Abstract
Microfluidic devices can be thought of as comprising interconnected miniaturized compartments performing multiple experimental tasks individually or in parallel in an integrated fashion. Due to its small size, portability, and low cost, attempts have been made to incorporate detection assays into microfluidic platforms for diseases such as cancer and infection. Some of these technologies have served as point-of-care and sample-to-answer devices. The methods for detecting biomarkers in different diseases usually share similar principles and can conveniently be adapted to cope with arising health challenges. The COVID-19 pandemic is one such challenge that is testing the performance of both our conventional and newly-developed disease diagnostic technologies. In this mini-review, we will first look at the progress made in the past few years in applying microfluidics for liquid biopsy and infectious disease detection. Following that, we will use the current pandemic as an example to discuss how such technological advancements can help in the current health challenge and better prepare us for future ones.
Keywords: Microfluidics, Liquid biopsy, Infectious diseases, COVID-19
1. Introduction
Certain human diseases do leave unique traceable biomarkers in our circulatory system. As such, this has made blood test one of the most common methods of diagnosis. In recent years, circulating tumor cells (CTCs), circulating exosomes, and circulating tumor DNAs (ctDNAs) have emerged as novel diagnostic markers for cancer (Vaidyanathan et al., 2018). The efficient detection of these small and rare cancer-associated factors requires devices that can handle a small amount of blood samples with high spatial–temporal accuracy and in a high throughput manner. Microfluidic technology provides a perfect platform that can meet such requirements. The integration of different detection methods onto microfluidic chips has led to a boom in this emerging field where cancer detection can be performed on blood samples collected (liquid biopsies). The success mostly comes from the detection and isolation of CTCs at the very beginning, and then extended to detecting extracellular vesicles (EVs) such as exosomes and also ctDNAs. Some of these technologies have been commercialized and performing a routine blood test for cancer diagnosis and monitoring has become a reality.
Recent advancement in nucleic acid separation and enrichment in microfluidic chips, together with isothermal amplification of DNAs and RNAs, has enabled ctDNA and pathogen nucleic acid detection in an integrated microfluidic system. These achievements in using microfluidics for disease detection have provided us with the necessary toolbox for tackling emerging health challenges. During this COVID-19 pandemic, some of the existing diagnostic technologies have enabled rapid confirmation of the pathogen with the detection kits made available just shortly after the disease has emerged. This came from our experience gained during the previous SARS and other epidemics in the last 20 years. However, the ongoing pandemic also exposed the limitations of the conventional PCR-based disease detection methods, which can only be performed by trained personnel in a laboratory setting. We believe microfluidics is a key component to overcome these challenges.
In the subsequent discussion, we will go through several recent advancements of using microfluidic-based platforms for disease diagnosis. We will skip the technical details which have been extensively reviewed before (Jiang and Lim, 2019, Vaidyanathan et al., 2018). Instead, we will discuss the topic from a developmental perspective and share our opinions on how technological advances can meet the current and future health challenges. We will use COVID-19 as a recent example to demonstrate the significance and potential of microfluidic-based platforms for current and future disease detection and diagnosis.
2. Microfluidics in liquid biopsy
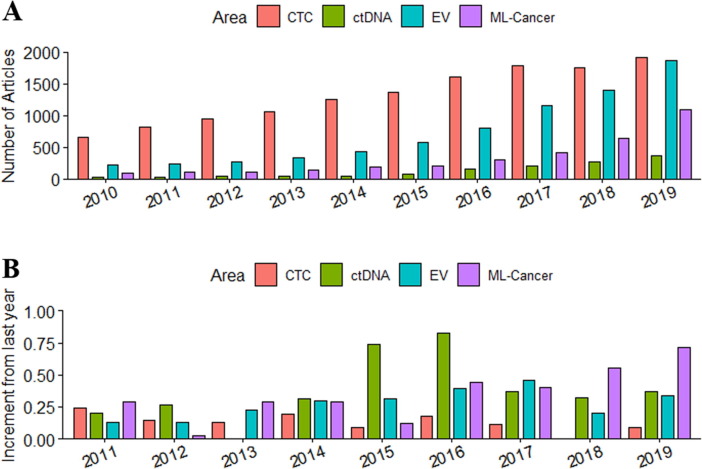
As a tumor grows, it develops its own blood supply system through angiogenesis to meet its high demand for nutrients. The bloodstream flowing inside the tumor takes tumor-associated materials into the systematic circulation of the human body. These rare circulating tumor biomarkers that carry information about the tumor, can be retrieved and isolated using a less-invasive, low risk, and inexpensive method for detecting cancers. We summarized the trend in the development of different liquid biopsy methods in Fig. 1 involving both CTCs and ctDNAs.
Fig. 1.
The number of research articles in the discussed cancer diagnostics areas (A) and their yearly increment (B). EV: Extracellular vesicles; ML-Cancer: Machine learning in cancer studies.
