Abstract
An epidemic caused by COVID-19 in China turned into pandemic within a short duration affecting countries worldwide. Researchers and companies around the world are working on all the possible strategies to develop a curative or preventive strategy for the same, which includes vaccine development, drug repurposing, plasma therapy, and drug discovery based on Artificial intelligence. Therapeutic approaches based on Computational biology and Machine-learning algorithms are specially considered, with a view that these could provide a fast and accurate outcome in the present scenario. As an effort towards developing possible therapeutics for COVID-19, we have used machine-learning algorithms for the generation of alignment kernels from diverse viral sequences of Covid-19 reported from India, China, Italy and USA. Using these diverse sequences we have identified the conserved motifs and subsequently a peptide library was designed against them. Of these, 4 peptides have shown strong binding affinity against the main protease of SARS-CoV-2 (Mpro) and also maintained their stability and specificity under physiological conditions as observed through MD Simulations. Our data suggest that these evolutionary peptides against COVID-19 if found effective may provide cross-protection against diverse Covid-19 variants.
Abbreviations: SARS-CoV-2, Severe Acute Respiratory Syndrome – Coronavirus; MERS-CoV, Middle East Respiratory Syndrome - Coronavirus; ACE2, angiotensin-converting enzyme 2; PLpro, papain like protease; Mpro, Main Protease; 3CLpro, 3-chymotrypsin-like protease; RTC, replicase-transcriptase complex; RBD, receptor binding domain; ML, Machine Learning; DEE, Dead End Elimination; MD, Molecular Dynamics; RMSD, root mean square deviation; RMSF, root mean square fluctuation
Keywords: Covid-19, Artificial intelligence, Evolutionary peptides, Computational biology, SARS-CoV-2
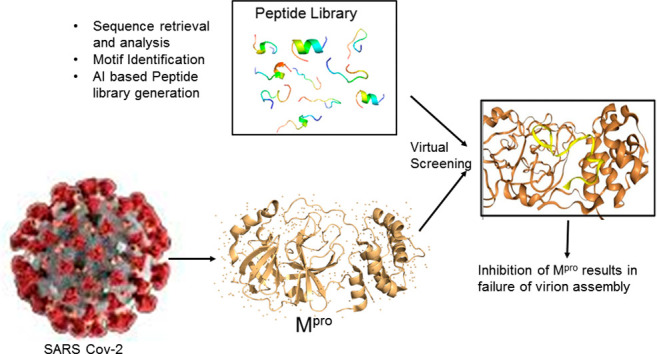
Graphical abstract
Highlights
-
•
Machine-learning algorithm was employed to analyze diverse viral sequences of Covid-19.
-
•
Alignment kernels were generated which were subsequently used for motif prediction.
-
•
Optimized algorithm helped in designing library of evolutionary set of peptides.
-
•
MD Simulations revealed the stability and selectivity of the selected top 4 peptides with main protease Mpro.
1. Introduction
A novel positive-sense, single-stranded RNA virus of family Coronaviridae named SARS-CoV-2 has become a serious threat to life emerging from China and spreading worldwide [1]. It has been declared a global health emergency by the World health organization (WHO). No effective treatment regimen is available against this virus. Affected nations are trying all the possible strategies for treatment and prevention of the spread of this deadly infection. Guidelines of WHO, along with a suggestion for isolation of suspected and confirmed patients, recommended supportive care for individuals infected with SARS-CoV-2 is by oxygen, fluid therapy and for secondary infections, antibiotics have been suggested.
The major ongoing treatments include antiviral drugs used against human immunodeficiency virus (HIV), anti-malarial drugs, and few compounds preventing the viral replication and convalescent plasma [[2], [3], [4]]. Anti-viral therapy included Lopinavir/ritonavir (Kaletra) which are protease inhibitors interfering with the replication and synthesis of HIV. Few reports suggest that IFN-γ could also be considered as a treatment strategy because it interferes with viral replication and promotes both arms of immune response i.e. Innate and adaptive. Cumulatively, we can say that the treatment strategies adopted against Covid-19 are going on three fronts viz., drug repurposing, designing new drugs, and ongoing studies for developing vaccines against Covid-19 [5].
Herein, our efforts are to report epidemiology of the disease, various systemic treatment methodologies under pursue and our Artificial Intelligence rationale-based treatment approach for Covid-19 and its therapeutics.
1.1. Epidemiology of COVID-19
Family of coronavirus including SARS-2003, MERS-2012, and COVID-19 has affected a larger population in the last 2 decades. COVID-19 caused by SARS-CoV-2 which started from Wuhan in China has spread rapidly in many countries around the globe, turning into a deadly pandemic and more than three million confirmed cases have been reported [6,7]. After the initiation of disease at the end of 2019, nearly 84,900 cases have been reported from China, Wuhan being the epicenter. It was jointly reported by the World Health Organization (WHO)-China fact-finding mission that the peak of the epidemic was observed in January and February 2020 and the emergence of new cases decreased by March. Although by the end of March; 938,113 total cases and 44,779 total deaths have been reported all over the world. Presently, data confirms cases from all the continents except Antarctica. After China, Italy was the second country to be affected badly by Covid-19 with a very high fatality ratio to the number of cases closely followed by USA, India and Brazil. The case fatality rate of USA was found to be highest among all the countries with more than 195,239 deaths till September 9, 2020. All over the world, 908,000 deaths have been reported as of September 9, 2020. All 50 states of the United States have been affected by the pandemic. India has also reported nearly 75,091 deaths with 87,700 average new cases every day (September 9, 2020). The total number of cases has been more than 4.46 million. Johns Hopkins data reports 6.54 million cases in the USA as of September 9, 2020.
Initially, when the disease originated, the transmission was found to be associated with the seafood market which sells live animals in Wuhan [8]. However, after the progression of the disease, the main transmission mode is found to be individual-to-individual transmission, which is the reason social-distancing is being followed by affected countries very strictly. This individual to the individual transmission of SARS-CoV-2 usually occurs by respiratory droplets, similar to influenza when the infected individual coughs, sneezes, or talks in close contact with other person directly reaching the mucous membrane [9]. A study was conducted using experimentally generated aerosol to evaluate the stability of SARS-CoV-2 in aerosols using a Bayesian regression model for the decay rate. They found that SARS-CoV-2 could exist in aerosols for 3 h. They also suggested that the stability of the virus was more on plastic and stainless steel than on copper and cardboard, and virus although in low titer was found nearly up to 72 h on these surfaces [2].
A study based on data from two hospitals in Wuhan found that the isolation wards and ventilated patient room had very less viral RNA and was found to be highly increased in the toilet area. Moreover, the area of medical staff was also found to be initially having a high concentration of viral RNA which was later on reduced by rigorous sanitization [10]. It has been suggested by few studies that these respiratory droplets could be carried in a gas cloud and their horizontal trajectories were estimated upto 6 ft. Overall studies suggest that recommendations on airborne settings could vary but precautions for avoiding airborne transmission must be universally followed, mainly where the generation of the aerosol is possible [11].
Along with respiratory samples, the presence of SARS-CoV-2 has also been studied in various other samples such as stool, blood, ocular secretions. Real-time PCR was conducted to analyze the presence of the virus in the pharyngeal swab, blood, and anal swab. Data indicated the presence of the virus in both blood and anal swab. Data confirmed that viral RNA could be present in extra-pulmonary sites [12,13].
Another study with 205 patients found live viruses in feces which suggested that the fecal route could also be a mode of viral transmission [14]. A 65 years old female having a travel history from Wuhan to Italy represented with nonproductive cough, sore throat, coryza, and bilateral conjunctivitis. Along with nasal swab, her ocular swab was also collected due to persistent conjunctivitis and viral RNA was detected in the ocular swab also. Surprisingly, SARS-CoV-2 RNA was detected in ocular swabs even after it was not detected in nasal swabs. This further strengthens the need for control measures like avoiding touching face, eyes, and nose. However, more studies need to confirm fecal-oral transmission [15].
The other major issue is for long the individual once infected with Covid-19 is infectious. It is speculated that the transmission of the virus could occur even before the symptoms appear as well as during disease. In 17 patients after onset of symptoms, a high viral load was detected in the nose as compared to the throat. It was suggested that nucleic acid shedding of the virus was similar to the influenza virus. Moreover, the viral load was similar in asymptomatic and symptomatic patients suggesting that asymptomatic patients have the equal potential of transmission and transmission could occur in the early stage of infection [16]. A cohort study conducted in two hospitals in Hong Kong suggested that viral load was initially high during the first week after the symptoms appear but declined subsequently with time, even in one patient viral load was detected after 25 days of onset of symptoms [17]. These suggest that viral RNA levels are higher in the upper respiratory tract after the onset of symptoms as compared to later illness [18].
