Abstract
Background
RT-qPCR is the reference test for identification of active SARS-CoV-2 infection, but is associated with diagnostic delay. Antigen detection assays can generate results within 20 min and outside of laboratory settings. Yet, their diagnostic test performance in real life settings has not been determined.
Methods
The diagnostic value of the Panbio™ COVID-19 Ag Rapid Test (Abbott), was determined in comparison to RT-qPCR (Seegene Allplex) in community-dwelling mildly symptomatic subjects in a medium (Utrecht, the Netherlands) and high endemic area (Aruba), using two concurrently obtained nasopharyngeal swabs.
Findings: 1367 and 208 subjects were enrolled in Utrecht and Aruba, respectively. SARS-CoV-2 prevalence, based on RT-qPCR, was 10.2% (n = 139) and 30.3% (n = 63) in Utrecht and Aruba respectively. Specificity of the Panbio™ COVID-19 Ag Rapid Test was 100% (95%CI: 99.7–100%) in both settings. Test sensitivity was 72.6% (95%CI: 64.5–79.9%) in the Netherlands and 81.0% (95% CI: 69.0–89.8%) in Aruba. Probability of false negative results was associated with RT-qPCR Ct-values, but not with duration of symptoms. Restricting RT-qPCR test positivity to Ct-values <32 yielded test sensitivities of 95.2% (95%CI: 89.3–98.5%) in Utrecht and 98.0% (95%CI: 89.2–99.95%) in Aruba.
Interpretation
In community-dwelling subjects with mild respiratory symptoms the Panbio™ COVID-19 Ag Rapid Test had 100% specificity, and a sensitivity above 95% for nasopharyngeal samples when using Ct-values <32 cycles as cut-off for RT-qPCR test positivity. Considering short turnaround times, user friendliness, low costs and opportunities for decentralized testing, this test can improve our efforts to control transmission of SARS-CoV-2.
Research in context.
Evidence before this study
We searched PubMed and MedRxiv for articles published with titles that included the search terms (“covid*”or “coronavirus”or “SARS-CoV-2″) AND title or abstracts that included (antigen test” or “POCT”). Different rapid antigen tests have been developed with usually high specificity but varying sensitivity. The manufacturer of the Panbio™ COVID-19 Ag rapid test reported a very high sensitivity. Real-life data is only available from a very small Spanish cohort in a hospital setting. There is no data yet on the diagnostic performance of this test in individuals with mild symptoms within the community.
Added value of this study
Testing and subsequent isolation and contact tracing is crucial for the global efforts to halt the ongoing SARS-CoV-2 pandemic. Rapid diagnostic tests have the potential to benefit testing strategies as they have short turnaround times, are cheap, and can be used in decentralized testing. Their potential hinges on their diagnostic performance, which has not been sufficiently determined to date. Our findings show that the Panbio™ COVID-19 Ag rapid test reliably identifies SARS-CoV-2 infected individuals with high viral load in nasopharyngeal samples, in a cohort of community-dwelling subjects with mild symptoms of respiratory tract infection. Specificity of this test was 100%. Although the sensitivity is lower than the RT-qPCR, false negative rapid test results were all due to low viral loads in nasopharyngeal samples.
Implications of all the available evidence
Due to the lower sensitivity of the Panbio™ COVID-19 Ag rapid test, RT-qPCR would be the preferred diagnostic test of choice for clinical purposes in a hospital setting. However, for surveillance of SARS-CoV-2 within the community, this rapid antigen test reliably and rapidly identifies individuals with high potential of further transmission, and could therefore be an essential new tool in our testing strategies to control transmission of SARS-CoV-2.
Alt-text: Unlabelled box
Introduction
The SARS-CoV-2 pandemic has extensive impact on healthcare globally, with over 37 million confirmed cases and currently more than one million deaths [1]. Rapid diagnosis of SARS-CoV-2 infection and subsequent contact tracing are essential in the containment of transmission [2].