2.1. Circulating tumor cells
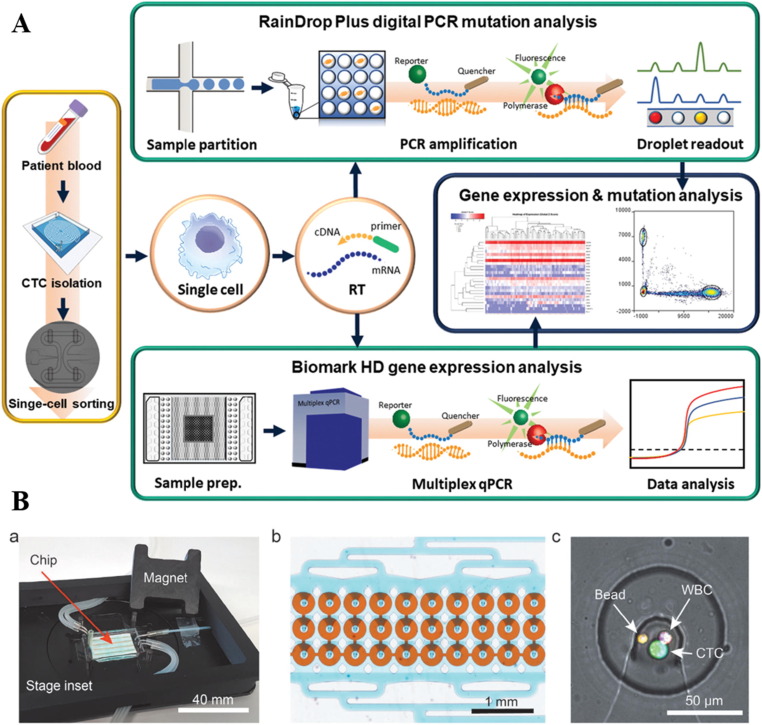
The earliest focus of liquid biopsy was mainly on detecting, enumerating, and separating CTCs from cancer patients’ blood for prognosis and disease monitoring (Hou et al., 2011). Antibody-tagged magnetic beads for immunomagnetic separation of CTCs was employed by the only FDA-approved technology for CTC detection - the CellSearch System (Riethdorf et al., 2007). This separation method was soon adapted into microfluidic-based systems for achieving higher throughput, specificity, and sensitivity (Nagrath et al., 2007). Embedding different categories of active and passive separation strategies into microfluidic platforms for CTC isolation followed thereafter. As antibody-labeling may select only a specific subgroup of CTCs, negative enrichment by labeling and removing other blood cells to enrich CTC was also effectively demonstrated (Fachin et al., 2017). Apart from affinity-based methods, separating CTCs by their unique biophysical properties has also been extensively explored and these have helped overcome several shortcomings associated with using antibodies (Che et al., 2017). Cell size (Hou et al., 2013) and deformability (Tan et al., 2009) have been shown to be good markers for separating CTCs from blood. The performance of different microfluidic-based CTC separation techniques has been greatly optimized for clinical applications through development in the past decade. Several commercialized systems have been able to serve the large market of CTC-based research and clinical investigations (Vaidyanathan et al., 2018). The development of separation techniques has enabled large-scale downstream studies of the biological properties and clinical value of CTCs. Mutational features (Owen et al., 2020)(Fig. 2 A), gene expression patterns (Lim et al., 2019), and cellular phenotypes (Lang et al., 2018) have been explored for their association with different aspects of tumor physiology. While isolation techniques are getting mature, the new challenge is whether we can perform analysis on the same chip that we used for isolating the cells. Culturing CTCs for drug screening (Khoo et al., 2018), performing secretome analysis (Armbrecht et al., 2020)(Fig. 2B), and single-cell western blot (Sinkala et al., 2017) on a chip have been explored which greatly emphasized the potential of CTCs for clinical use. We believe integrated CTC isolation and analysis systems based on microfluidic platforms for cancer precision medicine are moving closer to practice especially with advances in combining artificial intelligence (AI) for diagnosis.
Fig. 2.
Integrated microfluidic system for CTC studies. A. An integrated workflow for isolation and sorting of CTCs from patient whole blood for molecular analysis. Reproduced with permission from (Owen et al., 2020). B. An integrated system for capture and on-chip secretome analysis of CTCs. Reproduced with permission from (Armbrecht et al., 2020).
2.2. Machine learning in liquid biopsy
Incorporating machine learning into disease detection is a new trend that effectively minimizes human errors and can potentially achieve detection accuracy beyond the limits of the human eye. Recent attempts to complement microfluidic devices with machine learning has been very successful. Chen et. al. for instance, demonstrated that their deep neural network, trained on time-stretch quantitative phase microscopy data from a microfluidic platform, could achieve high throughput label-free classification of single cells with high specificity and sensitivity (Chen et al., 2016). Digital holographic microscopy is another sensing modality that has been successfully combined with machine learning to enable high throughput label-free screening of cells in liquid biopsies (Singh et al., 2017). Additionally, advances in microchip design and data analytics have enabled molecular diagnosis to shift from analyzing discrete biomarkers to signature-based analysis where complex non-linear combinations of biomarkers are learned and utilized for improved diagnosis (Molinski et al., 2020). Such a fusion of technologies will also pave the way into the era of personalized treatment (Chowdury et al., 2019). Machine learning-powered smart microfluidic devices have the potential of delivering new generation of scalable and cost-effective diagnostic strategies that can be rapidly adapted and deployed in the face of emerging health emergencies.
