Abstract
Since the emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), tremendous efforts have been made to sequence the viral genome from samples collected throughout the world. Here, we evaluate how various countries have performed in sequencing from the perspectives of “fraction”, “timeliness”, and “openness”. We found that high proportions of samples were sequenced in the UK, the USA, Australia, and Iceland; sequencing was performed promptly in Iceland, the Netherlands, and the Democratic Republic of the Congo; and data were shared timely from the Netherlands, the USA, Iceland, and the UK. Although many developing countries have high numbers of SARS-CoV-2 infected cases but few published sequences, we observed good performance on sequencing efforts for some low- and middle-income countries. Further strengthening of the sequencing capacity at a global level would help in the fight against not only the current pandemic but also future outbreaks of viral diseases.
Keywords: SARS-CoV-2, COVID-19, Genome, Sequencing, Molecular epidemiology, Global health, Evolution
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was identified as a causative agent of coronavirus disease 2019 (COVID-19), and its genomic data first became available from China on January 10, 2020. Since then, tremendous efforts have been made to sequence the viral genome from samples collected throughout the world. Genomic data can be utilized for epidemiological investigations at both local and global levels. For example, a study in the Netherlands on a large cluster of COVID-19 cases applied combined conventional and molecular epidemiology analyses using viral genomic data and identified multiple introductions of the virus from a community into healthcare facilities (Sikkema et al., 2020). Phylogeographic analysis using genomic data has revealed the transmission dynamics of the virus, including from where and when the virus was imported and how it has been spreading (Fauver et al., 2020, Worobey et al., 2020). Collecting viral sequence data is also important for conducting an evolutionary analysis to infer the origin of the virus (Boni et al., 2020), detect mutations that may affect the pathogenicity and infectivity of the virus (Korber et al., 2020), and identify a conserved region to target for future vaccine development (Day et al., 2020). Such analyses rely both on the viral sequence data collected locally and on the abundance of publicly available sequence data from throughout the world (Hadfield et al., 2018). Thanks to global solidarity and the trend of “open data”, genomic sequences of SARS-CoV-2 from many parts of the world are reported, shared, and publicly available. Here, we analyze and report the virus sequencing efforts by country during the pandemic.
Methods
We obtained data on the number of COVID-19 cases in each country from the World Health Organization website (https://covid19.who.int/), and we acquired SARS-CoV-2 sequence data along with metadata, such as the reporting country, sample collection date, and data submission date, from the GISAID database (Shu and McCauley, 2017); accessed on September 6, 2020. Sequence data longer than 20,000 nucleotides were regarded (near-complete) genomic sequences and included in the further analysis. The quality of sequence data was not considered for the inclusion criteria.
Results
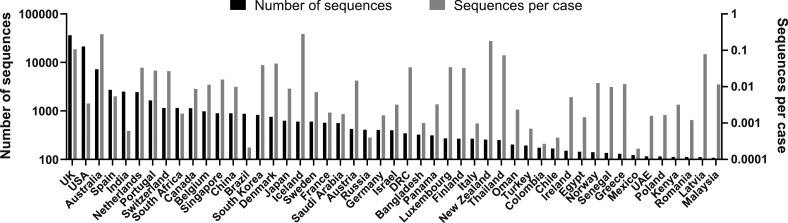
Forty-nine countries have published >100 genomic sequences. The UK (38.9%) and the USA (22.7%) accounted for the majority of all published genomic sequences (N = 93,817) (Figure 1 ). The rate of the number of SARS-CoV-2 genomic sequences per reported COVID-19 case varied widely among countries. Iceland sequenced the highest proportion of reported cases (up to 30% of all cases). Because epidemiological situations and timelines differ among countries, we analyzed each country’s genomic sequencing efforts of SARS-CoV-2 from the perspectives of “fraction”, “timeliness”, and “openness” at a relatively early stage of the epidemic (Figure 2 ).
Figure 1.
Number of genomic sequences of SARS-CoV-2.
Countries in which more than 100 genomic sequences had been published as of September 2020 are listed in order of the number of sequences. The number of SARS-CoV-2 genomic sequences per reported COVID-19 case in each country is also shown. UK, the United Kingdom; USA, the United States of America; DRC, the Democratic Republic of the Congo; UAE, the United Arab Emirates.
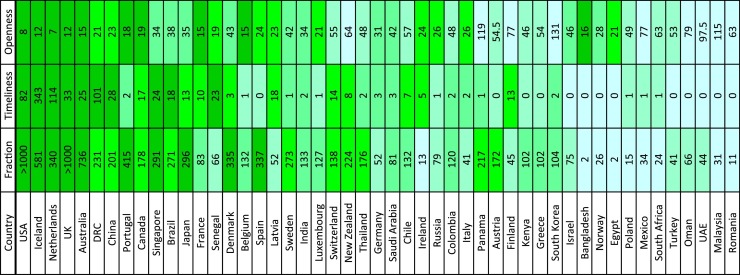
Figure 2.
Sequencing effort for SARS-CoV-2.
Countries are ranked by three indicators describing sequencing efforts. “Fraction” is the number of viral sequences of samples collected by the time the cumulative number of COVID-19 cases had reached 1000 in each country. Because samples were collected before disease confirmation and positive samples could be identified retrospectively, the value could be larger than 1000. “Timeliness” is the number of sequences that had been published by the time the cumulative number of COVID-19 cases had reached 1000 in each country. “Openness” is the time gap (days) between sample collection and sequence data submission for the first 100 sequences in each country. Sequence data for which the collection and submission dates are unknown were excluded from the analysis. Darker colors indicate better performances.
“Fraction” was assessed using the number of viral sequences of samples collected by the time the cumulative number of COVID-19 cases had reached 1000 in each country. The UK, the USA, Australia, and Iceland sequenced more than 50% of the first 1000 cases in each country. “Timeliness” was assessed by how many sequences had been published by the time the cumulative number of COVID-19 cases had reached 1000 in each country. Iceland, the Netherlands, and the Democratic Republic of the Congo published more than 100 sequences by the designated time point. Finally, we analyzed “openness”, noting that it is difficult to assess this point because the quantity of “unpublic” data remains unknown. Therefore, we used the time gap between sample collection and sequence data submission as a surrogate to gauge willingness to make data open. There is a caveat that this indicator can also be affected by the sequencing capacity of each country. We calculated the median days of the time gap for the first 100 sequences in each country and found that the Netherlands, the USA, Iceland, and the UK released sequence data within two weeks of sample collection.
Discussion
Overall, the USA, Iceland, the Netherlands, the UK, and Australia showed great performance in the three indicators for sequencing efforts. The number of SARS-CoV-2 genomic sequences deposited in the GISAID database has been substantially increasing day by day. Sequencing efforts keep improving in many countries, although the present study focused only on the early phase of the epidemic in each country. Another caveat is that we did not check the quality of sequence data such as a Q-score and ambiguous nucleotides. Unfortunately, there are a lot of low-quality sequence data in the database that would affect evolutionary and phylogenetic analyses (De Maio et al., 2020). That point should be also investigated to evaluate sequencing efforts in the future.
Importantly, a lower ranking in Figure 2 does not indicate that those countries exhibited poor performance. Although we listed 49 countries in which more than 100 sequences were deposited in the public database as of September 2020, there are many more countries with high numbers of cases but few, or no, sequence data available. Such missing data would create bias in a phylogeographic analysis to elucidate the global transmission dynamics of SARS-CoV-2. While the cost of sequencing has decreased and mobile sequencing machines have become available in the last few years, genomic sequencing is still technically, logistically, and financially challenging in resource-limited settings. International and domestic collaboration among public health authorities, healthcare facilities, academia, and industries must address these issues.
Simultaneously, we observed good performance of sequencing efforts for some low- and middle-income countries including the Democratic Republic of the Congo, Brazil, Senegal, India, and Thailand (Figure 2). This finding encourages further strengthening of sequencing capacity at the global level, which can lead to the development of an effective response strategy against not only the current pandemic but also future outbreaks of viral diseases.
Funding
This study was funded in part by the Leading Initiative for Excellent Young Researchers (grant number 16809810) from the M inistry of Education, Culture, Sports, Science and Technology in Japan. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest
The author declares no conflict of interest.
Author’s contribution
YF conceived the study design, performed data collection and analysis, and wrote the manuscript.
Ethical approval
Not required.
References
- Boni M.F., Lemey P., Jiang X., Lam T.T.-Y., Perry B.W., Castoe T.A., et al. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat Microbiol. 2020;5(11):1408–1417. doi: 10.1038/s41564-020-0771-4. [DOI] [PubMed] [Google Scholar]
- Day T., Gandon S., Lion S., Otto S.P. On the evolutionary epidemiology of SARS-CoV-2. Curr Biol. 2020;30(15):R849–R857. doi: 10.1016/j.cub.2020.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maio N., Walker C., Borges R., Weilguny L., Slodkowicz G., Goldman N. 2020. Issues With SARS-CoV-2 Sequencing Data.https://virological.org/t/issues-with-sars-cov-2-sequencing-data/473 [Google Scholar]
- Fauver J.R., Petrone M.E., Hodcroft E.B., Shioda K., Ehrlich H.Y., Watts A.G., et al. Coast-to-coast Spread of SARS-CoV-2 during the early epidemic in the United States. Cell. 2020;181:990–996. doi: 10.1016/j.cell.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., et al. NextStrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34(23):4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., et al. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data–from vision to reality. Eurosurveillance. 2017;22(13):30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikkema R.S., Pas S.D., Nieuwenhuijse D.F., O’Toole Á., Verweij J., van der Linden A., et al. COVID-19 in health-care workers in three hospitals in the south of the Netherlands: a cross-sectional study. Lancet Infect Dis. 2020;20(11):1273–1280. doi: 10.1016/S1473-3099(20)30527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worobey M., Pekar J., Larsen B.B., Nelson M.I., Hill V., Joy J.B., et al. The emergence of SARS-CoV-2 in Europe and North America. Science (80-) 2020;370(6516):564–570. doi: 10.1126/science.abc8169. [DOI] [PMC free article] [PubMed] [Google Scholar]