The reference test for detection of acute SARS-CoV-2 infection is reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) [3]. RT-qPCR requires the use of expensive laboratory instrumentation as well as dedicated lab supplies and trained personnel, which causes significant challenges to generate sufficient testing capacity and short turnaround times.
Lateral Flow Assay (LFA)-based point of care tests (POCT) for rapid antigen detection using antibodies are cheap, simple to perform, do not require laboratory instrumentation and generate results within 20 min [4]. Several different rapid antigen tests have been developed with usually high specificity but varying sensitivity [[5], [6], [7], [8], [9], [10]]. These tests have the potential to alter testing strategies worldwide [8]. Recently the WHO approved the first rapid diagnostic POCT and initiated a global partnership to pledge 120 million tests to low- and middle-income countries [11]. However, the diagnostic performance of individual POCTs in real-life community settings is unknown [10].
We evaluated the Abbott Panbio™ COVID-19 Ag rapid test in community testing locations in both a medium- and high endemic population and compared results to RT-qPCR and determined associations with duration of symptoms and risk of exposure.
Materials and methods
Populations and study period
All individuals visiting COVID-19 community testing centers, located at the University Medical Center Utrecht (UMCU) in the Netherlands (September 22nd to October 6th 2020) and the Horacio Oduber Hospital on Aruba (September 23rd to October 9th 2020), aged 16 and over were asked to participate in this prospective evaluation. These settings were chosen based on the different SARS-CoV-2 prevalence at onset of the study (approximately 4% in Utrecht compared to approximately 30% in Aruba), while using the same platform for the reference RT-qPCR test.
In both study sites, subjects were first sampled for routine RT-qPCR testing, using a combined throat/nasopharyngeal swab. Study participants received an additional nasopharyngeal swab. Participants at the Utrecht study site were asked to fill out a questionnaire regarding (onset of) symptoms and risk of exposure to SARS-CoV-2. Inclusion of participants was continued until the target of 100 positive LFA results, as recommended by the National Institute for Public Health and the Environment (RIVM), was obtained.
Diagnostic tests
RT-qPCR
PCR was conducted in a certified clinical laboratory and all procedures were validated according to the ISO 15,189 standard. After collection, swabs were transferred into 3 ml Universal transport medium until further processing. Nucleic acid extraction, RT-PCR and results interpretation were performed according the instructions of the manufacturer (Seegene, South-Korea). In short, RNA was isolated and purified using the MagC extraction kit (Seegene, South-Korea) on an automatic nucleic acid extractor Hamilton MicroLAB StartLET (Bonaduz, Switzerland). Subsequently, cDNA was generated and amplification was performed in a single tube assay using the Allplex 19-nCoV multiplex platform for detection of SARS-CoV-2 (Seegene, South-Korea), and results were interpreted with Seegene Viewer data analysis software. The assay uses fluorescent Taqman® probes for three SARS-CoV-2 genes (E [Envelope], N [Nucleocapsid]-, and RdRP [RNA dependent RNA Polymerase]genes). Amplification and detection were performed for 45 cycles on a Biorad CFX96 thermocycler (Biorad Laboratories, the Netherlands), the threshold Cycle (Ct) was automatically determined by the manufacturer's software. A positive result was defined as amplification of any of the three SARS-CoV-2 genes. If not all targets showed a positive result, this always corresponded with high Ct-values, suggesting low levels of SARS-CoV-2 RNA. If viral RNA levels are very low and around the limit of detection of the assay, amplification of these targets is more subject to stochasticity which can result in positive results in only one or two targets. Based on our experience within clinical practice and results from viral culture studies,[12] we use a cut-off Ct-value of 32 to determine clinically relevant levels of SARS-CoV-2 RNA.