2.3. Exosomes
Circulating tumor exosomes are extracellular vesicles (EVs) present in the blood of cancer patients and serve as messengers from tumor cells (Kalluri and LeBleu, 2020). These membrane encapsulated vesicles are found to contain nucleic acids and proteins that may play a crucial role in cell–cell communication and the modification of tumor microenvironment during tumor progression (Kalluri and LeBleu, 2020). Some evidence showed that the chemical factors carried by exosomes might help modulate the cellular environment at a distant site to form a pre-metastatic niche for subsequent metastatic seeding of cancer cells (Peinado et al., 2017, Yokoi et al., 2017). Traditional ways to detect exosomes include ultracentrifugation (Théry et al., 2006) and other size-based methods (Yeo et al., 2018), which are usually non-specific and the products are often in low purity. Leveraging on these biophysical properties and using antibody-based capturing are two mainstream methods that are currently being used for exosome separation.
Unlike CTCs, capturing and counting exosomes alone are not as informative in revealing the disease status of cancer. Instead, performing molecular profiling of the exosomes is more essential to derive clinically valuable information. For example, some surface proteins or miRNAs carried by exosomes are found specific to the disease stage in different cancers which can be potential disease markers for diagnosis (Kalluri and LeBleu, 2020). Thus, much effort has recently been put into creating an integrated system that can perform exosome capture and analysis on the same chip (Fang et al.,2017, Li et al., 2020a, Li et al., 2020b). For example, exosomes captured by immuno-functionalized surfaces (Zhang et al., 2019a, Zhang et al., 2019b) or nano-herringbone structured surfaces (Zhang et al., 2019a, Zhang et al., 2019b) can be fluorescence-labeled in-situ to detect different surface proteins or detect tumor-specific exosomes from patient blood samples, for exosome-based diagnosis. Capturing and analyzing tumor-secreted exosomes is currently growing rapidly with an increasing number of papers unveiling more and more physiological processes that are mediated by exosomes and their unique therapeutic potential (Ferguson and Nguyen, 2016, Kalluri and LeBleu, 2020) as they do play a role in immunosuppression (Chen et al., 2018) and cancer dissemination (Yokoi et al., 2017). These advancements in understanding the functional roles and molecular contents of exosomes will inspire new integrative systems that target exosomes for more informative cancer diagnostics.
2.4. ctDNA
ctDNA is another category of liquid biopsy biomarker and target. Cell-free DNAs (cfDNAs) have been observed in human blood for a long time, but are only thought to be from the dead cells (Schwarzenbach, Hoon and Pantel, 2011). The importance of these circulating cfDNAs is realized when cancer-associated mutations are detected from cfDNAs in the blood from cancer patients (Vasioukhin et al., 1994). As genomic mutations are highly specific markers for the presence of tumors, this finding revealed a potential strategy to detect and monitor tumors from blood samples. Due to the low signal-to-noise level of ctDNAs in the background of cfDNAs from other normal cells and the technological limitation in detecting low number DNA transcripts, the development and understanding of cfDNAs were progressing rather slowly over the years (Fig. 1). Recently, there is increased research interest in ctDNA due to the advancement and commercialization of next-generation sequencing (NGS) which have made ctDNA research cheaper and more accessible than before. However, another major challenge arising is how to distinguish the low-frequency mutations (<0.1% in early cancer) from those that might arise from clonal hematopoiesis (Umer et al., 2018). This problem has recently been tackled by sequencing the white blood cells (WBCs) simultaneously with the cfDNAs in plasma to distinguish mutations arising from clonal hematopoiesis (Chabon et al., 2020). In this study, the clinical relevance and the association between ctDNA and tumor biology have also been established in a large cohort of non-small cell lung cancer (NSCLC) patients. It is noteworthy that the early detection of cancers in high-risk populations is proven feasible through a simple blood test. Despite the recent progression in ctDNA detection, it is also noticeable that the cost and complexity of the sequencing-based method are currently not practical or economical to support the wide adoption of such methodology for cancer screening. In NSCLC, the overall performance of the test is no better than traditional low-dose CT screening but the cost is far higher.
Microfluidic devices have shown values during the target enrichment process and could reduce the steps required for sample preparation in ctDNA detection (Gwak et al., 2019). It is also possible to detect single mutations on microfluidic platforms (Pekin et al., 2011, Jin et al., 2018) with specific probes. However, separate sequencing steps are still necessary for ctDNA-based clinical research due to the low abundance and multiple targets required for guaranteeing detection sensitivity. The current technology is not able to support sequencing directly in an integrative microfluidic system, however, with increasing knowledge on what are the valuable targets for clinical utility, for example, AI-assisted methods have shown potential to identify those mutations that are more relevant for detecting early-stage lung cancer in patients (Chabon et al., 2020), the recent achievement in using integrative microfluidic systems for nucleic acid detection can bring us towards one-step ctDNA screening in clinical settings (Gauri and Ahmad, 2020).
2.5. Summary
The range of biochemical workflows that can be efficiently performed on microfluidic devices is growing exponentially (Riordon et al., 2019). Single-cell proteomics (Su, Shi and Wei, 2017), nucleic acid sequencing (Lan et al., 2017, Zilionis et al., 2017), Polymerase Chain Reaction (Ahrberg et al., 2016, Zhang and Jiang, 2016, Salman et al., 2020), and secretome analysis (Castiello et al., 2016, Hu et al., 2018) are some examples that have been extensively validated and are directly translatable into diagnostic devices. Furthermore, advances in microfluidic technologies have enabled the addition of biophysical discrimination as possible high-throughput diagnostic strategies – capture and analysis of circulating tumor cells (Nagrath et al., 2007, Vaidyanathan et al., 2018), motility-based sperm sorting (Cho et al., 2003), and hydrodynamic stretching-based mechanical phenotyping (Gossett et al., 2012), just to name a few. The clinical usages of different circulating biomarkers have further evolved through recent developments. For example, CTCs are shown to be more effective when used as companion diagnostics (Punnoose and Lackner, 2012, Chowdury et al., 2019) instead of for early detection. On the other hand, exosomes are shown to be better candidates as early diagnostic markers through detecting the miRNAs (Bhome et al., 2018, Sun et al., 2018, Kohama et al.,2019) or surface protein markers carried by them (Kalluri and LeBleu, 2020, Makler and Asghar, 2020). Apart from this, with the increasing amount of multi-parametric data being generated by microfluidic-based technologies, machine learning will become a natural component of various microfluidic-based diagnostic platforms and these collective efforts will lead the detection of cancer to an new era.
