Sir,
despite tremendous efforts of containing SARS-CoV-2 spreading in Europe through a test-and-trace system, at the beginning of November most European Countries experienced a second wave of pandemic, posing serious challenges to the civil society and health systems.
Recent research indicates that symptoms screening alone will not enable to contain COVID-19 outbreaks, because an estimated 40 % of COVID-19 cases are asymptomatic and 50 % of transmissions results from asymptomatic subjects [1].
An important breakthrough would be identifying a testing system allowing for multiple screening and early detection, thus changing the timing and effectiveness of tracing asymptomatic spreaders. Moreover, autumn-winter season is characterized by other common respiratory infections with overlapping COVID-19 symptoms with an obvious economical and logistic burden for the National Health System. This aspect is of relevance among the pediatric population as young children, especially pre-school toddlers, have recurrent viral infections, and mitigation measures (distancing, masks) are difficult to implement, all resulting in more frequent testing. Moreover, SARS-CoV-2 transmission dynamics in children is not fully elucidated.
Recently, Wyllie et al. reported that saliva and nasopharyngeal swab samples have similar sensitivity in detecting SARS-CoV-2 in symptomatic patients, and that SARS-CoV-2 can be detected in saliva, before nasopharyngeal swab (NPS), of asymptomatic individuals [2].
To address this issue, starting from the FDA emergency approved SalivaDirect™ protocol of Yale University [3], we optimized a saliva collection method able to guarantee a correct self-sampling or caregiver-guided sampling. Briefly, a sterile dental cotton roll was kept for two minutes in the vestibular space next to the lower premolar-molar area, then under the tongue close to the Wharton duct opening for additional two minutes. Once duly soaked with saliva, the dental roll was preserved in a sterile 50 mL tube, at room temperature until use (up to 7 days). Saliva was then recovered under sterile conditions using a 10 mL syringe to squeeze the cotton roll. Collected saliva was processed as recommended by Vogels et al. [3], with a modification of the thermal cycler profile: 5′ at 95 °C followed by 5′ at 4 °C. Five microliters of the processed saliva was used directly for qPCR using N1 (FAM, BHQ-1 labeled probe) and RP (Cy5, BHQ-1) primers/probes published by the Centers for Disease Control (https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html) and Applied Biosystem 7500 Fast instrument. Samples were considered positive upon detection of N1 (Ct<40). Invalid samples were assessed by RP (Ct>35).
Here we report the results of the comparison between paired samples of self-collected saliva and NPS. This analysis is part of a prospective study at Vittore Buzzi Children Hospital and San Paolo Hospital in Milan. This study was carried out in accordance with the recommendations of the Comitato Etico Interaziendale Milano Area A, protocol approval numbers N. 31554 (adults) and N. 0050308 (children). All subjects/caregivers gave written informed consent in accordance with the Declaration of Helsinki.
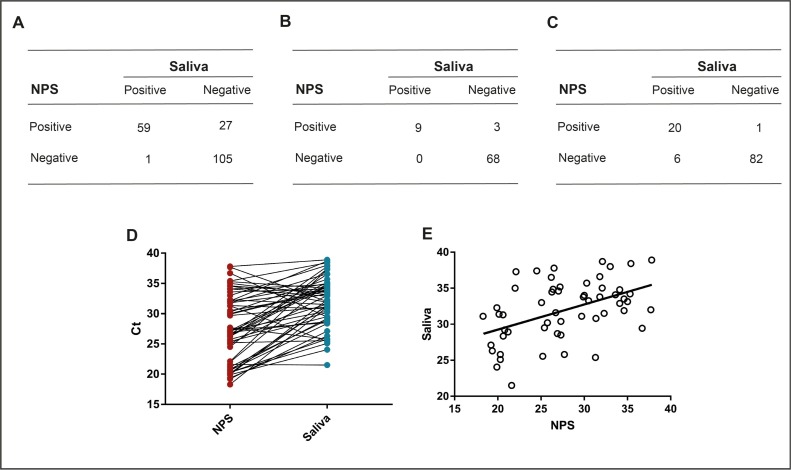
For assessing this method reliability, first we tested paired nasopharyngeal swabs and saliva samples in 192 adults (age 18–85, 52.2 % females). Concordance between tests was assessed using the Kappa statistic (Cohen’s unweighted Kappa, k) as: poor (k = 0), slight (0.01 < k < 0.20), fair (0.21 < k < 0.40), moderate (0.41 < k < 0.60), substantial (0.61 < k < 0.80), almost perfect (0.81 < k < 1.00), or perfect (k = 1). Overall, concordance was 0.85 and k coefficient was 0.69, indicating a substantial concordance (Fig. 1 A). When analyzing asymptomatic subjects, concordance rose to 0.96 with k equal to 0.83, indicating an almost perfect concordance (Fig. 1B).
Fig. 1.
SARS-CoV-2 RNA detection concordance tables for adult cohort (A), asymptomatic subjects (B), and children (C). Cycle Threshold (Ct) Values for 55 matched NPS and saliva positive samples (D), and linear regression for Pearson’s r (E).
Having confirmed the reliability of the test in the adult population, especially in asymptomatic individuals, we then coupled the saliva test to NPS in a pediatric population (N. 109; age: 0–17, 46.4 % females). Concordance was analyzed using the Kappa statistics and overall concordance was 0.94 and k was 0.81, considered almost perfect (Fig. 1C).