LFA
The Panbio™ COVID-19 Ag rapid test device by Abbott (Lake Country, IL, U.S.A) is a membrane-based immunochromatography assay which detects the nucleocapsid protein of SARS-CoV-2 in nasopharyngeal samples. Collected swabs were transferred into dedicated sample collection tubes containing a sampling buffer and transported to the laboratory. The laboratory is located within 5 min of walking distance from the sampling location. All samples were analysed within a maximum of 2 h after collection, and in practice the delay was much shorter (between 30 and 45 min on average), during which time the samples were kept at ambient temperature, which should have very little effect on relatively stable viral proteins. The PCR samples are processed similarly with no perceivable negative effect. Collected samples were subsequently processed in a level 2 biosafety cabinet in accordance with the manufacturer's protocol. Test results were recorded after 15 min of assay initiation, by two independent observers (blinded to each other and to the PCR results). Single lot LFA testing devices were used: lot 41ADF011A.
Ethical approval
The medical research ethics committee (MREC) of Utrecht decided the study is not subject to the Medical Research Involving Human Subjects Act (WMO) and did not require full review by an accredited MREC. The ethical committee of the hospital board of Aruba approved the study. All participants have provided written informed consent.
Statistical analysis
Population characteristics are reported as mean (Standard Deviation, SD) or median [InterQuartile Range, IQR] values. Difference testing for comparisons of groups was performed by Chi-square testing for categorical variables, independent samples Student's t-tests with Welch's correction for continuous normally distributed variables and by using Mann-Whitney U tests for not non-normally distributed variables.
Specificity and sensitivity with 95% confidence intervals, and positive and negative predictive value of the LFA were calculated using the RT-qPCR results as reference test. Factors associated with LFA results were determined using logistic regression, using Nagelkerke's pseudo R2 as a measure of goodness-of-fit. Data was analysed using the free, open-source software environment R [13].
Role of the funding source
This study was investigator initiated. No external funding was received.
Results
Population characteristics
At the Utrecht study site 1369 subjects were included, of which 139 tested positive for SARS-CoV-2 by RT-qPCR (prevalence: 10.2%). The mean (SD) Ct-values for E-gene, N-gene and RdRP-gene were 24.74 (5.73), 27.51 (6.01) and 26.35 (5.60), respectively.
At the Aruba study site 208 subjects were included, of which 63 tested positive for SARS-CoV-2 (prevalence: 30.3%). The mean (SD) Ct-values for E-gene, N-gene and RdRP-gene at the Aruba site were 25.69 (5.96), 26.56 (6.41) and 26.26 (6.36), respectively.
Estimated participation rates were 50% at the Utrecht study site and 25% at the Aruba study site, based on the total number of samples processed at each site during the study period. Samples from two participants were excluded, due to inappropriate application of the nasopharyngeal swab and laboratory mislabelling. Symptom registration forms were absent from nine subjects and only incomplete data on duration of symptoms was available from 201 subjects (14.7% of total subjects, 11.9% of SARS-CoV-2 positive subjects, p = 0.439).
Individuals at the Utrecht study site were more often female (61.7%) and were largely between 20 and 50 years of age (Table 1). Nearly all individuals reported symptoms (97.3%), most frequently coryza (69.0%), sore throat (66.3%) and cough (57.1%). Duration of symptoms were 1–3 days in 387 subjects (33.2%), 4–7 days in 560 subjects (48.0%), and more than a week in 191 subjects (16.4%). Of all individuals, 17% reported prior contact with a confirmed SARS-CoV-2 positive individual.
Table 1.
Population characteristics (Utrecht study site).
