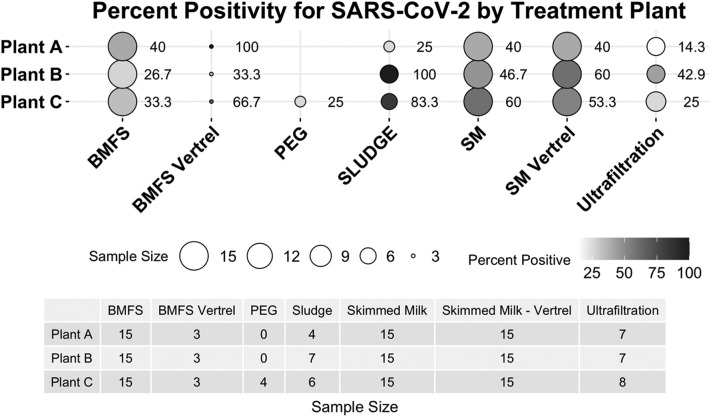
Detection of SARS-CoV-2 differed across wastewater treatment plants and between samples. For example, Plant A had less detection using direct sludge extraction compared to skimmed milk flocculation (Fig. 6 ), while the other two treatment plants had more detection for sludge extraction. This indicates that the between-plant differences in handling of influent wastewater and biosolids can substantially affect detection of SARS-CoV-2. Additionally, investigators noted differences in wastewater consistency and free suspended solids in samples from different treatment plants. This demonstrates that differences in influent and variability in SARS-CoV-2 detection cannot be differentiated.
Fig. 6.
SARS-CoV-2 detection by wastewater treatment plant. Percent positivity includes undiluted and 10−1 diluted RT-qPCR assays for each method and treatment plant. Detection of SARS-CoV-2 varied by method for each treatment plant.
Across the three SARS-CoV-2 assays, there was a substantial amount of variability in detection. While the N1 assay had fewer non-detects (Fig. 4) compared N2 and N3, the mean Cq value for all three assays is similar (37.0, 37.1, and 36.8, respectively) (SI Fig. A5) indicating some variability of performance amongst the assays or degradation in the target genetic material. Although, several samples (weeks 19, 23, 24, 27 and 28) amplified across all three assays at quantifiable ranges using a standard curve, many samples are non-quantifiable. This indicates that SARS-CoV-2 is at or near detectable levels in the majority of these samples due to dilution in the wastewater, low persistence, and/or the presence of inhibitors.
Since these WWTPs serve hundreds of thousands of people in King County, most of detectable SARS-CoV-2 samples were often out of quantifiable ranges, and template degradation is likely, it is not feasible to use this data as precise measure of infections in the community. However, these results serve as an indicator of wide-spread community transmission and can track trends in infections at the community level. Additionally, these results can be used to determine effective sampling and processing techniques that allow for the detection of endemic SARS-CoV-2 on a smaller scale such as in wastewater conveyance systems, pump stations, or residential communities.
4. Conclusion
These results stress the importance of validating sampling and concentration methods in different water matrices using seeded recovery controls. Other studies have utilized surrogate viruses to control for recovery and compare methods (Jafferali et al., 2020; Ahmed et al., 2020b), but this is the first study to conduct a methods comparison of SARS-CoV-2 at multiple treatment plants for consecutive weeks. Additionally, this is the first methods comparison published using another human respiratory coronavirus as the recovery control organism for SARS-CoV-2. Although all methods tested here and previously published were shown effective at detecting SARS-CoV-2 and the seeded recovery control (Jafferali et al., 2020; Ahmed et al., 2020b), a method that is feasible in one setting and environment may not be feasible elsewhere. Skimmed milk flocculation was chosen for continued surveillance in our lab because of its detection consistency and simplicity. Because skimmed milk flocculation does not require extensive laboratory resources, it is a promising method for wastewater surveillance in resource limited settings. It does not rely on hard to acquire consumables and can therefore allow groups to conduct uninterrupted surveillance. For wastewater surveillance to effectively supplement clinical surveillance, methods must be validated and selected using both logistical and performance considerations. Given the national shortage in clinical SARS-CoV-2 tests, wastewater surveillance has the potential to effectively prevent new outbreaks in communities without cases, as was reported at a residential hall at the University of Arizona (Peiser, 2020). Additionally, wastewater surveillance may help understand changes in pandemic trends in the water catchment area, such as reductions or increases in cases (Water Research Foundation, 2020). Using effective sampling methods is critical for wastewater surveillance to serve as a leading indicator of clinical infection and to accurately describe community transmission of SARS-CoV-2.
CRediT authorship contribution statement
Sarah E. Philo: methodology, software, validation, formal analysis, investigation, writing – original draft, writing – review & editing, visualization. Erika K. Keim: methodology, investigation, writing – original draft, writing – review & editing, visualization. Rachael Swanstrom: resources, investigation, writing – original draft. Angelo Q. W. Ong: validation, investigation, writing – original draft. Elisabeth A. Burnor: validation, investigation. Alexandra L. Kossik: methodology, investigation, supervision. Joanna C. Harrison: methodology, investigation. Bethel A. Demeke: resources, investigation. Nicolette A. Zhou: conceptualization, methodology, formal analysis, writing – review & editing, visualization, supervision, project administration, funding acquisition. Nicola K. Beck: conceptualization, methodology, resources, supervision, project administration funding acquisition. Jeffry H. Shirai: resources, supervision, project administration, funding acquisition. J. Scott Meschke: conceptualization, methodology, resources, formal analysis, writing – review & editing, supervision, project administration, funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
Thanks to our collaborators at the wastewater treatment plants who helped facilitate sample collection, particularly the West Point Process lab; the South Treatment Plant collection team including Wieslawa Wakulak, Jason Karlstrom, Rebecca Hee, Neila Glidden, and Rachael Dyda; and the Brightwater Operations Staff.
Funding
This work was supported by the University of Washington Population Health Initiative. This project was also supported by funds provided by the Department of Environmental & Occupational Health Sciences at the University of Washington.
Editor: Jay Gan
Footnotes
Bag-mediated filtration system (BMFS), polyethylene glycol (PEG), Human coronavirus OC43 (OC43), World Health Organization (WHO), Middle East Respiratory Syndrome (MERS), wastewater treatment plants (WWTP), Reverse-Transcriptase quantitative-Polymerase Chain Reaction (RT-qPCR).
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2020.144215.
Appendix A. Supplementary data
Supplementary material
References
- Ahmed W., Angel N., Edson J. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. (PMID – 32387778) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P., Bivins A. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139960. (PMID – 32758945) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd E.M. Validation of laboratory-developed molecular assays for infectious diseases. Clin. Microbiol. Rev. 2010;23(3):550–576. doi: 10.1128/cmr.00074-09. (PMID – 20610823) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calgua B., Mengewein A., Grunert A. Development and application of a one-step low cost procedure to concentrate viruses from seawater samples. J. Virol. Methods. 2008;153(2):79–83. doi: 10.1016/j.jviromet.2008.08.003. (PMID – 18765255) [DOI] [PubMed] [Google Scholar]
- Fagnant C.S., Sánchez-Gonzalez L.M., Zhou N.A. Improvement of the bag-mediated filtration system for sampling wastewater and wastewater-impacted waters. Food and Environmental Virology. 2018;10(1):72–82. doi: 10.1007/s12560-017-9311-7. (PMID – 28674934) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falman J.C., Fagnant-Sperati C.S., Kossik A.L., Boyle D.S., Meschke J.S. Evaluation of secondary concentration methods for poliovirus detection in wastewater. Food and Environmental Virology. 2019;11(1):20–31. doi: 10.1007/s12560-018-09364-y. (PMID – 30612304) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fani M., Teimoori A., Ghafari S. Comparison of the COVID-2019 (SARS-CoV-2) pathogenesis with SARS-CoV and MERS-CoV infections. Futur. Virol. 2020;15(5):317–323. doi: 10.2217/fvl-2020-0050. [DOI] [Google Scholar]
- He X., Lau E.H.Y., Wu P. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020;26(5):672–675. doi: 10.1038/s41591-020-0869-5. (PMID – 32296168) [DOI] [PubMed] [Google Scholar]
- WHO Director-General's opening remarks at the media briefing on COVID-19 - 11 March 2020. 2020. https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 (March 11)
- Jafferali M.H., Khatami K., Atasoy M., Birgersson M., Williams C., Cetecioglu Z. Benchmarking virus concentration methods for quantification of SARS-CoV-2 in raw wastewater. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.142939. (PMID – 33121776) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.-j., Dong X., Cao Y.-y. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy. 2020;75(7):1730–1741. doi: 10.1111/all.14238. (PMID – 32077115) [DOI] [PubMed] [Google Scholar]
- Klymus K.E., Merkes C.M., Allison M.J. Reporting the limits of detection and quantification for environmental DNA assays. Environmental DNA. 2020;2(3):271–282. doi: 10.1002/edn3.29. [DOI] [Google Scholar]
- Lambert F, Jacomy H, Marceau G, Talbot* PJ. SARS- and Other Coronaviruses, Laboratory Protocols. Methods in molecular biology (Clifton, NJ). 2008;454:93–102. doi: 10.1007/978-1-59745-181-9_8 PMID - 19057861. [DOI] [PMC free article] [PubMed]
- Lewis G.D., Metcalf T.G. Polyethylene glycol precipitation for recovery of pathogenic viruses, including hepatitis A virus and human rotavirus, from oyster, water, and sediment samples. Appl. Environ. Microbiol. 1988;54(8):1983–1988. doi: 10.1128/aem.54.8.1983-1988.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loge F.J., Thompson D.E., Call D.R. PCR detection of specific pathogens in water: a risk-based analysis. Environmental Science & Technology. 2002;36(12):2754–2759. doi: 10.1021/es015777m. (PMID – 12099475) [DOI] [PubMed] [Google Scholar]
- Mallapaty S. How sewage could reveal true scale of coronavirus outbreak. Nature. 2020;580(7802):176. doi: 10.1038/d41586-020-00973-x. [DOI] [PubMed] [Google Scholar]
- Medema G, Heijnen L, Elsinga G, Italiaander R, Brouwer A. Presence of SARS-Coronavirus-2 in sewage. medRxiv. 2020:2020.03.29.20045880. doi: 10.1101/2020.03.29.20045880 [DOI] [PubMed]
- Mendez I.I., Hermann L.L., Hazelton P.R., Coombs K.M. A comparative analysis of Freon substitutes in the purification of reovirus and calicivirus. J. Virol. Methods. 2000;90(1):59–67. doi: 10.1016/s0166-0934(00)00217-2. (PMID – 11011081) [DOI] [PubMed] [Google Scholar]
- Peccia J, Zulli A, Brackney DE, et al. SARS-CoV-2 RNA concentrations in primary municipal sewage sludge as a leading indicator of COVID-19 outbreak dynamics. medRxiv. 2020:2020.05.19.20105999. doi: 10.1101/2020.05.19.20105999 [DOI]
- Peiser J. The University of Arizona Says It Caught a Dorm's Covid-19 Outbreak Before It Started. Its Secret Weapon: Poop. The Washington Post. 2020.
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- US Centers for Disease Control and Prevention. n.d. CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel. Accessed 3 March, 2020.
- Vijgen L., Keyaerts E., Moës E., Maes P., Duson G., Ranst M.V. Development of one-step, real-time, quantitative reverse transcriptase PCR assays for absolute quantitation of human coronaviruses OC43 and 229E. J. Clin. Microbiol. 2005;43(11):5452–5456. doi: 10.1128/jcm.43.11.5452-5456.2005. (PMID – 16272469) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. (PMID – 32159775) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Water Research Foundation. Wastewater surveillance of the COVID-19 genetic signal in sewersheds. 2020. https://www.waterrf.org/sites/default/files/file/2020-06/COVID-19_SummitHandout-v3b.pdf.
- World Health Organization Pneumonia of unknown cause - China. https://www.who.int/csr/don/05-january-2020-pneumonia-of-unkown-cause-china/en/
- Zhou N.A., Fagnant-Sperati C.S., Komen E. Feasibility of the bag-mediated filtration system for environmental surveillance of poliovirus in Kenya. Food and Environmental Virology. 2019:1–13. doi: 10.1007/s12560-019-09412-1. (PMID – 31679104) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material