3. Microfluidic diagnosis of infectious diseases
Coronavirus disease 2019 (COVID-19) is the most serious human infectious disease in the past 100 years. Until December 2020, the SARS-CoV-2 virus has infected more than 66 million people with over 1.5 million deaths. The daily increase of new cases increased from 200,000 a day in June 2020 to over 600,000 a day in December 2020. Despite good news of the development of vaccines with very high efficacy, how this pandemic will developed in 2021 is still uncertain at the current stage. Actively detecting and isolating infected individuals and asymptomatic carriers will be the most effective way to limit the spread of this highly infectious disease and to save lives. SARS-CoV-2 is an RNA virus that can spread rapidly among people in close contact through the respiratory system. A nucleic acid test is the standard screening method used for disease detection in this pandemic due to its high sensitivity and specificity. However, the conventional reverse transcriptase PCR (RT-PCR) method is labor-intensive and time-consuming, making it a rate-limiting step for disease screening in most countries at the beginning of the outbreak (Li et al., 2020a, Li et al., 2020b). This pandemic is a vivid lesson urging us to improve on the disease detection techniques for coping with the current situation and be prepared for future outbreaks. An inexpensive, portable, easy to operate, rapid point-of-care (POC) assay will be the most ideal solution that meets our need. In the past two decades, microfluidic-based technology has been extensively studied for POC infectious disease detection and on-going efforts are trying to adapt them to the current pandemic (Berkenbrock et al., 2020, Lin et al., 2020).
3.1. Conventional laboratory-based screening
Before we discuss more on the current pandemic, let us first look at how microfluidic technologies are increasingly being used for current infectious disease detection. A good example to start with is malaria, a life-threatening parasitic disease caused by the species of Plasmodium which infects red blood cells and is transmitted through the Anopheles mosquito. The gold-standard detection method for malaria diagnosis is a blood smear check under an optical microscope to look for the parasites. This method while being labor-intensive and prone to human error is currently still the most widely used method (The malERA Consultative Group on Diagnoses, 2011, Gitta and Kilian, 2020). The gold-standard methods for other infectious disease diagnoses in the clinic include cell culture for bacterial detection, immunology-based methods for antigen detection, and PCR–based nucleic acid detection for the virus. These techniques usually involve a centralized laboratory, trained personnel, and bulky specialized equipment and facilities. Such shortcomings, from time to time, have only been considered as a problem in resource-poor communities of developing countries. However, the COVID-19 pandemic has shown us that these conventional testing methods can be highly deficient even in the most developed areas when the need for rapid and accurate screening gets overwhelmed.
3.2. Point-of-care diagnosis
Substitutes for these laboratory-based detections have been explored with microfluidic technologies (Nasseri et al., 2018). For example, by exploring the biophysical properties of infected red blood cells (Lee and Lim, 2007) or the size difference between parasites and blood cells (Warkiani et al., 2015), different microfluidic assays have been developed to enrich the parasites, detect infect RBC or separate the parasites directly from the blood (Tay et al., 2016). The exploration of molecular-based fast POC detection for malaria has also started long ago. Apart from the microscopy-based parasites screening, a rapid detection test (RDT) which employs lateral flow immunoassay (LFA), is an inexpensive alternative that has been approved by the US FDA in 2007. LFA incorporates capillary-driven fluid flow on a cellulose membrane and antibody-antigen reaction for colorimetric visualization of the test result. This technology has been widely adopted for serology testing or screening using other bodily fluids (e.g. urine in early pregnancy test) due to its convenience and relatively low cost. It is currently one of the major players in POC diagnostics market. The limitation of LFA arises from the quality of immobilized reagents on the assay and the batch difference of membranes from different manufacturers which may result in inaccurate test results. Other limitations of LFA include the need for larger sample volume and the detection limit is relatively low. In some cases, LFA could not reflect the disease state, and it is also hard to achieve a simultaneous testing on multiple targets due to the limited control of fluid flow.