| Total | SARS-CoV-2 Negative* | SARS-CoV-2 Positive* | p-Value | |
|---|---|---|---|---|
| n | 1367 | 1228 | 139 | |
| Sex = F (%) | 844 (61.7) | 772 (63.1) | 72 (52.2) | 0.016 |
| Age in years (median [IQR]) | 36.41 [27.0, 49.6] | 36.59 [27.3, 49.6] | 33.73 [23.4, 49.3] | 0.034 |
| Contact with Confirmed Positive | 233 (17.0) | 185 (15.1) | 48 (34.5) | <0.001 |
| Asymptomatic | 37 (2.7) | 34 (2.8) | 3 (2.2) | 0.875 |
| Duration of symptoms: | 0.019 | |||
| 1–3 days | 387 (33.2) | 353 (33.6) | 34 (29.6) | |
| 4–7 days | 560 (48.0) | 493 (46.9) | 67 (58.3) | |
| >7 days | 191 (16.4) | 181 (17.2) | 10 (8.7) | |
| Symptoms: | ||||
| Fever (%) | 221 (16.2) | 170 (13.9) | 51 (36.7) | <0.001 |
| Chills (%) | 279 (20.4) | 225 (18.5) | 54 (38.8) | <0.001 |
| Sore Throat (%) | 907 (66.3) | 839 (68.8) | 68 (48.9) | <0.001 |
| Cough (%) | 780 (57.1) | 692 (56.8) | 88 (63.3) | 0.165 |
| Shortness of Breath (%) | 274 (20.0) | 250 (20.5) | 24 (17.3) | 0.429 |
| Coryza (%) | 943 (69.0) | 867 (71.1) | 76 (54.7) | <0.001 |
| Altered Smell or Taste (%) | 202 (14.8) | 159 (13.0) | 43 (30.9) | <0.001 |
| General Malaise (%) | 365 (26.7) | 317 (26.0) | 48 (34.5) | 0.041 |
| Abdominal Pain (%) | 108 (7.9) | 97 (8.0) | 11 (7.9) | 1.000 |
| Vomiting (%) | 32 (2.3) | 26 (2.1) | 6 (4.3) | 0.189 |
| nausea (%) | 115 (8.4) | 100 (8.2) | 15 (10.8) | 0.380 |
| Diarrhea (%) | 137 (10.0) | 122 (10.0) | 15 (10.8) | 0.887 |
| Headache (%) | 601 (44.0) | 520 (42.7) | 81 (58.3) | 0.001 |
| Rash (%) | 25 (1.8) | 22 (1.8) | 3 (2.2) | 1.000 |
| Eye Infection (%) | 31 (2.3) | 26 (2.1) | 5 (3.6) | 0.426 |
| Muscle Ache (%) | 247 (18.1) | 195 (16.0) | 52 (37.4) | <0.001 |
| Joint Ache (%) | 111 (8.1) | 83 (6.8) | 28 (20.1) | <0.001 |
| Tiredness (%) | 565 (41.3) | 491 (40.3) | 74 (53.2) | 0.004 |
| Reduced Appetite (%) | 132 (9.7) | 111 (9.1) | 21 (15.1) | 0.035 |
| E-gene Ct-value (mean (SD)) | 24.74 (5.7) | |||
| N-gene Ct-value (mean (SD)) | 27.51 (6.0) | |||
| RdRP-gene Ct-value (mean (SD)) | 26.35 (5.6) |
based on RT-qPCR results.
Compared to subjects who tested negative for SARS-CoV-2 in RT-qPCR, SARS-CoV-2 positive subjects were younger (p = 0.034), more likely male (p = 0.016), more frequently had prior contact with a confirmed SARS-CoV-2 positive individual (p<0.001), and more frequently reported fever, chills, an altered sense of smell or taste, or joint- or muscle ache (p<0.001 for all symptoms, see table 1). The most frequently reported symptoms, coryza and sore throat, were negatively associated with detection of SARS-CoV-2 by RT-qPCR (p<0.001).
LFA results
At the Utrecht study site, 101 subjects tested positive by LFA yielding an overall sensitivity of 72.6% (95% Confidence Interval, CI: 64.5 – 79.9%) (Table 2). False positive LFA results were not observed (specificity 100%, 95% CI: 99.7 – 100%). Similar results were obtained at the Aruba study site, with an overall sensitivity of 81.0% (95% CI: 69.0 – 89.8%) and specificity of 100% (95% CI: 97.5 – 100%) (Table 2). We observed no inter-rater variability in interpretation of test bands and no bands were classified as unclear by the independent observers.
Table 2.
Test characteristics of the LFA compared to the RT-qPCR for the Utrecht study site and the Aruba study site.
| Study site | LFA result | PCR result |
Specificity | Sensitivity | ||
|---|---|---|---|---|---|---|
| Positive (Ct<32) | Positive (Ct ≥32) | Negative | ||||
| Utrecht | Positive | 101 | 0 | 0 | 100% (99.7–100) | Overall: 72.6% (64.5–79.9) Ct<32: 95.2% (89.3–98.5) |
| Negative | 5 | 33 | 1228 | |||
| Aruba | Positive | 48 | 3 | 0 | 100% (97.5–100) | Overall: 81.0% (69.0–89.9) Ct<32: 98.0% (89.2–99.95) |
| Negative | 1 | 11 | 145 | |||
Sensitivity and specificity are reported with 95% CI.
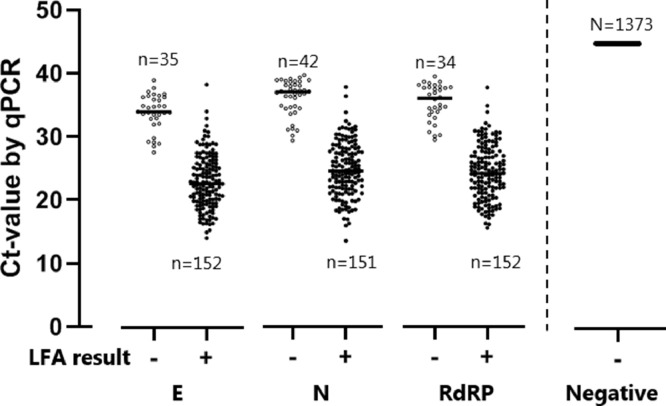
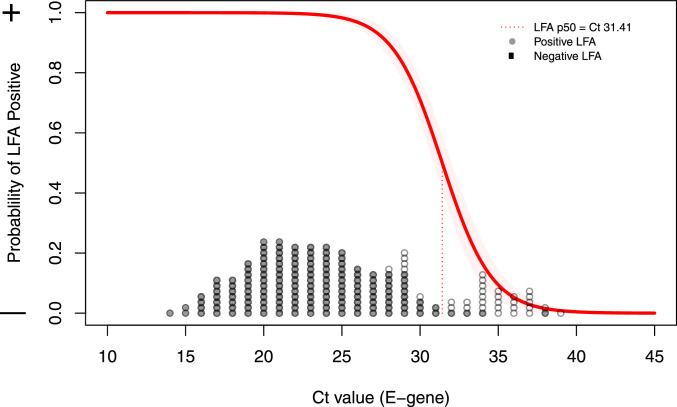
False negative LFA results were mostly observed in subjects with high RT-qPCR Ct-values, reflecting low viral load levels in nasopharyngeal material (Fig. 1). Using logistic regression, the likelihood of a false negative LFA result was associated with the RT-qPCR Ct-value (R2 = 0.77, p<0.0001, Fig. 2). When defining RT-qPCR Ct positivity on a cut-off Ct-value of 32 LFA sensitivity was 95.2% (95% CI: 89.3 – 98.5%) at the Utrecht site and 98.0% (89.2 – 99.95%) at the Aruba study site.
Fig. 1.
PCR and LFA results of all subjects. All PCR results for the three targets are shown by Ct-value on the y-axis (left side: positive PCR results per target; right side: negative PCR results), grouped based on the LFA result on x-axis.
Fig. 2.
Association between Ct-value and LFA test result. All dots reflect positive PCR results, shown on the x-axis at the observed Ct-value of the E-gene. gray dots reflect positive LFA samples, white dots negative LFA samples. The red line reflects the probability of a positive LFA based per Ct-value, the red dotted line denotes the point where 50% of LFAs are expected to become positive.
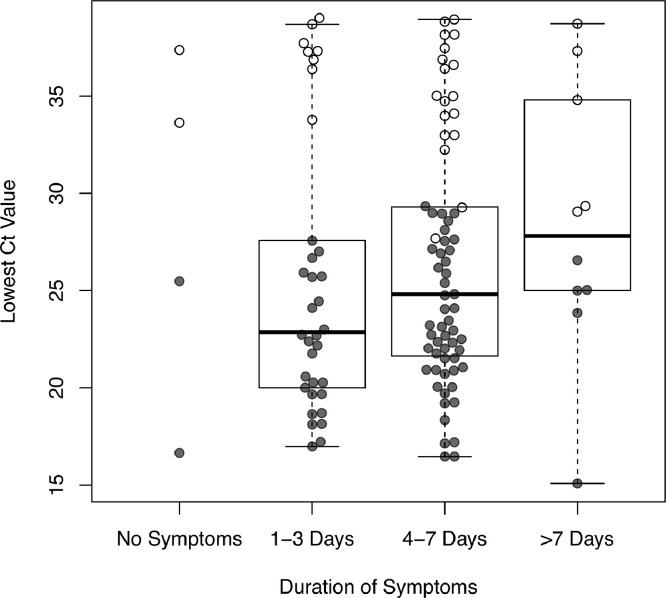
Relation with symptoms
The duration of symptoms was not associated with Ct-values (p = 0.46) or with the occurrence of false negative LFA results (p = 0.30) (Fig. 3). When including only symptoms positively associated with COVID-19 (i.e., fever, chills, loss of taste/smell, muscle- or joint ache), duration of symptoms was weakly associated with Ct-values (p = 0.02), but no such association was found with LFA results (p = 0.45). Restricting analyses to individuals with symptoms for less than 7 days did not change sensitivity of LFA neither for all subjects positive with RT-qPCR (74.3%; 95%CI 64.6 – 82.4%) or when applying a cut-off at Ct>32 (97.4%; 95%CI 90.9 – 99.7%).
Fig. 3.
Ct-value of positive subjects grouped by duration of symptoms. The dots represent individuals with positive PCR results shown on the y-axis based on the lowest observed Ct-value in any of the three targets. The dots are groups based on the duration of symptoms. gray dots represent positive LFA results, white dots negative LFA results. Horizontal lines of the boxes are median and IQR; whiskers extend to the minimum and maximum.
Discussion
In this real-life evaluation of the Panbio™ COVID-19 Ag rapid test in community-dwelling subjects with mild symptoms of respiratory tract infection, the assay reliably identified SARS-CoV-2 infected subjects with low Ct-values by RT-qPCR (i.e. infections with a high viral load in nasopharyngeal samples). In our study cohorts specificity was 100%, overall sensitivity was 72.6% and 95.2% when using a Ct-value of 32 as cut-off.
This new LFA test has not been evaluated extensively. The manufacturer reported a higher sensitivity (93.3%; 95CI 83.8–98.2), obtained in a high endemic setting in Brazil, testing individuals with symptoms for less than seven days only [14]. In another cohort of 257 patients (both symptomatic and asymptomatic) enrolled at the emergency department and primary health care setting in Spain, overall sensitivity was 73.3%, and 86.5% among individuals with symptoms for less than seven days [9].
In our study cohort, false negative results were observed only at high Ct-values, i.e. with low viral load in nasopharyngeal material. This may occur very early in the infection (presymptomatic stage) before viral replication peaks, or in a late stage of infection when replication has decreased. Individuals with symptoms for more than 7 days may, therefore, be more likely to have a low viral load in nasopharyngeal swabs than those tested shortly after symptom onset. Indeed, a longer duration of symptoms associated with SARS-CoV-2 infection, was associated with higher Ct-values, but no such association was observed between false negative LFA results and duration of symptoms. As a result, test sensitivity did not substantially increase when analyses were restricted to individuals with symptoms for less than a week. It must be noted that the study period corresponded with high prevalence of seasonal rhinovirus and other respiratory infections in the Netherlands,[15,16] thereby possibly obscuring the association of upper respiratory tract symptoms with SARS-CoV-2 infection.
From a public healthcare perspective, missed infections with the LFA in patients with high Ct-values in a late stage of infection may have limited impact, as these individuals are less likely to contribute to transmission. This is supported by findings that culturing of SARS-CoV-2 appeared not possible at Ct-values above 29 [12].
Yet, false negative LFA results were also observed among individuals with a very short duration of symptoms and high Ct-values. A missed infection in the early stage may have consequences for transmission as these individuals may become infectious [[17], [18], [19]]. Furthermore, the majority of individuals in our cohort reported (mild) symptoms. Studies are needed to determine the sensitivity of this LFA test in asymptomatic subjects with detectable SARS-CoV-2 in RT-qPCR. These studies could also provide more insight in the performance of this LFA in the presymptomatic stage of infection.
For symptomatic persons requiring hospital admission a diagnostic test with high sensitivity is needed to establish a definitive clinical diagnosis [20]. In this clinical context, we recommend to use RT-qPCR, as its sensitivity is superior to this LFA, allowing detection of individuals in the presymptomatic and late stage of infection which are both relevant in this context. However, in the context of community-based surveillance, tests need to identify symptomatic and asymptomatic infections in short time to stop onward spread. Transmission of SARS-CoV-2 is considered to occur mainly around symptom onset, when viral load peaks [21,22]. This LFA, therefore, appears to reliably identify those patients that are most likely to contribute to onward transmission and could therefore be an essential new tool in our testing strategies. As the virus remains infectious in the assay sampling buffer (data not shown), biosafety should be taken into account when implementing the use of the Panbio™ COVID–19 LFA outside a laboratory setting.
In the design of the present study many variables that may affect test outcome and interpretation were controlled as much as possible. For instance, all tests were conducted in laboratory conditions, with controlled temperature and lighting; interpreted by a limited pool of trained operators, and a single lot of assays was used throughout the study. This limits the generalizability of the results to some extent, as widespread, decentralized community testing will potentially occur under less well-controlled circumstances. A surprising finding is the perfect inter-rater agreement observed in this study, which occurred as there were no inconclusive bands observed. This finding could be due to the controlled laboratory conditions or the use of a single lot.
Considering the short turnaround time, user friendliness, opportunity for decentralized testing and low costs we believe that in the context of community-based surveillance of symptomatic individuals, these advantages outweighs the lower sensitivity of LFA compared to RT-qPCR. Modeling studies may provide further reassurance on the safety of such an approach.
Declaration of Competing Interest
Dr. Wensing reports grants from ViiV Healthcare, Gilead, grants from Janssen, Gilead, Merck, grants from Janssen, Gilead, Merck, ViiV Healthcare, grants from ARK diagnostics, outside the submitted work. All other authors report no conflict.
Acknowledgments
Acknowledgements
The authors would like to thank all participating employees from the GGD Utrecht and the department of Public Health in Aruba and the participating students from the UMCU for their help with data collection, as well as all participating volunteers.
Funding
This study was investigator initiated. No external funding was received.
Data sharing statement
All de-identified participant available data can be made available upon request per email to the corresponding author.
Footnotes
The Panbio™ COVID-19 Ag Rapid Test (distributed by Abbott) was provided by the Ministry of Health, Welfare and Sport (VWS). UMCU and LABHOH, Aruba.