Ct values for N1 SARS-CoV-2 gene of 55 matched saliva (blue dots) and NPS (red dots) samples (Fig. 1D) were used to calculate Pearson correlation coefficient (Fig. 1E) that resulted 0.47, indicating a statistically significant moderate positive correlation (p = 0.0003).
Among the differing results in the adult cohort having a positive NPS and negative saliva, two subjects already developed IgG antibodies against SARS-CoV-2, three individuals underwent the second NPS test 14 days following the COVID-19 diagnosis (according to the Italian protocol), ten subjects reported symptoms onset 8–15 days earlier.
Among the differing results in the pediatric cohort, in one case the saliva test was negative and NPS positive, while in six cases the saliva test detected SARS-CoV-2 RNA in asymptomatic individuals who resulted negative with the NPS. In two instances, saliva tested positive two days before NPS, suggesting oropharynx as the first site for viral replication.
Our data are in accordance with Yokota and colleagues [4] arguing that saliva molecular testing could be a more sensitive tool for contact-tracing.
The saliva sampling as an alternative for NPS has been proposed since the first SARS-CoV-2 wave [5] albeit two limiting factors: i) the infection dynamic within different body fluids is still debated [6]; ii) saliva results are highly dependent on the tested cohort and on the presence/absence of sputum, which in our hands is strongly confounding. In light of our data, we believe further experiments should be directed towards settling whether long positive NPS subjects with no virus in saliva are to be considered infectious.
In conclusion, we feel that this tool represents an opportunity for testing SARS-CoV-2 infection in between pandemic/epidemic wave peaks. In addition, an active surveillance in close communities based on multiple saliva testing could be envisaged. Further, using broadly available and inexpensive cotton dental rolls would allow for boosting sampling capacity for low-compliant subjects and in low-income countries.
UNIMI SAL Study group: Stefano Centanni, Silvia Bianchi, Anna Caretti, Elena Canciani, Elisa Adele Colombo, Elisabetta Di Fede, Cristina Gervasini, Paolo Grazioli, Dolaji Henin, Elena Lesma, Antonella Lettieri, Giulia Marchetti, Gaia Pellegrini, Mariachiara Perrotta, Carlotta Pipolo.
Declaration of Competing Interest
All the authors declare no conflict of interest.
References
- 1.Paltiel A.D., Zheng A., Walensky R.P. Assessment of SARS-CoV-2 screening strategies to permit the safe reopening of college campuses in the United States. JAMA Netw. Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.16818. e2016818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P., Warren J.L., Geng B., Muenker M.C., Moore A.J., Vogels C.B.F., Petrone M.E., Ott I.M., Lu P., Venkataraman A., Lu-Culligan A., Klein J., Earnest R., Simonov M., Datta R., Handoko R., Naushad N., Sewanan L.R., Valdez J., White E.B., Lapidus S., Kalinich C.C., Jiang X., Kim D.J., Kudo E., Linehan M., Mao T., Moriyama M., Oh J.E., Park A., Silva J., Song E., Takahashi T., Taura M., Weizman O.-E., Wong P., Yang Y., Bermejo S., Odio C.D., Omer S.B., Dela Cruz C.S., Farhadian S., Martinello R.A., Iwasaki A., Grubaugh N.D., Ko A.I. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N. Engl. J. Med. 2020;383:1283–1286. doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vogels C.B.F., Watkins A.E., Harden C.A., Brackney D., Shafer J., Wang J., Caraballo C., Kalinich C.C., Ott I., Fauver J.R., Kudo E., Lu P., Venkataraman A., Tokuyama M., Moore A.J., Muenker M.C., Casanovas-Massana A., Fournier J., Bermejo S., Campbell M., Datta R., Nelson A., Team Y.I.R., Dela Cruz C., Ko A., Iwasaki A., Krumholz H.M., Matheus J., Hui P., Liu C., Farhadian S., Sikka R., Wyllie A.L., Grubaugh N. SalivaDirect: A simplified and flexible platform to enhance SARS-CoV-2 testing capacity. MedRxiv. 2020 doi: 10.1101/2020.08.03.20167791. 2020.08.03.20167791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yokota I., Shane P.Y., Okada K., Unoki Y., Yang Y., Inao T., Sakamaki K., Iwasaki S., Hayasaka K., Sugita J., Nishida M., Fujisawa S., Teshima T. Mass screening of asymptomatic persons for SARS-CoV-2 using saliva. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iwasaki S., Fujisawa S., Nakakubo S., Kamada K., Yamashita Y., Fukumoto T., Sato K., Oguri S., Taki K., Senjo H., Sugita J., Hayasaka K., Konno S., Nishida M., Teshima T. Comparison of SARS-CoV-2 detection in nasopharyngeal swab and saliva. J. Infect. 2020;81 doi: 10.1016/j.jinf.2020.05.071. e145–e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riccò M., Ranzieri S., Peruzzi S., Valente M., Marchesi F., Balzarini F., Bragazzi N.L., Signorelli C. RT-qPCR assays based on saliva rather than on nasopharyngeal swabs are possible but should be interpreted with caution: results from a systematic review and metaanalysis. Acta Biomed. 2020;91:1–15. doi: 10.23750/abm.v91i3.10020. [DOI] [PMC free article] [PubMed] [Google Scholar]