3.3. Emerging microfluidics-based rapid point-of-care devices
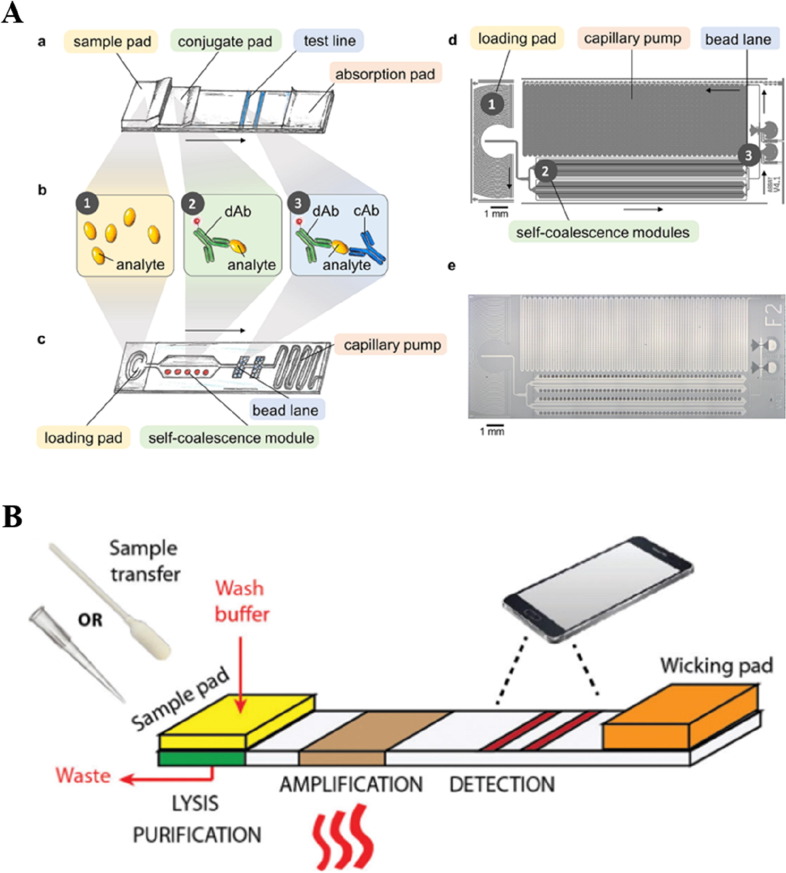
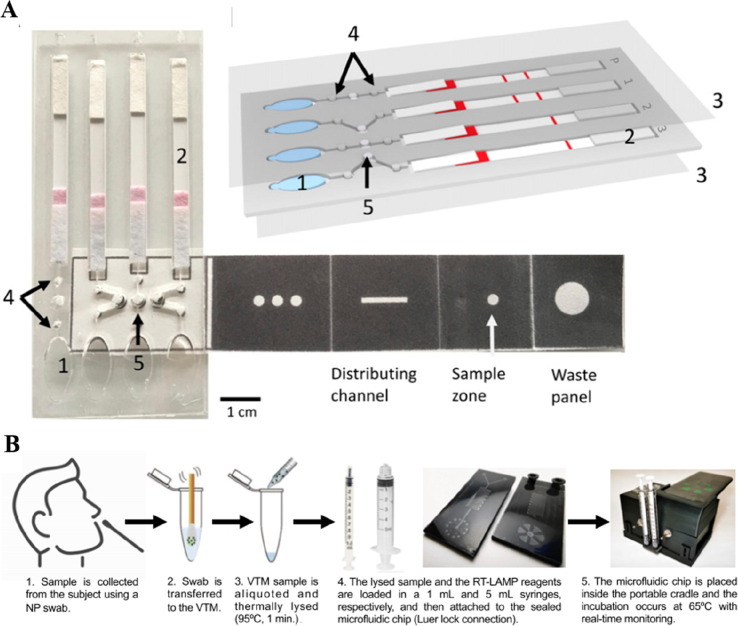
Recent investigations have shown the possibility to substitute LFA with microfluidic channels for improved disease detection performance (Carrell et al., 2019, Hemmig et al., 2020). The fluid flow can be self-driven on a microfluidic chip through capillary force and high spatial–temporal control of the reaction is achieved recently through a self-coalescence module (Gökçe et al., 2019) (Fig. 3 A). Immuno-based antigen detection and recombinase polymerase amplification (RPA) for detecting nucleic acid of HPV are successfully demonstrated on such assays (Hemmig et al., 2020). Identifying different pathogen strains is a new challenge in malaria and other infectious diseases which influences subsequent treatment strategies. This is currently only achievable through nucleic acid detection. In virus detection, screening for pathogen nucleic acid is also the most reliable and sensitive approach. For a long time, a laboratory-based PCR test is the only option for nucleic acid detection-based diagnosis. The development of loop-mediated isothermal amplification (LAMP) and other isothermal methods provide a toolbox that does not require PCR to amplify and detect nucleic acids (Basiri et al., 2020). These isothermal nucleic acid detection methods also make it possible to realize the entire nucleic acid sample preparation and screening workflow on a microfluidic chip. It has been successfully shown in a recent report to integrate LAMP to microfluidic platforms for achieving field detection of malaria with a sensitive and specificity performance better than both RDT and microscopy test (Reboud et al., 2019)(Fig. 4 A). Although the assay needs a larger sample size for validating its applicability, it demonstrated great promise to bring nucleic acid-based POC tests into infectious disease detection, especially, for detecting viruses (Yao et al., 2020, Zhuang et al., 2020).
Fig. 3.
A. Schematics of using microfluidic channels to replace the traditional LFA assays for immunoaffinity-based screening. Reproduced from (Hemmig et al., 2020) with permission. B. Schematics of an integrated paper-based microfluidic device for nucleic acid amplification and detection. Reproduced from (Kaur and Toley, 2018) with permission.
Fig. 4.
Microfluidic-based nucleic acid detection. A. Illustration of a paper-based microfluidic device for malaria DNA detection. Reproduced with permission from (Reboud et al., 2019). B. Schematics of using RT-LAMP for SARS-CoV-2 detection using an integrative microfluidic system. Reproduced with permission from (Ganguli et al., 2020).