References
- 1.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. The Lancet Infect Dis. 2020;5:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kretzschmar M.E., Rozhnova G., Bootsma M.C.J., van Boven M., van de Wijgert J.H.H.M., Bonten M.J.M. Impact of delays on effectiveness of contact tracing strategies for COVID-19: a modelling study. Lancet Public Heal. 2020;5(8):e452–e459. doi: 10.1016/S2468-2667(20)30157-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25(3):2000045 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grant B.D., Anderson C.E., Williford J.R., Alonzo L.F., Glukhova V.A., Boyle D.S. SARS-CoV-2 coronavirus nucleocapsid antigen-detecting half-strip lateral flow assay toward the development of point of care tests using commercially available reagents. Anal Chem. 2020;92(16):11305–11309. doi: 10.1021/acs.analchem.0c01975. [DOI] [PubMed] [Google Scholar]
- 5.Finddx.org. Access date October 13 2020, [Internet]. Available from: https://www.finddx.org/covid-19/pipeline/
- 6.Hirotsu Y., Maejima M., Shibusawa M., Nagakubo Y., Hosaka K., Amemiya K. Comparison of automated SARS-CoV-2 antigen test for COVID-19 infection with quantitative RT-PCR using 313 nasopharyngeal swabs, including from seven serially followed patients. Int J Infect Dis. 2020;99:397–402. doi: 10.1016/j.ijid.2020.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scohy A., Anantharajah A., Bodéus M., Kabamba-Mukadi B., Verroken A., Rodriguez-Villalobos H. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J Clin Virol. 2020;129:104455 doi: 10.1016/j.jcv.2020.104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guglielmi G. Fast coronavirus tests: what they can and can’t do. Nature. 2020;585(7826):496–498. doi: 10.1038/d41586-020-02661-2. [DOI] [PubMed] [Google Scholar]
- 9.Linares M., Pérez Tanoira R., Romanyk J., Pérez García F., Gómez-Herruz P., Arroyo T. Panbio antigen rapid test is reliable to diagnose SARS-CoV-2 infection in the first 7 days after the onset of symptoms. J. Clin. Virol. 2020;133:104659 doi: 10.1016/j.jcv.2020.104659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dinnes J., Deeks J.J., Adriano A., Berhane S., Davenport C., Dittrich S. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst Rev. 2020;8:CD013705 doi: 10.1002/14651858.CD013705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.WHO.com. Access date October 13 2020, [Internet]. Available from: https://www.who.int/news/item/28-09-2020-global-partnership-to-make-available-120-million-affordable-quality-covid-19-rapid-tests-for-low–and-middle-income-countries
- 12.Kampen JJA van, Vijver DAMC van de, Fraaij P.L.A., Haagmans B.L., Lamers M.M., Okba N. Shedding of infectious virus in hospitalized patients with coronavirus disease-2019 (COVID-19): duration and key determinants. medRxiv. 2020 doi: 10.1101/2020.06.08.20125310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.R Development Core Team. A language and environment for statistical computing R foundation for statistical computing. 2018. Available from: http://www.r-project.org/. Access date October 13, 2020.
- 14.globalpointofcare.abbott. Access date October 13 2020.
- 15.van der Linden L., Bruning A.H.L., Thomas X.V., Minnaar R.P., Rebers S.P.H., Schinkel J. A molecular epidemiological perspective of rhinovirus types circulating in Amsterdam from 2007 to 2012. Clin Microbiol Infect. 2016;22(12):1002.e9–1002.e14. doi: 10.1016/j.cmi.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edridge A.W.D., Kaczorowska J., Hoste A.C.R., Bakker M., Klein M., Loens K. Seasonal coronavirus protective immunity is short-lasting. Nat Med. 2020;26(11):1691–1693. doi: 10.1038/s41591-020-1083-1. [DOI] [PubMed] [Google Scholar]
- 17.Li R., Pei S., Chen B., Song Y., Zhang T., Yang W. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2) Science. 2020;368(6490):489–493. doi: 10.1126/science.abb3221. 80- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bai Y., Yao L., Wei T., Tian F., Jin D.Y., Chen L. Presumed Asymptomatic Carrier Transmission of COVID-19. JAMA - J Am Med Assoc. 2020;323(14):1406–1407. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C. Transmission of 2019-NCOV infection from an asymptomatic contact in Germany. New Engl J Med. 2020;382(10):970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mina M.J., Parker R., Larremore D.B. Rethinking Covid-19 Test Sensitivity — A Strategy for Containment. N Engl J Med. 2020 doi: 10.1056/NEJMp2025631. [DOI] [PubMed] [Google Scholar]
- 21.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 22.He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26(5):672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]