Clinical data of viral shedding was compared with incubation period, epidemiological data and interval between cases in the transmission chain for a conclusive inference on the profile of infectivity. The group reported a temporal pattern of viral shedding in 94 patients with confirmed Covid-19 infection and the infectiousness profile of Covid-19 was modeled from a separate 77 transmission pairs. The highest viral load was found in throat swabs at the onset of symptoms and it was inferred that infectivity reached very high before the onset of symptoms. It was calculated that nearly 44% of secondary cases got an infection during the pre-symptomatic stage of index cases [19]. Most of these studies suggest that the infected individual has a higher possibility of causing transmission at the initial stage, which needs to be further validated.
1.2. Life cycle of SARS-CoV-2
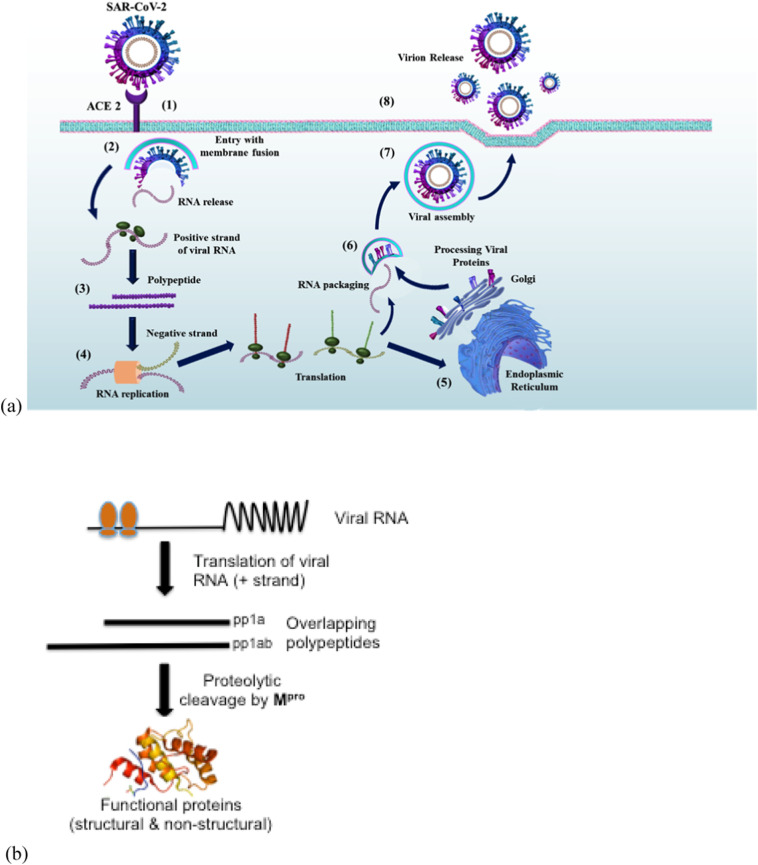
The exact life cycle and disease proliferation of SARS-CoV-2 is yet to be deciphered, but due to its similarity with other coronaviruses, like SARS-CoV and MERS-CoV, its believed to share similar life cycle. The replication of coronavirus occurs inside host cell and different stages of its life cycle are shown in Fig. 1a. As previously mentioned, coronavirus is a positive-sense RNA virus with a genome size of ~30 kb which acts as mRNA for translation of replicase polyproteins. Of this genome, only one-third encodes for structural and accessory protein, while the rest genome encodes for non-structural proteins. The virus comprises of mainly four structural proteins; S (Spike) protein, E (Envelope) protein, M (Membrane) protein and N (Nucleocapsid) protein [20]. The S protein initiates the attachment of virion with the receptor angiotensin-converting enzyme 2 (ACE2) of host cell. Upon entering the host cell, S protein is cleaved by TMPRRS2 protease exposing fusion peptide, which in turn is responsible for binding of cellular membranes and subsequently release of viral RNA into cytoplasm [21,22].
Fig. 1.
(a) Life cycle of SARS-CoV-2. (1) The replication of SAR-CoV-2 begins with the binding of its spike protein (S) with receptor angiotensin-converting enzyme 2 (ACE2) on the cell surface molecules of the host. (2) The viral S protein undergoes conformational changes leading to a fusion between the viral envelope with the plasma or endosomal membranes, entering the cell's cytoplasm, and releasing RNA. (3) The positive-strand viral RNA serves as mRNA for translation of ORF 1a and 1b, encoding units of the RNA-dependent RNA polymerase. (4) RNA polymerase transcribes full-length complementary negative-sense RNA. Viral proteins (S, M, E, and N) get translated (5) The proteins are further processed in the Endoplasmic Reticulum and Golgi Apparatus. (6) Packaging of viral proteins and positive-strand RNA. (7) Assembling complete virion surrounded by the plasma membrane. (8) Apoptosis of cells with the release of the virion. (b) Role of Main protease Mpro in SARS-CoV-2.
The viral RNA released in cytosol now translates the replicase gene encoding two ORFs, rep1a and rep1b, which further expresses two overlapping polyproteins, pp1a and pp1ab. Coronaviruses family usually has 2–3 proteases for cleaving these polyproteins. They may be papain like protease (PLpro) or 3-chymotrypsin-like protease (3CLpro). 3CLpro is the main protease, also known as Mpro cleaving at 11 distinct sites (Fig. 1b). Post processing, these polyproteins assemble at replicase – transcriptase complex (RTC), serving as negative strand RNA for the actual replication and transcription of positive strand viral RNA [[23], [24], [25]]. The newly transcribed positive strand mRNAs expresses the structural and non-structural accessory proteins including viral proteinases. These viral proteins translocate to a zone between ER and Golgi apparatus that acts as an assembly site for new virions. Post maturation, new virions bud off from golgi apparatus as vesicles and are released from the host cell [26,27].
1.3. Therapies against Covid-19
The major issue for urgently designing therapies against Covid-19 is no background data available. The reason being, SARS-CoV-2 the causative organism of the disease was never heard and studied for any type of therapeutic strategy [28]. However, the genetic sequence of SARS-CoV-2 was deciphered in January 2020 in a very fast manner, which triggered scientists around the world to identify various methodologies to fight it, which is the need of time visualizing the scenario in the world, due to the pandemic. All researchers are targeting one of the four major pathways to block the spread of disease, i.e., inhibiting the interaction of virus with human cell receptors, or counteracting the synthesis and replication of viral RNA, or re-establishing and boosting host innate immunity, or inhibiting host specific receptors or enzyme [29].
1.3.1. Vaccine technologies deployed against Covid-19
A successful vaccine is essential for the prevention of future infections and mortalities [30]. This will not only reduce pressure on the healthcare system but also on the economy, which is badly affected by this pandemic.
Various vaccination strategies including whole virus vaccine, subunit vaccine, nucleic acid vaccines are in pipeline initiated by multinational companies like Johnson and Johnson along with research organizations in many nations. Johnson and Johnson in collaboration with Beth Israel Deaconess Medical Center, part of Harvard Medical School initiated their efforts in January 2020 once the sequence of the virus was deciphered. Their vaccines are based on Janssen's proven AdVac® and PER.C6® technology which the company has used to design Zika, RSV, and HIV vaccines which are mostly in Phase2 or Phase 3 trials. Its estimated availability is in early 2021 [31]. A live-attenuated vaccine has been developed by Serum Institute of India in partnership with US-based biotech drug research company Codagenix against SARS-CoV-2 and is under the pre-clinical trial phase [32].
Various other strategies, based on inducing neutralizing antibodies against surface-exposed spike (S) glycoprotein or S protein are underway to develop the Covid-19 vaccine [33]. Most of these spike protein vaccines are based on the rationale of inducing a robust immunological response against the spike protein to avoid its interaction with the ACE2 receptor. Full length S protein or S1-receptor binding domain is being expressed in viral-like particles, DNA, or any other suitable expression vector. Clover bio-pharmaceuticals, a clinical-stage, Biotechnology Company based on research, are in process to develop a recombinant subunit vaccine for SARS-CoV-2. The company is using its patented Trimer-Tag© technology for designing a subunit vaccine, which is the S protein subunit-trimer vaccine (S-Trimer) and the expression system used is rapid mammalian cell-culture based expression system. Once a successful vaccine candidate is obtained, Clover can speed up the production of vaccines as they have in-house cGMP bio-manufacturing capabilities [3]. Texas Children's Hospital Center for Vaccine Development at Baylor College of Medicine has been working on the development and testing of a subunit vaccine which comprises only the receptor-binding domain (RBD) of the SARS-CoV S-protein. This antigen has undergone cGMP manufacture and therefore can undergo clinical trials soon. It was found that this vaccine candidate did not induce significant eosinophilia or eosinophilic lung pathology in the mouse challenge model supporting its safety [34].