3.4. Microfluidics-based detection in COVID-19
During this on-going COVID-19 pandemic, apart from the PCR-based screening, LFA-based fast detection assays have been widely used for disease screening in low-risk populations from as early as April in China (Li et al., 2020a, Li et al., 2020b). Multiple companies have quickly developed and released these products which require only a finger-prick of blood for rapid result readout (Demey et al., 2020, Lin et al., 2020). This can greatly complement and identify potential asymptomatic carriers and unidentified former infections. However, antibody-based screening cannot replace the nucleic acid-based method which can better detect and diagnose current infection. A faster, portable, and precise one-step nucleic acid detection kit will thus be an essential need during any phase of a pandemic (Raimondi et al., 2020). Integrative microfluidic-based nucleic acid detection systems have undergone great technological advancement in the past couple of years (Yeh et al., 2020, Zhuang et al., 2020). There are successful demonstrations of performing sample preparation, nucleic acid detection, and result visualization on a simple microfluidic-based platform (Kaur and Toley, 2018) (Fig. 3B). Such devices have been explored for detecting nucleic acid from SARS-CoV-2 patient samples and demonstrated enhanced performance as compared to LFA-based assays (Dao Thi et al., 2020, Ganguli et al., 2020, Huang et al., 2020) (Fig. 4B), most of which are based on reverse transcriptase LAMP (RT-LAMP). Although the relevant techniques are still undergoing laboratory development, some trials have been carried out to validate the possibility of applying these platforms for assisting current clinical screening (Dao Thi et al., 2020). We believe this is an unprecedented time for the emerging microfluidic nucleic acid detection technologies to demonstrate their true capability and to further improve their performance in clinical settings for onsite POC testing.
4. Perspectives
Microfluidic technology has made a significant impact on biomedicine since being introduced to biomedical engineering in the early 2000s. Like a motherboard for electronics, it provides a convenient platform for integrating different compartmentalized experiments and can achieve automation for some labor-intensive and time-consuming tasks. Conventional techniques in both infectious diseases and cancer detection usually involve centralized laboratories, bulky specialized equipment, and trained personnel. From the trends of development in liquid biopsy and infectious disease detection, we are seeing microfluidic-based systems being widely adopted and integrated into nearly all disease detection techniques for improving their portability and achieving POC applications. Detecting pathogens, molecules, and nucleic acid sequences have all been shown possible on a small microfluidic chip. The revolutionized update on smartphones and other smart electronics have also provided perfect detection platforms for some applications requiring digitalized quantification (Kim et al., 2020, Sun et al., 2020).
The COVID-19 pandemic has exposed the limitations of the current disease detection method even in the most developed areas of the world. While one can perform reliable diagnosis, how one can make the process faster to increase detection throughput still needs to be improved upon. The experience and achievements in developing integrative liquid biopsy platforms and microfluidic POC tests for infectious disease detection have inspired possible solutions. The fight with the current pandemic might not end soon, but now is a good time to test whether these emerging detection systems are the answers we are looking for. More importantly, COVID-19 is unlikely to be the last pandemic that we are going to see. Getting technologies ready so that they can be easily adapted for our next such challenge is a lesson we must learn from the current one.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
K. Jiang thanks the Mechanobiology Institute at the National University of Singapore for the Ph.D. Fellowship. C.T. Lim acknowledges funding support by the Singapore Ministry of Health’s National Medical Research Council under its COVID-19 Research Fund (COVID19RF) (COVID19RF3-0024) and the National University of Singapore COVID-19 Research Seed Grant (NUSCOVID19RG-34).
References
- Ahrberg C.D., Manz A., Chung B.G. Polymerase chain reaction in microfluidic devices. Lab Chip. 2016;16(20):3866–3884. doi: 10.1039/c6lc00984k. [DOI] [PubMed] [Google Scholar]
- Armbrecht L., Rutschmann O., Szczerba B.M., Nikoloff J., Aceto N., Dittrich P.S. Quantification of Protein Secretion from Circulating Tumor Cells in Microfluidic Chambers. Adv. Sci. 2020;7(11):1903237. doi: 10.1002/advs.v7.1110.1002/advs.201903237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basiri A., et al. Microfluidic devices for detection of RNA viruses. Rev. Med. Virol. 2020;2019(April):1–11. doi: 10.1002/rmv.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkenbrock J.A., Grecco-Machado R., Achenbach S. Arsenal of microfluidic testing devices may combat COVID-19 pandemic. MRS Bull. 2020;45(7):511–514. doi: 10.1557/mrs.2020.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhome R., Del Vecchio F., Lee G.-H., Bullock M.D., Primrose J.N., Sayan A.E., Mirnezami A.H. Exosomal microRNAs (exomiRs): Small molecules with a big role in cancer. Cancer Lett. 2018;420:228–235. doi: 10.1016/j.canlet.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrell C., Kava A., Nguyen M., Menger R., Munshi Z., Call Z., Nussbaum M., Henry C. Beyond the lateral flow assay: A review of paper-based microfluidics. Microelectron. Eng. 2019;206:45–54. [Google Scholar]
- Castiello F.R., Heileman K., Tabrizian M. Microfluidic perfusion systems for secretion fingerprint analysis of pancreatic islets: applications, challenges and opportunities. Lab Chip. 2016;16(3):409–431. doi: 10.1039/c5lc01046b. [DOI] [PubMed] [Google Scholar]
- Chabon J.J., Hamilton E.G., Kurtz D.M., Esfahani M.S., Moding E.J., Stehr H., Schroers-Martin J., Nabet B.Y., Chen B., Chaudhuri A.A., Liu C.L., Hui A.B., Jin M.C., Azad T.D., Almanza D., Jeon Y.-J., Nesselbush M.C., Co Ting Keh L., Bonilla R.F., Yoo C.H., Ko R.B., Chen E.L., Merriott D.J., Massion P.P., Mansfield A.S., Jen J., Ren H.Z., Lin S.H., Costantino C.L., Burr R., Tibshirani R., Gambhir S.S., Berry G.J., Jensen K.C., West R.B., Neal J.W., Wakelee H.A., Loo B.W., Jr, Kunder C.A., Leung A.N., Lui N.S., Berry M.F., Shrager J.B., Nair V.S., Haber D.A., Sequist L.V., Alizadeh A.A., Diehn M. Integrating genomic features for non-invasive early lung cancer detection. Nature. 2020;580(7802):245–251. doi: 10.1038/s41586-020-2140-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che J., Yu V., Garon E.B., Goldman J.W., Di Carlo D. Biophysical isolation and identification of circulating tumor cells. Lab Chip. 2017;17(8):1452–1461. doi: 10.1039/c7lc00038c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.L., Mahjoubfar A., Tai L.-C., Blaby I.K., Huang A., Niazi K.R., Jalali B. Deep Learning in Label-free Cell Classification. Sci. Rep. 2016;6(1) doi: 10.1038/srep21471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Huang A.C., Zhang W., Zhang G., Wu M., Xu W., Yu Z., Yang J., Wang B., Sun H., Xia H., Man Q., Zhong W., Antelo L.F., Wu B., Xiong X., Liu X., Guan L., Li T., Liu S., Yang R., Lu Y., Dong L., McGettigan S., Somasundaram R., Radhakrishnan R., Mills G., Lu Y., Kim J., Chen Y.H., Dong H., Zhao Y., Karakousis G.C., Mitchell T.C., Schuchter L.M., Herlyn M., Wherry E.J., Xu X., Guo W. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560(7718):382–386. doi: 10.1038/s41586-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho B.S., Schuster T.G., Zhu X., Chang D., Smith G.D., Takayama S. Passively Driven Integrated Microfluidic System for Separation of Motile Sperm. Anal. Chem. 2003;75(7):1671–1675. doi: 10.1021/ac020579e. [DOI] [PubMed] [Google Scholar]
- Chowdury, M.A., et al., 2019. Biomicrofluidic Systems for Hematologic Cancer Research and Clinical Applications. SLAS Technology. SAGE Publications Inc., pp. 457–476. http://doi.org/10.1177/2472630319846878. [DOI] [PubMed]
- Dao Thi V.L., Herbst K., Boerner K., Meurer M., Kremer L.PM., Kirrmaier D., Freistaedter A., Papagiannidis D., Galmozzi C., Stanifer M.L., Boulant S., Klein S., Chlanda P., Khalid D., Barreto Miranda I., Schnitzler P., Kräusslich H.-G., Knop M., Anders S. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020;12(556):eabc7075. doi: 10.1126/scitranslmed.abc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demey B., Daher N., François C., Lanoix J.-P., Duverlie G., Castelain S., Brochot E. Dynamic profile for the detection of anti-SARS-CoV-2 antibodies using four immunochromatographic assays. J. Infect. 2020;81(2):e6–e10. doi: 10.1016/j.jinf.2020.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fachin F., Spuhler P., Martel-Foley J.M., Edd J.F., Barber T.A., Walsh J., Karabacak M., Pai V., Yu M., Smith K., Hwang H., Yang J., Shah S., Yarmush R., Sequist L.V., Stott S.L., Maheswaran S., Haber D.A., Kapur R., Toner M. Monolithic Chip for High-throughput Blood Cell Depletion to Sort Rare Circulating Tumor Cells. Sci. Rep. 2017;7(1) doi: 10.1038/s41598-017-11119-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, S. et al., 2017). Clinical application of a microfluidic chip for immunocapture and quantification of circulating exosomes to assist breast cancer diagnosis and molecular classification. PLoS ONE, 12, 4, pp. 7–9. http://doi.org/10.1371/journal.pone.0175050. [DOI] [PMC free article] [PubMed]
- Ferguson S.W., Nguyen J. Exosomes as therapeutics: The implications of molecular composition and exosomal heterogeneity. J. Control. Release. 2016;228:179–190. doi: 10.1016/j.jconrel.2016.02.037. [DOI] [PubMed] [Google Scholar]
- Ganguli A., Mostafa A., Berger J., Aydin M.Y., Sun F.u., Ramirez S.A.S.d., Valera E., Cunningham B.T., King W.P., Bashir R. Rapid isothermal amplification and portable detection system for SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020;117(37):22727–22735. doi: 10.1073/pnas.2014739117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauri S., Ahmad M.R. ctDNA Detection in Microfluidic Platform: A Promising Biomarker for Personalized Cancer Chemotherapy. J. Sens. 2020;2020:1–10. [Google Scholar]
- Gitta B., Kilian N. Diagnosis of Malaria Parasites Plasmodium spp . in Endemic Areas: Current Strategies for an Ancient Disease. BioEssays. 2020;42(1):1900138. doi: 10.1002/bies.v42.110.1002/bies.201900138. [DOI] [PubMed] [Google Scholar]
- Gökçe O., Castonguay S., Temiz Y., Gervais T., Delamarche E. Self-coalescing flows in microfluidics for pulse-shaped delivery of reagents. Nature. 2019;574(7777):228–232. doi: 10.1038/s41586-019-1635-z. [DOI] [PubMed] [Google Scholar]
- Gossett D.R., Tse H.T.K., Lee S.A., Ying Y., Lindgren A.G., Yang O.O., Rao J., Clark A.T., Di Carlo D. Hydrodynamic stretching of single cells for large population mechanical phenotyping. Proc. Natl. Acad. Sci. 2012;109(20):7630–7635. doi: 10.1073/pnas.1200107109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwak H., Kim J., Cha S., Cheon Y., Kim S.-I., Kwak B., Hyun K.-A., Jung H.-I. On-chip isolation and enrichment of circulating cell-free DNA using microfluidic device. Biomicrofluidics. 2019;13(2):024113. doi: 10.1063/1.5100009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmig E., Temiz Y., Gökçe O., Lovchik R.D., Delamarche E. Transposing Lateral Flow Immunoassays to Capillary-Driven Microfluidics Using Self-Coalescence Modules and Capillary-Assembled Receptor Carriers. Anal. Chem. 2020;92(1):940–946. doi: 10.1021/acs.analchem.9b03792. [DOI] [PubMed] [Google Scholar]
- Hou H.W., Warkiani M.E., Khoo B.L., Li Z.R., Soo R.A., Tan D.-W., Lim W.-T., Han J., Bhagat A.A.S., Lim C.T. Isolation and retrieval of circulating tumor cells using centrifugal forces. Sci. Rep. 2013;3(1) doi: 10.1038/srep01259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J.-M., Krebs M., Ward T., Sloane R., Priest L., Hughes A., Clack G., Ranson M., Blackhall F., Dive C. Circulating Tumor Cells as a Window on Metastasis Biology in Lung Cancer. Am. J. Pathol. 2011;178(3):989–996. doi: 10.1016/j.ajpath.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q., Luni C., Elvassore N. Microfluidics for secretome analysis under enhanced endogenous signaling. Biochem. Biophys. Res. Commun. 2018;497(2):480–484. doi: 10.1016/j.bbrc.2018.02.025. [DOI] [PubMed] [Google Scholar]
- Huang W.E., Lim B., Hsu C.-C., Xiong D., Wu W., Yu Y., Jia H., Wang Y., Zeng Y., Ji M., Chang H., Zhang X., Wang H., Cui Z. RT‐LAMP for rapid diagnosis of coronavirus SARS‐CoV‐2. Microb. Biotechnol. 2020;13(4):950–961. doi: 10.1111/1751-7915.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K, Lim C.T. Dynamics of Blood Cell Suspensions in Microflows. CRC Press; 2019. Microfluidic biotechnologies for hematology: separation, disease detection and diagnosis; pp. 343–370. [Google Scholar]
- Jin C.E., Koo B., Lee T.Y., Han K., Lim S.B., Park I.J., Shin Y. Simple and Low-Cost Sampling of Cell-Free Nucleic Acids from Blood Plasma for Rapid and Sensitive Detection of Circulating Tumor DNA. Adv. Sci. 2018;5(10):1800614. doi: 10.1002/advs.v5.1010.1002/advs.201800614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R., LeBleu V.S. The biology, function, and biomedical applications of exosomes. Science. 2020;367(6478) doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur N., Toley B.J. Paper-based nucleic acid amplification tests for point-of-care diagnostics. Analyst. 2018;143(10):2213–2234. doi: 10.1039/c7an01943b. [DOI] [PubMed] [Google Scholar]
- Khoo B.L., Grenci G., Lim Y.B., Lee S.C., Han J., Lim C.T. Expansion of patient-derived circulating tumor cells from liquid biopsies using a CTC microfluidic culture device. Nat. Protoc. 2018;13(1):34–58. doi: 10.1038/nprot.2017.125. [DOI] [PubMed] [Google Scholar]
- Kim D.W., Jeong K.Y., Yoon H.C. Smartphone Based Medical Diagnostics. Elsevier; 2020. pp. 103–128. [DOI] [Google Scholar]
- Kohama, I., et al., 2019. An Insight into the Roles of MicroRNAs and Exosomes in Sarcoma. Cancers, 11, 3. http://doi.org/10.3390/cancers11030428. [DOI] [PMC free article] [PubMed]
- Lan F., Demaree B., Ahmed N., Abate A.R. Single-cell genome sequencing at ultra-high-throughput with microfluidic droplet barcoding. Nat. Biotechnol. 2017;35(7):640–646. doi: 10.1038/nbt.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang J.E., Ring A., Porras T., Kaur P., Forte V.A., Mineyev N., Tripathy D., Press M.F., Campo D. RNA-Seq of Circulating Tumor Cells in Stage II–III Breast Cancer. Ann Surg Oncol. 2018;25(8):2261–2270. doi: 10.1245/s10434-018-6540-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G.Y.H., Lim C.T. Biomechanics approaches to studying human diseases. Trends Biotechnol. 2007;25(3):111–118. doi: 10.1016/j.tibtech.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Li G., Tang W., Yang F. Cancer Liquid Biopsy Using Integrated Microfluidic Exosome Analysis Platforms. