Abstract
Microbiological response of SARS-CoV-2 to remdesivir in immunocompromised patients has not been evaluated. We present the case of a severely immunocompromised patient with persistent replication of SARS-CoV-2, who required different courses of remdesivir. Short courses of remdesivir might be insufficient in immunocompromised patients due to prolonged viral clearance.
Keywords: Remdesivir, COVID-19, SARS-CoV-2, Immunosuppression, Lymphoma
More than 37 million people worldwide have been infected and one 1 million have died since the first cases of COVID-19 were described (WHO, 2020). However, data about the efficacy of different treatment agents against SARS-CoV-2 is still scarce (Sanders et al., 2020). Remdesivir is the only antiviral agent that has demonstrated efficacy against COVID-19 in terms of shortening length of hospital stay in a randomized placebo-controlled trial (Beigel et al., 2020). Another randomized clinical trial (RCT) has shown that a 5-day course of remdesivir is as effective as a 10-day regimen in patients with severe COVID-19 not requiring mechanical ventilation (Goldman et al., 2020). However, there is no evidence whether these results can be applied to special populations such as immunocompromised patients. Here we present the case of a severely immunocompromised patient with persistent replication of SARS-CoV-2 who required different courses of remdesivir.
A 37-year-old woman presented to the hospital with fever. During the previous months, the patient had received 3 cycles of R-ESHAP (rituximab, etoposide, cisplatin, cytarabine and methylprednisolone) due to a relapse of a stage IV-A follicular lymphoma. A partial response was observed by PET-CT and a salvage therapy was about to start when the patient was diagnosed with an upper respiratory tract infection by Influenza A virus, requiring treatment with oseltamivir for 10 days two weeks before the present admission.
In March 2020, the patient presented to the hospital with a 3-day history of fever, malaise, anosmia and dysgeusia. At admission, oxygen saturation at room air was 98% and physical exam was unremarkable. Blood tests showed thrombocytopenia and increased D-dimer levels, with no elevation of the other acute-phase reactants and a normal lymphocyte count. Initial chest X-ray did not show any infiltrates. SARS-CoV-2 polymerase-chain reaction (PCR), detecting the envelope (E) and the open reading frame 1a (ORF1a) genes (Roche Diagnostic), performed on a nasopharyngeal swab resulted positive, confirming the diagnosis of COVID-19. Following hospital protocols, treatment with lopinavir/ritonavir, hydroxychloroquine and azithromycin was initiated, but lopinavir/ritonavir was withdrawn after 24 h due to moderate diarrhoea.
The patient showed clinical worsening with persistent fever, diaphoresis, dyspnoea and a slight but painful spleen enlargement. Under the suspicion of neoplastic disease progression, 60 mg prednisone and 1000 mg cyclophosphamide, as well as broad-spectrum antibiotics, were administered. In the following days, oxygen needs increased and PET-CT ruled out neoplastic progression and showed signs of organizing pneumonia. Treatment with 250 mg methylprednisolone per day for 3 days followed by 60 mg prednisone daily was then started. However, ferritin levels increased, thus anakinra was also indicated. Bronchoalveolar lavage samples confirmed positive SARS-CoV-2 PCR with a high viral load and discarded other causes of infection. Based on microbiological results and clinical condition, 4 weeks after the onset of symptoms, the patient was enrolled in an RCT being allocated to receive remdesivir (200 mg continued with 100 mg/day). In the following days, the patient presented an excellent clinical evolution, being discharged after 8 days of treatment.
Three days after discharge, the patient started again with fever and cough. Blood tests showing lymphopenia with reduction in all lymphocyte subpopulations and decrease in all immunoglobulin levels confirmed cellular and humoral immunosuppression. A CT-scan showed new bilateral infiltrates and a complete resolution of the previous signs of organizing pneumonia. A new SARS-CoV-2 nasopharyngeal PCR was positive, thus diagnosis of COVID-19 relapse was assumed and treatment with hydroxychloroquine, azithromycin and darunavir/cobicistat was started. Accidentally, a single dose of remdesivir was administered to the patient, becoming afebrile for 24 h afterwards. Concomitantly, a decreasing steroids scheme was continued and, given the severe hypogammaglobulinemia, non-specific intravenous immunoglobulins (IVIG) were administered. Despite the different treatments prescribed, an increase in nasopharyngeal SARS-CoV-2 viral load was observed. Consequently, the patient was included into a second RCT and treated with remdesivir at the same previous doses for 10 additional days. The patient rapidly improved, fever resolved and PCR in nasopharyngeal sample became negative. Given that antibody tests seeking for IgA, IgM, and IgG against SARS-CoV-2 (VITROS® Immunodiagnostic anti-SARS-CoV-2-Total) performed 42 and 64 days after the onset of symptoms resulted negative, another infusion of IVIG and COVID-19 convalescent plasma were also administered.
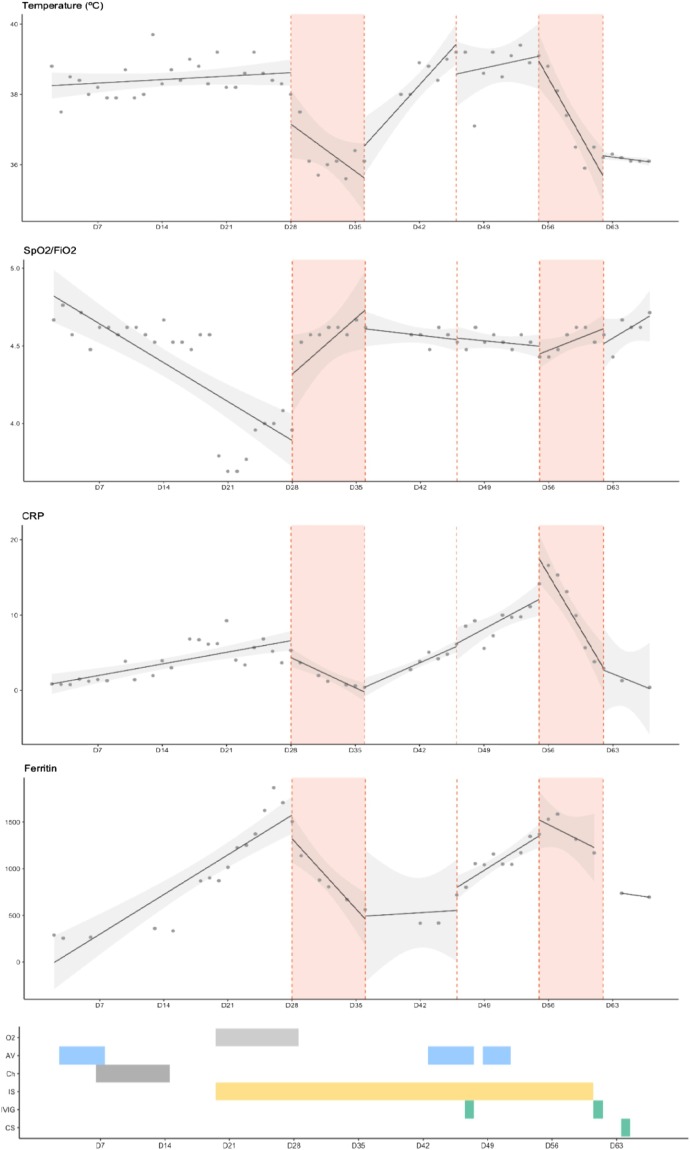
Fig. 1 shows the evolution of SARS-CoV-2 adjusted viral load in serial nasopharyngeal swabs and the different antiviral treatments received (Lescure et al., 2020). Adjusted SARS-CoV-2 viral load initially increased up to 1,1 × 107copies/1000cells. Coinciding with first remdesivir administration, viral load decreased to 3,1copies/1000cells. However, a new peak on the viral load was observed (4,1 × 103copies/1000cells) 54 days after symptoms initiation. After a second cycle of remdesivir, a reduction and negativization of viral load was finally achieved (63 days after the onset of symptoms). In this case, remdesivir showed an important antiviral effect in an immunocompromised patient, significantly reducing SARS-CoV-2 viral load, which was not observed with the other antiviral treatments. Remarkably, in our case, the antiviral effect of remdesivir was consistent with different clinical aspects like the resolution of fever, improvement of respiratory insufficiency and decreasing of acute-phase reactants (Fig. 1).
Fig. 1.
Microbiological and clinical evolution of SARS-CoV-2 infection.
AV: Antiviral treatment (lopinavir/ritonavir, hydroxychloroquine, azithromycin, darunavir/cobicistat). Ch: chemotherapy (prednisone, cyclophosphamide). CRP: C-reactive protein. CS: convalescent serum. IVIG: intravenous immunoglobulin. IS: immunosupression (steroids, anakinra). O2: oxygen support. SpO2/FiO2: oxygen saturation to fraction of inspired oxygen ratio.
First graph shows changes on SARS-CoV-2 adjusted viral load in serial nasopharyngeal swabs and the different antiviral treatments received. Adjusted viral load was calculated by adjusting cycle threshold (Ct) for the number of cells per sample and is represented as a logarithm of the number of copies per 1000 cells (log10 copies/1000 cells). Red areas and dashed lines represent time under remdesivir treatment. Grey areas represent time under other antiviral treatment.
The following graphs show the evolution of temperature (ºC), SpO2/FiO2, CRP and ferritin.
Red areas and dashed lines represent time under remdesivir treatment. The different treatments received are presented in coloured bars.
Notably, the patient presented a positive SARS-CoV-2 PCR 60 days after symptoms initiation. Median time of viral RNA clearance in nasopharyngeal samples is 20 days but it has been detected after more than 5 weeks (Zhou et al., 2020). Although some authors suggested possible longer duration of positive PCR in immunocompromised patients, RNA dynamics in this population are unknown (Han et al., 2020, Helleberg et al., 2020, Choi et al., 2020). In this case, the increase in the viral load 54 days after symptoms initiation and the clinical worsening suggested persistent replication of SARS-CoV-2 and supports the hypothesis of persistent replication of SARS-CoV-2 in immunocompromised hosts (Helleberg et al., 2020, Choi et al., 2020).
Finally, although an RCT showed no benefit of 10-day courses of remdesivir compared with 5 days, no specific evaluation of participants with underlying immunosuppression conditions was reported (Goldman et al., 2020). Consequently, these results may not be extrapolated to special high-risk populations like severe immunocompromised patients in whom persistent viral replication can play an essential role on the pathogenesis of the disease (Goldman et al., 2020, Han et al., 2020, Helleberg et al., 2020, Choi et al., 2020). In our patient, a late peak on SARS-CoV-2 viral load was detected despite an 8-day regimen of remdesivir. This suggests that short courses of remdesivir might be insufficient for treating high-risk populations such as severely immunocompromised patients, especially those with humoral immune deficiency (Helleberg et al., 2020).
Declarations
The patient signed an informed consent form and the study was approved the Drug Research Ethics Committee of the Hospital Clinic of Barcelona (CEIm), Barcelona, Spain.
The authors of the article declare no conflicts of interest. The authors received no financial support for the research or publication of this article.
The authors of the article want to thank Gilead for allowing reporting the case of a participant included in RCT.
References
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C. Remdesivir for the treatment of Covid-19 — final report. New Engl J Med. 2020;(Oct) doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi B., Qiu X., Solomon I.H., Sparks J.A., Padera R.F., Solomon I.H. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N Engl J Med. 2020;383:2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman J.D., Lye D.C.B., Hui D.S., Marks K.M., Bruno R., Montejano R. Remdesivir for 5 or 10 days in patients with severe Covid-19. New Engl J Med. 2020;(May) doi: 10.1056/NEJMoa2015301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Jiang M., Xia D., He L., Lv X., Liao X. COVID-19 in a patient with long-term use of glucocorticoids: a study of a familial cluster. Clin Immunol. 2020;214(May):108413. doi: 10.1016/j.clim.2020.108413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleberg M., Utoft Niemann C., Sommerlund Moestrup K., Kirk O., Lebech A.M., Lane C. Persistent COVID-19 in an immunocompromised patient temporarily responsive to two courses of remdesivir therapy. J Infect Dis. 2020;222:1103–1107. doi: 10.1093/infdis/jiaa446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescure F.X., Bouadma L., Nguyen D., Parisey M., Wicky P.H., Behillil S. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect Dis. 2020;20(6):e116. doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders J.M., Monogue M.L., Jodlowski T.Z., Cutrell J.B. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;(April) doi: 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- WHO Coronavirus disease (COVID-19) situation report. 12 October 2020. (https://www.who.int/docs/default-source/coronaviruse/situation-reports/20201012-weekly-epi-update-9.pdf).
- Zhou F., Yu T., Du R., Fan F., Liu Y., Liu Z. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;6736(20):1–9. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]