Texas Children's Hospital has developed one vaccine based on yeast-derived (Pichia pastoris) recombinant protein comprised of the receptor-binding domain (RBD) of the SARS-CoV which has been formulated on alum is termed as SARS-CoV 219-N1. This vaccine may be a potential heterologous vaccine. The Australian group has developed a vaccine that includes synthetic viral proteins and an additional stabilization domain called the molecular clamp. This is based on the idea that synthetic proteins to be held in the conformation and viral fusion proteins be maintained before they are merged with the host cells. This molecular clamp was used so that vaccines could be recognized by the host immune system more efficiently [35].
Novavax has also identified a SARS-CoV-2 Recombinant Spike Protein Nanoparticle vaccine, NVX-CoV2373 which has shown high immunogenicity and neutralizing antibodies were produced in high levels, as the pre-clinical studies data indicates. The nano-particle technology used is the propriety of Novavax. In the animal studies wherein, they were immunized with one dose, a high level of antibodies specific to spike protein with Angiotensin-converting enzyme 2 (ACE-2) human receptor binding domain blocking activity and neutralizing antibodies for SARS-CoV-2 wild-type virus were observed. Eight folds increase in antibody titer was observed after the second immunization dose [36].
This is the first vaccine against COVID-19 which would be tested outside of the USA, and as for clinical trials are concerned, it is third in the world, conducted by Australia's largest Phase 1 clinical trials specialist Nucleus Network. Nucleus will commence its Phase 1 trial in Melbourne and Brisbane clinics in the coming weeks. Inovio is also using nucleic acid-based technology but instead of RNA, they are using DNA as a base for the vaccine. Their concept also involves the ability of the body to produce proteins that mimic the SARS-CoV-2 spike protein. Their vaccine relies on electroporation of plasmid DNA into human cells. Phase 1 trial is in pipeline for this candidate vaccine [37].
Various companies based on biotech research are using advanced nucleic acid-based vaccine platform for Covid-19. Various companies like Inovio Pharmaceuticals, Moderna therapeutics are involved using this strategy. Moderna has come up with an mRNA-1273 based platform for the vaccine in close association with the National Institute of Health. These RNAs when administered will direct the cells of the recipient to produce protein mimicking “spike protein” and elicit an immune against the Covid-19 virus. They are about to initiate Phase 1 clinical trials.
The above data suggests that there are many vaccines in the pipeline with different strategies involved. However, there is a lengthy process of phase trials including manufacturing and testing toxicity. The coming few months are very critical for determining the fate of this pandemic. We urgently require vaccines and even FDA or other regulatory bodies approve it, after evaluating the data of clinical trials for safety and efficacy, another major hurdle to prevail would be to generate a bulk supply of the candidate vaccine to meet the need of the large population.
1.3.2. Passive antibody therapy
The general strategy in this therapy is to draw blood from an individual who was sick with COVID-19 and has recovered for the screening of virus-neutralizing antibodies [38,39]. The samples in which neutralizing antibodies are found in high levels; these serum samples with virus-neutralizing antibodies could be administered prophylactically in individuals at high risk such as health care professionals, close contacts of confirmed cases of COVID-19 patients and to individuals who are having vulnerable health conditions. Trials to use the convalescent serum for patients also have been initiated, to reduce the symptoms and mortality. In a small clinical study conducted in China recruiting 10 severely infected patients and it was found that one dose of convalescent plasma was well tolerable by patients and neutralizing antibodies levels were significantly increased or levels were maintained. Viremia was found to disappear after 7 days with improvement in clinical and paraclinical symptoms suggesting its potential efficacy to treat patients [40,41].
FDA has initiated guidelines for using convalescent plasma therapy and Methodist Hospital in Houston recruited plasma donors a few days back and initiated the first plasma transfusions to COVID-19 patient the following day [[42], [43], [44]]. The Drug Controller General of India (DCGI) has also approved clinical trials using convalescent plasma therapy. According to The Print, the US has started a clinical trial for evaluating the efficacy of gimsilumab which is an artificially synthesized monoclonal antibody and targets granulocyte macrophage-colony stimulating factor, a key driver in lung hypertension [45,46].
1.3.3. Drug based therapeutic approaches
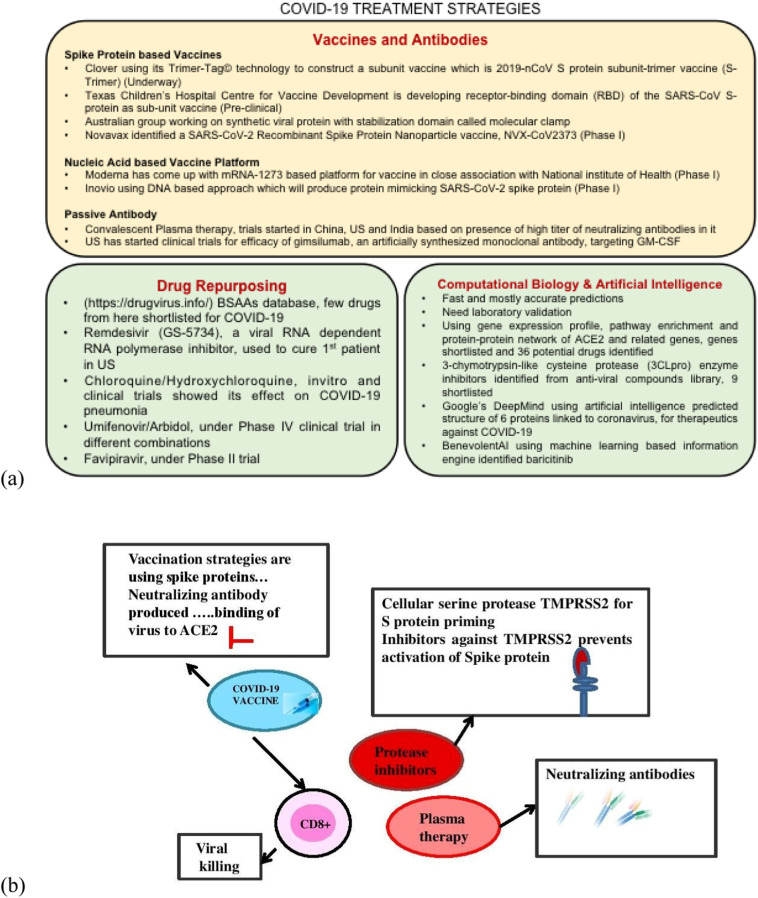
This arm of treatment against COVID-19 is bifurcated into two strategies, one is drug repurposing and other is search for novel drug molecules based on structure-based drug designing using computational biology and Artificial intelligence. FDA and many other big organizations are insisting researchers for long, for the repurposing of drugs. Broad-spectrum antiviral agents (BSAAs), which are small molecules inhibiting spectrum of human viruses have been considered as safe in humans by early phase clinical trials. A database has been created for these nearly 120 drugs wherein they suggested that few can be effective against COVID-19 (https://drugvirus.info/). Various therapeutic strategies and their proposed outcomes are presented in Fig. 2 .
Fig. 2.
Various therapeutic strategies underway against COVID-19 (a) and their proposed outcomes (b).
Remdesivir (GS-5734), which is a viral RNA-dependent RNA polymerase inhibitor, has been under study for early-stage SARS-CoV-2 and is in Phase III clinical trial (ClinicalTrials.gov Identifier: NCT04252664; NCT04280705). It has shown potential against SARS-CoV and the Middle East respiratory syndrome (MERS-CoV) [47,48]. Intravenous remdesivir was used to treat the first case of SARS-CoV-2 patient in the USA when his condition deteriorated. However, to evaluate safety and efficacy, randomized controlled trials are needed [49].
Chloroquine is in use against malaria and some autoimmune diseases [50]. Potential of hydroxychloroquine has been studied in combination therapy for viral pneumonia (ClinicalTrials.gov Identifier: NCT04261517). In vitro studies for SARS-CoV-2 showed that chloroquine is potentially active in suppressing it. Hydroxychloroquine clinical trial for COVID-19 indicated that in patients treated with hydroxychloroquine, the total time of recovery was shortened and it also helped in resolving pneumonia [4,51]. A study coordinated by The Méditerranée Infection University Hospital Institute in Marseille suggested that viral load was reduced or it disappeared after treatment with COVID-19 and this effect was reinforced by azithromycin [13]. U.S. National Institutes of Health has also initiated a clinical trial for hydroxychloroquine for COVID-19.
Umifenovir/Arbidol is the drug used against influenza infection in Russia and China and is membrane fusion inhibitor which targets the entry of virus [52]. Few pieces of evidence along with a retrospective cohort study suggest that Umifenovir alone or along with antiviral drugs is beneficial against COVID-19 pneumonia and various randomized clinical controlled trials are in progress to study the efficacy of Arbidol on COVID-19. Umifenovir with another antiviral drug lopinavir/ritonavir in different combinations is currently considered for Phase IV clinical trial for pneumonia associated with COVID-19 (ClinicalTrials.gov ID: NCT04255017) [53].
Phase II clinical trials are ongoing for Favipiravir which is an inhibitor for viral RNA polymerase in combination for pneumonia associated with COVID-19 (Chinese Clinical Trial Registry Identifier: ChiCTR2000029544). Japan-based company Fujifilm is working on Phase III clinical trials of Favipiravir in Japan and initiated Phase II trials in the US [54]. Other therapeutic approaches are discussed in other communications [5,[55], [56], [57], [58], [59], [60]].
Another protein acting as potential drug target is the main protease of SARS-CoV-2, Mpro. Mpro is a 33.8 kDa protein which cleaves polypeptides formed from translation of viral RNA. These polypeptides are usually two overlapping products pp1a and pp1ab and their proteolytic cleavage results in functional polypeptides (Fig. 1b), which are then recruited in RTC region wherein translation of other viral structural and non-structural proteins occur. The main protease Mpro, also known as 3CLpro (3-chymotrypsin-like protease) cleaves polypeptides at 11 distinct sites. Mpro protein, being functionally important in the survival and replication of SARS-CoV-2 virus and with the absence of homologues in human, makes them a potential drug target in fighting against SARS-CoV-2 [61]. Zhenming et al. have demonstrated that N3 is a potential inhibitor of Mpro not only for SARS-CoV-2 but also for SARS-CoV and MERS-CoV using structure-based ab initio drug design, virtual screening and high-throughput screening methodologies [62]. Kanhed et al. have screened a FDA approved drug library and the Asinex BioDesign Library to determine potential molecules which can be useful as therapeutic agents for COVID-19 [61]. Even various bioactives from medicinal plants have been screened for potential candidacy against Mpro [63]. Attempts have been made to design protein specific peptides but due to their small size (8 aa residues) they were unable to maintain secondary structure thus were less effective [64,65]. Here, in this study, we have used machine-learning approach to design protein specific peptides inhibiting SARS-CoV-2-Mpro, which would be able to provide effective cross-protection against various COVID-19 variants.
2. Materials and methodology
2.1. Sequence retrieval
Nucleotide Sequence database downloaded from GISAID server [66] for Covid-19. Nucleotide Sequence data set consists of 2765 sequences of different Covid-19 patients from different countries and/or region. We analyzed sequence data of mainly four regions including Wuhan, Italy, USA and India.
2.2. Sequence alignment
The sequence alignment of all sequences belonging to 4 regions (India, Wuhan, Italy, USA) was performed using European Bioinformatics Institute (EBI) ClustalW [67]. The parameters were defined as DEALIGN INPUT = NO, MBED-LIKE CLUSTERING = Yes, NUMBER OF COMBINED = 0, MAX GUIDE TREE = DEFAULT, MAX HMM = 1 and order = aligned. In addition, the alignment results were analyzed using phylogenetic tree, aligned sequences and scoring matrix.
2.3. Generation of alignment kernel and classification
Once alignment is done, we proceeded further with defining sequence kernels on neighborhoods by taking into account the most conserved central residue of the sequence fragment. Thereafter, we created matrices by assuming a fixed serial ordering of the residues. Let P and Q be m x k and m x l matrices respectively. Let R and S be m x m and l x k matrices, such that R and extension of S to n x n matrices, n = max (m, k, l), which is a positive semi definite matrix. Then, (P, Q) = tr (PT R Q) S) is a pair sum and direct alignment kernel. Generalizing this result, hereafter, we defined our generated sets of sequence-based kernels on neighborhoods. We took minimalistic set of neighborhoods that cover all residues in an alignment.
- Training dataset: Here, we divided sequence of the training data set, as mentioned in equation below:
into groups according to their class labels. We obtained frequent occurring subsequences as seed sequences. Thereafter, identified frequent subsequences from each clustered aligned group and then took the merged subsequences sets together as the seed set S′. For each seed SɛS′ and for each A in the training dataset, we used fs to define the feature vector. For two sets of sequences, A, A′, we constructed the complete weighted bipartite sequences and computed the kernel Ka(A,A′) using equation:
where, π : V[A] → V[A′] denotes an alignment of sequence A and A′.and f(V) is a set of features associated with the sequence.
Thereafter, we trained the predictive model obtained through the above equation using a kernel classifier.
Testing dataset: We used the model which was trained to forecast about sequence in the test data set. Here, for each seed SɛS' and for each A in the testing data set, fs is used to label the sequence in A and thereby create feature vectors as performed above for training data set. The above equation illustrated is a representation, wherein, the kernel function Ka(A,A′) is computed for each sequence A in the test data set and for each sequence S′ in the training data set. Hereafter, we have used kernel classifier and thereby models were trained to obtain prediction accuracy of the test data set. The obtained sets of kernel functions were used as an input for the generation of motifs.
2.4. Motif generation
Motifs one of a kind for the main protease of SARS-CoV-2 (Mpro) was recognized by the use of MEME suite v4.11.2 [68]. A motif recognizable proof measure incorporates any number of repetition sites of motifs within the given set of input arrangements. The number of motifs that ought to be found was set as 6 with a least length of 6 buildups to a most extreme length of 20 buildups were distinguished. The motif shortlisting was done based on their secondary structure and from that point 6 motifs were shortlisted.
2.5. Peptide library design
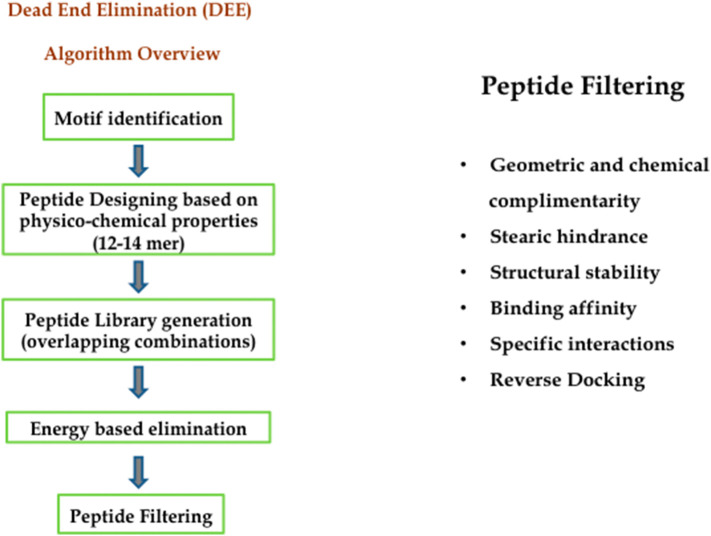
A synthetic library of peptides was designed where the 6 shortlisted motifs, as mentioned above, served as template. For the peptide designing and optimization purpose, a deterministic search approach in consideration with physiochemical properties was implemented. This algorithm is known as Dead End Elimination (DEE) Algorithm. Its main motive is to design novel sequences with an exhaustive searching to identify and eliminate those who doesn't fit in global minimum energy conformation. The total energy of a given pair is represented as:
where, ETot is the total energy of the system; Er is the initial energy of the template; ir is the rotamer state at position i; E(ir) is the self-energy of rotamer at position i; and E(ir, js) is the pairwise energy between rotamers at posiiton i and j [69]. A brief overview of the algorithm is shown in Fig. 3 . Contemplating with overlapping combinations and permutations, a library of 97 peptides was created with an average residue length of 12 to 14 amino acids. Applying various filters narrowed the peptides library search space down. The different filtering criteria's considered were structural stability of protein-peptide complex; geometric and chemical complementarity to ensure minimum stearic clashes between residues; degree and extent of interactions between receptor-ligand, high binding affinity. The concept of reverse docking was also implemented to minimize non-specific binding and toxic effects on humans. Based on these criteria; 4 peptides were shortlisted.
Fig. 3.
Overview of the algorithm used for peptide designing and filtering.
2.6. Structure prediction and docking
PEPstr [70] was used to predict the 3D structure of the designed peptides. From the library of 97 peptides, 4 peptides were chosen as potential candidates based on their structural geometry and chemical complementarity, stearic hindrance, structural stability, hydropathy index, net charges, binding affinity, specific interactions, etc.
AutoDock Vina [71] and MGL Tools 1.5.6. [72] respectively, were used for protein-peptide docking and analysis. While preparing the files for protein and peptides, Gasteiger charges and polar hydrogen atoms were added and subsequently saved as ‘pdbqt’ file. AutoDock follows empirical type of scoring method where total energy is calculated as:
The docking was achieved using Genetic Algorithm with rigid type of docking. PyMOL v1.7.4.5 (Delano Scientific) was used for the visualization of docked complexes and Ligplot+ [73] for the mapping of protein-peptide interactions.
2.7. Molecular dynamics simulation
The Molecular Dynamics (MD) Simulations were used to check the stability and extent of interactions maintained between protein-peptide complexes in physiological conditions using DESMOND 3.2 with maestro-v11.6 (D.E. Shaw Research) [74]. Schrodinger 2018-2 was used as GUI interface for 50 ns long MD Simulations. MD Simulation was done with Normal Pressure and Temperature (NPT) ensemble class at 300 K temperature and 1.01325 bar pressure with Nose-Hoover chain thermostat and Martyna-Tobias-Klein barostat methods. The results were analyzed using inbuilt analysis Simulation Interaction Diagrams. It provides information about protein-ligand interactions in a graphical form, RMSD (Root Mean Square Deviation), protein and ligands RMSF (Root Mean Square Fluctuation) and protein-ligand interactions (hydrophobic, ionic and water-bridge contacts).
RMSD is calculated as:
where, N: total number of atoms for which RMSD has to be calculated; tref: the reference time, (usually the first frame); and r′ is the position of the selected atoms in frame x after overlaying on the reference frame, where frame x is recorded at time tx. The procedure is iterated for every frame in the simulation trajectory.
RMSF is calculated as:
where, T: RMSF is calculated based on the trajectory time, tref: the reference time, ri: the position of residue i; r′: the location of atoms in residue i after superposition on the reference frame, and the angular brackets signifies the average of the square distance is taken over the selection of atoms in the residue.
The total energy of the system is calculated as:
where, Ebond is the bond length energy based on either Morse or LJ Potential, and is defined as:
Eangle is the bond angle energy and is defined as:
Etorsion is the torsion energy associated with the rotation of two atoms of a molecule relative to each other. It is defined as:
Eoop is the out of plane interactions and is defined as:
Ecross is the cross term between different energies like angle energy and torsion energy, etc.
Enonbond includes the energies of non-covalent bonds like van-der-Waals, Coulombs and hydrogen bonds [75].
3. Results
We know that algorithm based machine learning approach is not only fast but chances of failure of drug discovery in lab are reduced largely because so many filters and conditions are implicated while performing these algorithms. Therefore, this systematic approach aided by merit of reduced time required could show the way for a better outcome.
In pursuit of this aim, we defined our therapeutic strategy using machine learning algorithms based on two perspectives, one is selection of diverse viral sequences of Covid-19 reported from India, China, Italy and USA and using these diverse sequences to identify peptides with activity against diverse strain of COVID-19 virus worldwide. Once identified, these peptides were tested against Proteases of COVID-19 for their efficacy or based on diverse peptides identified, peptides of different amino-acid composition could be designed based on identified peptides and tested for cross protection against various viral strains worldwide. Viral sequences were taken from GISAID wherein more than 95,000 viral sequences have been submitted since the emergence of pandemic till date. We aligned these sequences using European Bioinformatics Institute (EBI) tool Clustal Omega and the alignment results were analyzed using phylogenetic tree, aligned sequences and scoring matrix. As we know that identification and designing of these peptides is resource consuming approach, thus, we relied on computational method of high accuracy for prediction of a library of these peptides (Supplementary Table 1) and hence used Dead End Elimination algorithm which is one of the most popular algorithm for model development and further identification of peptides from molecular diverse sequences. We selected proteases as a target because the main protease of SARS-CoV-2 (Mpro) is an important or can say key enzyme, which is involved in the process of replication and transcription of virus.
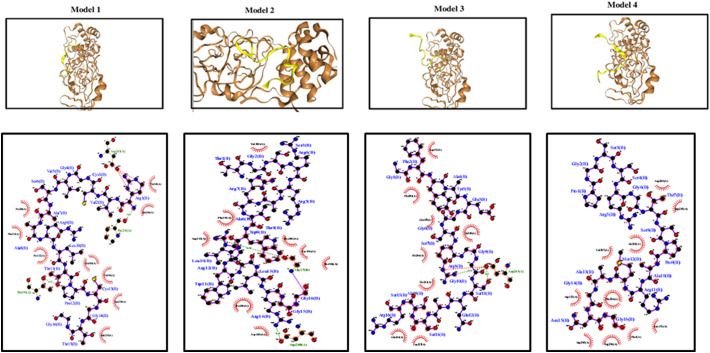
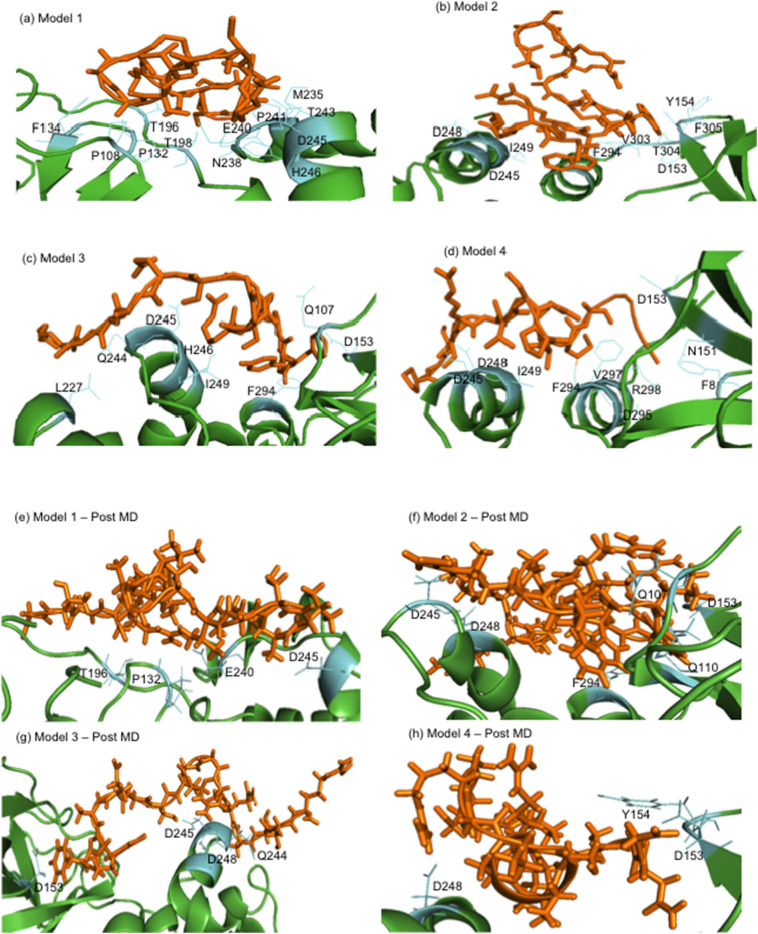
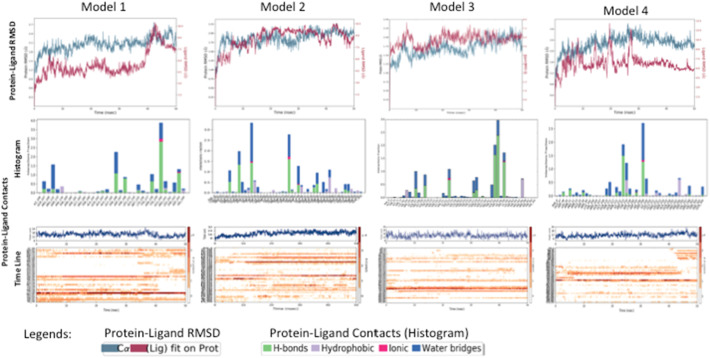
Through the virtual screening against the Mpro identified 4 peptides as the potential candidates in inhibiting protease function with strong binding activity (Table 1). Docking of these selected peptides along with their interactions against the Mpro and their docked poses are shown in Fig. 4, Fig. 5a–d and Table 2. We further carried out short scale MD Simulation of 50 ns time scale to check the stability, strength and amount of interactions being maintained between protein and peptide during simulation time. For model1 and model4, interactions increased by 3 fold and 4 fold respectively but for model2 and model3 interactions increased by more than 5folds during MD simulations (Table 3 and Fig. 6). In the Fig. 5e–h, only those residues that share more than 1 fraction of interactions, i.e. more than one atom of that residue remain in contact with peptide throughout simulation time, are highlighted. All pre-MD interactions were observed to remain during MD. It was also observed that during MD simulation, the negatively charged amino acids aspartate and glutamate (position 153, 240, 245, 248) forms strong-ionic interaction in all the four protein-peptide models indicating that these residues might be crucial while inhibiting the activity of Mpro. The MD Simulation trajectory of all four models in form movie is available as supplementary files (Movie M1, Movie M2, Movie M3, Movie M4). These peptides need further validation in laboratory conditions. In short, we can say we have used the approach of sequence alignment and machine learning algorithm for identifying peptides based on molecular diversity of COVID-19.
Table 1.
Identified nucleotide sequences for peptide designing and Total Energy score.
| S. No | Motif sequences | Motif E-value | Translated sequence | Total energy (kcal/mol) |
|---|---|---|---|---|
| Model 1 | CGGGTTTGCGGTGTAAGTGCAGCCCGTCTTACACCGTGCGGCACAGGCAC | 1.0e-038 | RVCGVSAARLTPCGTG | −23.060 |
| Model 2 | ACAGGTTCGCGACGTGCTCGTACGTGGCTTTGGAGACTCCGTGGAGGAGG | 5.5e-038 | TGSRRARTWLWRLRGG | −38.704 |
| Model 3 | GGCTTCTACGCAGAAGGGAGCAGAGGCGGCAGTCAAGCCTCTTCTCGTTC | 3.0e-037 | GFYAEGSRGGSQASSR | −37.035 |
| Model 4 | CCAGGCAGCAGTAGGGGAACTTCTCCTGCTAGAATGGCTGGCAATGGCGG | 3.0e-037 | PGSSRGTSPARMAGNG | −21.124 |
Fig. 4.
Docking results of identified sequences with proteases wherein peptide is in yellow color and receptor in brown. The upper panel shows the docked poses with the 4 shortlisted peptides (protein represented in brown ribbon form while peptide in yellow color) and the lower panel represents their respective protein-peptide interactions.
Fig. 5.
Mpro residues interacting with peptides. (a) to (d) are protein-peptide complexes before MD Simulations and (e) to (h) are protein-peptide complexes post MD Simulations. Peptide is represented in orange stick form while residues of Mpro protein interacting with peptides are shown in cyan color.
Table 2.
List of interacting residues with the shortlisted peptides.
| Model | Interacting residues |
|---|---|
| 1 | P108, P132, F134, T196, T198, M235, N238, E240, P241, T243, D245, H246 |
| 2 | D153, Y154, D245, D248, I249, F294, V303, T304, F305 |
| 3 | Q107, D153, L227, T243, Q244, D245, H246, I249, F294 |
| 4 | F8, N151, D153, D245, D248, I249, F294, D295, V297, R298 |
Table 3.
Number of interactions between protein-peptide pre-MD and during MD simulations.
| Model | Interactions pre-MD | Interactions during MD |
|---|---|---|
| 1 | 12 | 33 |
| 2 | 9 | 48 |
| 3 | 9 | 49 |
| 4 | 10 | 45 |
Fig. 6.
Short scale Molecular Dynamics Simulation studies to check the stability, strength and amount of interactions being maintained between protein and peptide. The first row represents the protein – ligand (here peptide) RMSDs with protein in blue and ligand in red color. The middle row represents the type and fraction of interactions maintained by various protein residues during simulation time. H-bond is represented in green, hydrophobic in purple, ionic in pink and water bridges in blue color. The bottom row also represents the extent of protein-peptide interactions for a given residue at a particular simulation time. In the timeline graph, darker the color more is the fraction of contacts between protein and peptide.
4. Discussion
We have extensively reviewed the pathogenesis mechanism of SARS-CoV-2 in the context of immunological features to find possible drug candidates. It is based on the strategy that such drugs will prevent the entry of viruses in cells via inhibition of endocytosis and combination therapy with antiviral drugs could reduce the infectivity of the virus and its replication along with preventing inflammatory responses. As the above data suggests, lot of efforts are being taken by both industry and research organizations for developing a therapeutic strategy that might be a positive click for treatment of COVID-19. However, most of the studies are underway, and scientific, medical, and regulatory authorities will not neglect the validation of these aspects, so rigorous validation of the hypothesis is equally important. Strategy along with having an advantage of a short period with accuracy is majorly focused on molecular diversity of pathogen, which is the main lacunae and dilemma for the efficacy of various strategies in process, to be effective against various strains of the virus. To add to the crux of the story, ML has provided us a novel and a promising technique for peptide design, calculating energies for training set of sequences and helps assign an energy criterion that is only applicable to one or few of the amino acid residues. Most importantly, it outscores the generation of possible rotamers and thereby rendering it to be combinatorial easier with fewer amino acid residues. Through this work, we augmented simple kernel representations recognizing important topological similarities and incorporating structural information into kernel functions, which actually helped in approximate matching of structure features of the peptide library created. We demonstrated machine learning algorithms to address biological applications in high-dimensionality datasets. Leveraging the novel framework implemented in this paper opens a newer dimension of generation of peptide library through kernel function. Finally, optimal generation of diverse sequences with favorable conformation has yielded the strategy adopted to be statistically equivalent to discrete design as we are able to design peptides with high selectivity towards their desired target that warrants experimental validation. Considering the evolutionary aspect of SARS-CoV-2, we have retrieved 2765 sequences containing wide variety of mutants from COVID-19 patients. This large dataset was used to generate alignment kernels, which further provided conserved motifs across all mutants. Against these motifs, a library of 97 peptides was designed. This tactic enabled us to cover a wide spectrum of existing as well as probable mutants of SARS-CoV-2 ensuring the efficient targeting of main protease Mpro. Based on binding energies, peptide conformation and strength and type of interactions with protein, 4 peptides were shortlisted. These 4 protein-peptide complexes were subsequently subjected to MD Simulation studies to ensure their stability in physiological conditions as well confirmed which peptide has maintained their potency. Of the 4 most efficient peptides obtained post screening of peptide library, four common interacting residues were found in model 2, 3 and 4. These residues were D153, D245, I249 and F294. Among these also D245 showed strong interaction with all the four peptides suggesting that these residues might be crucial for its functionality. During docking simulations we have observed that the hydrophobic residues viz., I106, H246, I249, P293, F204 and F305 showed good interactions strength with peptides on one side and on the other side the polar residues includes D153, N228, D245, D248, R298, Q306 of the amphipathic structure. These observed structural implications accounts for peptides selectivity towards the target and apparently their antiviral effect. Docking as well as MD simulation analysis shows that model2 and model3 with peptides 2 and 3 respectively, are the best candidates. However, further experimental validation is required to assess the effect of these peptides on the progression of SARS-CoV-2. Machine learning is far more precise than the traditional conventional approaches since it takes into consideration the important factors required for biological activity and selectivity of the peptides. It can also be perceived that the computational designed set of peptides can serve as antiviral therapeutics as well as immuno-modulatory agents because of its versatility as well as tenability. These peptides may also be used as templates for the design of other molecules guided by its hydrophobicity, net positive charge and amphiphilicity.
The following are the supplementary data related to this article.
Library of synthetic peptides along with their sequence, structure and hydropathy index.
M1-model1-0to50ns.mpeg.
M2-model2-0to50ns.mpeg.
M3-model3-0to50ns.mpeg.
M4-model4-0to50ns.mpeg.
Credit authorship contribution statement
All authors contributed to study design and conception. Ritika Kabra (RK): data curation, methodology implementation and writing the original draft. Shailza Singh (SS): Initialization and conceptualization, data curation, supervision, writing and editing the original draft. All authors agree with the submission of the final version of the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
We are thankful to the Director, National Centre for Cell Science (NCCS) for supporting the Bioinformatics and High Performance Computing Facility at NCCS, India.
References
- 1.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhai P., Ding Y., Wu X., Long J., Zhong Y., Li Y. The epidemiology, diagnosis and treatment of COVID-19. Int. J. Antimicrob. Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dhama K., Sharun K., Tiwari R., Dadar M., Malik Y.S., Singh K.P., Chaicumpa W. COVID-19, an emerging coronavirus infection: advances and prospects in designing and developing vaccines, immunotherapeutics, and therapeutics. Hum. Vaccines Immunother. 2020;16:1232–1238. doi: 10.1080/21645515.2020.1735227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Manli, Cao Ruiyuan, Zhang Leike, Yang Xinglou, Liu Jia, Xu Mingyue, Shi Zhengli, Hu Zhihong, Wu Zhong, Xiao Gengfu. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. https://www.nature.com/articles/s41422-020-0282-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naqvi A.A.T., Fatima K., Mohammad T., Fatima U., Singh I.K., Singh A., Atif S.M., Hariprasad G., Hasan G.M., Hassan M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim. Biophys. Acta Mol. basis Dis. 1866;2020 doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu J., Zhao S., Teng T., Abdalla A.E., Zhu W., Xie L., Wang Y., Guo X. Systematic comparison of two animal-to-human transmitted human coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses. 2020;12 doi: 10.3390/v12020244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mandal S., Bhatnagar T., Arinaminpathy N., Agarwal A., Chowdhury A., Murhekar M., Gangakhedkar R., Sarkar S. Prudent public health intervention strategies to control the coronavirus disease 2019 transmission in India: a mathematical model-based approach. Indian J. Med. Res. 2020;151:190–199. doi: 10.4103/ijmr.IJMR_504_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T.Y., Wu J.T., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu N., Li W., Kang Q., Xiong Z., Wang S., Lin X., Liu Y., Xiao J., Liu H., Deng D., Chen S., Zeng W., Feng L., Wu J. Clinical features and obstetric and neonatal outcomes of pregnant patients with COVID-19 in Wuhan, China: a retrospective, single-centre, descriptive study. Lancet Infect. Dis. 2020;20:559–564. doi: 10.1016/S1473-3099(20)30176-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prateek B., Doolan C., de Silva C., Chughtai A.A., Bourouiba L., MacIntyre R.C. Airborne or droplet precautions for health workers treating COVID-19? J. Infect. Dis. 2020;189 doi: 10.1093/infdis/jiaa189. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.W. World Health Organization Modes of transmission of virus causing COVID-19: implications for IPC precaution recommendations. Sci. Br. 2020:1–3. doi: 10.1056/NEJMoa2001316.5. [DOI] [Google Scholar]
- 12.Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science (80-. ) 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 13.Lau S.K.P., Woo P.C.Y., Li K.S.M., Huang Y., Tsoi H.W., Wong B.H.L., Wong S.S.Y., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U. S. A. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu J., Han B., Wang J. COVID-19: gastrointestinal manifestations and potential fecal–oral transmission. Gastroenterology. 2020;158:1518–1519. doi: 10.1053/j.gastro.2020.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colavita F., Lapa D., Carletti F., Lalle E., Bordi L., Marsella P., Nicastri E., Bevilacqua N., Giancola M.L., Corpolongo A., Ippolito G., Capobianchi M.R., Castilletti C. SARS-CoV-2 isolation from ocular secretions of a patient with COVID-19 in Italy with prolonged viral RNA detection. Ann. Intern. Med. 2020;173:242–243. doi: 10.7326/M20-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J., Guo Q., Song T., He J., Yen H.L., Peiris M., Wu J. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.To K.K.W., Tsang O.T.Y., Leung W.S., Tam A.R., Wu T.C., Lung D.C., Yip C.C.Y., Cai J.P., Chan J.M.C., Chik T.S.H., Lau D.P.L., Choi C.Y.C., Chen L.L., Chan W.M., Chan K.H., Ip J.D., Ng A.C.K., Poon R.W.S., Luo C.T., Cheng V.C.C., Chan J.F.W., Hung I.F.N., Chen Z., Chen H., Yuen K.Y. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X., Lau Y.C., Wong J.Y., Guan Y., Tan X., Mo X., Chen Y., Liao B., Chen W., Hu F., Zhang Q., Zhong M., Wu Y., Zhao L., Zhang F., Cowling B.J., Li F., Leung G.M. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 20.Zhao L., Jha B.K., Wu A., Elliott R., Ziebuhr J., Gorbalenya A.E., Silverman R.H., Weiss S.R. Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host Microbe. 2012;11:607–616. doi: 10.1016/j.chom.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng P.K.C., Wong D.A., Tong L.K.L., Ip S.M., Lo A.C.T., Lau C.S., Yeung E.Y.H., Lim W.W.L. Viral shedding patterns of coronavirus in patients with probable severe acute respiratory syndrome. Lancet. 2004;363:1699–1700. doi: 10.1016/S0140-6736(04)16255-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U. S. A. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baranov P.V., Henderson C.M., Anderson C.B., Gesteland R.F., Atkins J.F., Howard M.T. Programmed ribosomal frameshifting in decoding the SARS-CoV genome. Virology. 2005;332:498–510. doi: 10.1016/j.virol.2004.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 25.Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L.M., Guan Y., Rozanov M., Spaan W.J.M., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fehr A.R., Perlman S. Coronaviruses Methods Protoc. 2015. Coronaviruses: an overview of their replication and pathogenesis; pp. 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skariyachan S., Challapilli S.B., Packirisamy S., Kumargowda S.T., Sridhar V.S. Recent aspects on the pathogenesis mechanism, animal models and novel therapeutic interventions for middle east respiratory syndrome coronavirus infections. Front. Microbiol. 2019;10 doi: 10.3389/fmicb.2019.00569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le T. Thanh, Andreadakis Z., Kumar A., Gómez Román R., Tollefsen S., Saville M., Mayhew S. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19:305–306. doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- 29.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peeples L. Avoiding pitfalls in the pursuit of a COVID-19 vaccine. Proc. Natl. Acad. Sci. U. S. A. 2020;117:8218–8221. doi: 10.1073/pnas.2005456117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamey G., Schäferhoff M., Hatchett R., Pate M., Zhao F., McDade K.K. Ensuring global access to COVID-19 vaccines. Lancet. 2020;395:1405–1406. doi: 10.1016/S0140-6736(20)30763-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen W.H., Strych U., Hotez P.J., Bottazzi M.E. The SARS-CoV-2 vaccine pipeline: an overview. Curr. Trop. Med. Rep. 2020;7:61–64. doi: 10.1007/s40475-020-00201-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corey L., Mascola J.R., Fauci A.S., Collins F.S. A strategic approach to COVID-19 vaccine R&D. Science (80-. ) 2020;368:948–950. doi: 10.1126/science.abc5312. [DOI] [PubMed] [Google Scholar]
- 35.Graham B.S. Rapid COVID-19 vaccine development. Science (80-. ) 2020;368:945–946. doi: 10.1126/science.abb8923. [DOI] [PubMed] [Google Scholar]
- 36.Lurie N., Saville M., Hatchett R., Halton J. Developing covid-19 vaccines at pandemic speed. N. Engl. J. Med. 2020;382:1969–1973. doi: 10.1056/NEJMp2005630. [DOI] [PubMed] [Google Scholar]
- 37.Prompetchara E., Ketloy C., Palaga T. Immune responses in COVID-19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 2020;38:1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- 38.Duan K., Liu B., Li C., Zhang H., Yu T., Qu J., Zhou M., Chen L., Meng S., Hu Y., Peng C., Yuan M., Huang J., Wang Z., Yu J., Gao X., Wang D., Yu X., Li L., Zhang J., Wu X., Li B., Xu Y., Chen W., Peng Y., Hu Y., Lin L., Liu X., Huang S., Zhou Z., Zhang L., Wang Y., Zhang Z., Deng K., Xia Z., Gong Q., Zhang W., Zheng X., Liu Y., Yang H., Zhou D., Yu D., Hou J., Shi Z., Chen S., Chen Z., Zhang X., Yang X. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl. Acad. Sci. U. S. A. 2020;117:9490–9496. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou B., Zhong N., Guan Y. Treatment with convalescent plasma for influenza A (H5N1) infection [9] N. Engl. J. Med. 2007;357:1450–1451. doi: 10.1056/NEJMc070359. [DOI] [PubMed] [Google Scholar]
- 40.Tanne J.H. Covid-19: FDA approves use of convalescent plasma to treat critically ill patients. BMJ. 2020;368:m1256. doi: 10.1136/bmj.m1256. [DOI] [PubMed] [Google Scholar]
- 41.Ko J.H., Seok H., Cho S.Y., Ha Y.E., Baek J.Y., Kim S.H., Kim Y.J., Park J.K., Chung C.R., Kang E.S., Cho D., Müller M.A., Drosten C., Kang C.I., Chung D.R., Song J.H., Peck K.R. Challenges of convalescent plasma infusion therapy in Middle East respiratory coronavirus infection: a single centre experience. Antivir. Ther. 2018;23:617–622. doi: 10.3851/IMP3243. [DOI] [PubMed] [Google Scholar]
- 42.Cheng Y., Wong R., Soo Y.O.Y., Wong W.S., Lee C.K., Ng M.H.L., Chan P., Wong K.C., Leung C.B., Cheng G. Use of convalescent plasma therapy in SARS patients in Hong Kong. Eur. J. Clin. Microbiol. Infect. Dis. 2005;24:44–46. doi: 10.1007/s10096-004-1271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hung I.F.N., To K.K.W., Lee C.K., Lee K.L., Chan K., Yan W.W., Liu R., Watt C.L., Chan W.M., Lai K.Y., Koo C.K., Buckley T., Chow F.L., Wong K.K., Chan H.S., Ching C.K., Tang B.S.F., Lau C.C.Y., Li I.W.S., Liu S.H., Chan K.H., Lin C.K., Yuen K.Y. Convalescent plasma treatment reduced mortality in patients with severe pandemic influenza A (H1N1) 2009 virus infection. Clin. Infect. Dis. 2011;52:447–456. doi: 10.1093/cid/ciq106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang B., Liu S., Tan T., Huang W., Dong Y., Chen L., Chen Q., Zhang L., Zhong Q., Zhang X., Zou Y., Zhang S. Treatment with convalescent plasma for critically ill patients with severe acute respiratory syndrome coronavirus 2 infection. Chest. 2020;158:e9–e13. doi: 10.1016/j.chest.2020.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen L., Xiong J., Bao L., Shi Y. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect. Dis. 2020;20:398–400. doi: 10.1016/S1473-3099(20)30141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeng Q.-L., Yu Z.-J., Gou J.-J., Li G.-M., Ma S.-H., Zhang G.-F., Xu J.-H., Lin W.-B., Cui G.-L., Zhang M.-M., Li C., Wang Z.-S., Zhang Z.-H., Liu Z.-S. Effect of convalescent plasma therapy on viral shedding and survival in COVID-19 patients. J. Infect. Dis. 2020;222(1):38–43. doi: 10.1093/infdis/jiaa228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat L.W., Wang Q., Lou Z., Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science (80-. ) 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Wit E., Feldmann F., Cronin J., Jordan R., Okumura A., Thomas T., Scott D., Cihlar T., Feldmann H. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc. Natl. Acad. Sci. U. S. A. 2020;117:6771–6776. doi: 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., Feldt T., Green G., Green M.L., Lescure F.X., Nicastri E., Oda R., Yo K., Quiros-Roldan E., Studemeister A., Redinski J., Ahmed S., Bernett J., Chelliah D., Chen D., Chihara S., Cohen S.H., Cunningham J., D’Arminio Monforte A., Ismail S., Kato H., Lapadula G., L’Her E., Maeno T., Majumder S., Massari M., Mora-Rillo M., Mutoh Y., Nguyen D., Verweij E., Zoufaly A., Osinusi A.O., DeZure A., Zhao Y., Zhong L., Chokkalingam A., Elboudwarej E., Telep L., Timbs L., Henne I., Sellers S., Cao H., Tan S.K., Winterbourne L., Desai P., Mera R., Gaggar A., Myers R.P., Brainard D.M., Childs R., Flanigan T. Compassionate use of remdesivir for patients with severe Covid-19. N. Engl. J. Med. 2020;382:2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Colson P., Rolain J.M., Lagier J.C., Brouqui P., Raoult D. Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int. J. Antimicrob. Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., Li Y., Hu Z., Zhong W., Wang M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6 doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu Z., Lu Z., Xu T., Chen C., Yang G., Zha T., Lu J., Xue Y. Arbidol monotherapy is superior to lopinavir/ritonavir in treating COVID-19. J. Inf. Secur. 2020;81:e21–e23. doi: 10.1016/j.jinf.2020.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lian N., Xie H., Lin S., Huang J., Zhao J., Lin Q. Umifenovir treatment is not associated with improved outcomes in patients with coronavirus disease 2019: a retrospective study. Clin. Microbiol. Infect. 2020;26:917–921. doi: 10.1016/j.cmi.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cai Q., Yang M., Liu D., Chen J., Shu D., Xia J., Liao X., Gu Y., Cai Q., Yang Y., Shen C., Li X., Peng L., Huang D., Zhang J., Zhang S., Wang F., Liu J., Chen L., Chen S., Wang Z., Zhang Z., Cao R., Zhong W., Liu Y., Liu L. Experimental treatment with favipiravir for COVID-19: an open-label control study. Engineering. 2020 doi: 10.1016/j.eng.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Misra D.P., Agarwal V., Gasparyan A.Y., Zimba O. Rheumatologists’ perspective on coronavirus disease 19 (COVID-19) and potential therapeutic targets. Clin. Rheumatol. 2020;39:2055–2062. doi: 10.1007/s10067-020-05073-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cao X. COVID-19: immunopathology and its implications for therapy. Nat. Rev. Immunol. 2020;20:269–270. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu X., Han M., Li T., Sun W., Wang D., Fu B., Zhou Y., Zheng X., Yang Y., Li X., Zhang X., Pan A., Wei H. Effective treatment of severe COVID-19 patients with tocilizumab. Proc. Natl. Acad. Sci. U. S. A. 2020;117:10970–10975. doi: 10.1073/pnas.2005615117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kumar V., Kashav T., Hassan I. Pathol. Prev. Ther. Neurodegener. Dis. 2018. Amyotrophic lateral sclerosis: current therapeutic perspectives; pp. 207–224. [DOI] [Google Scholar]
- 59.Lung J., Lin Y.S., Yang Y.H., Chou Y.L., Shu L.H., Cheng Y.C., Te Liu H., Wu C.Y. The potential chemical structure of anti-SARS-CoV-2 RNA-dependent RNA polymerase. J. Med. Virol. 2020;92:693–697. doi: 10.1002/jmv.25761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shamsi A., Mohammad T., Anwar S., AlAjmi M.F., Hussain A., Tabish R. Md, Islam A., Imtaiyaz H. Md. Glecaprevir and Maraviroc are high-affinity inhibitors of SARS-CoV-2 main protease: possible implication in COVID-19 therapy. Biosci. Rep. 2020;40 doi: 10.1042/BSR20201256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kanhed A.M., Patel D.V., Teli D.M., Patel N.R., Chhabria M.T., Yadav M.R. Identification of potential Mpro inhibitors for the treatment of COVID-19 by using systematic virtual screening approach. Mol. Divers. 2020 doi: 10.1007/s11030-020-10130-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 63.Khaerunnisa S., Kurniawan H., Awaluddin R., Suhartati S. Potential inhibitor of COVID-19 main protease (M pro) from several medicinal plant compounds by molecular docking study. Preprints. 2020:1–14. doi: 10.20944/preprints202003.0226.v1. [DOI] [Google Scholar]
- 64.Du Q., Wang S., Wei D., Sirois S., Chou K.C. Molecular modeling and chemical modification for finding peptide inhibitor against severe acute respiratory syndrome coronavirus main proteinase. Anal. Biochem. 2005;337:262–270. doi: 10.1016/j.ab.2004.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Han Y., Král P. Computational design of ACE2-based peptide inhibitors of SARS-CoV-2. ACS Nano. 2020;14:5143–5147. doi: 10.1021/acsnano.0c02857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Challenges (Hoboken, NJ) 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J.D., Higgins D.G. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME suite: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Desmet J., De Maeyer M., Hazes B., Lasters I. The dead-end elimination theorem and its use in protein side-chain positioning. Nature. 1992;356:539–542. doi: 10.1038/356539a0. [DOI] [PubMed] [Google Scholar]
- 70.Kaur H., Garg A., Raghava G.P.S. PEPstr: a de novo method for tertiary structure prediction of small bioactive peptides. Protein Pept. Lett. 2007;14:626–631. doi: 10.2174/092986607781483859. [DOI] [PubMed] [Google Scholar]
- 71.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Morris G., Huey R. Vol. 30. 2009. AutoDock4 and AutoDockTools4: Automated Docking With Selective Receptor Flexibility; pp. 2785–2791. J. …. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Laskowski R.A., Swindells M.B. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 74.Bowers K.J., Sacerdoti F.D., Salmon J.K., Shan Y., Shaw D.E., Chow E., Xu H., Dror R.O., Eastwood M.P., Gregersen B.A., Klepeis J.L., Kolossvary I., Moraes M.A. Proc. 2006 ACM/IEEE Conf. Supercomput. - SC ’06. ACM Press; New York, New York, USA: 2006. Molecular dynamics—scalable algorithms for molecular dynamics simulations on commodity clusters; p. 84. [DOI] [Google Scholar]
- 75.Schneider R., Sharma A.R., Rai A. Lect. Notes Phys. 2008. Introduction to molecular dynamics; pp. 3–40. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Library of synthetic peptides along with their sequence, structure and hydropathy index.
M1-model1-0to50ns.mpeg.
M2-model2-0to50ns.mpeg.
M3-model3-0to50ns.mpeg.
M4-model4-0to50ns.mpeg.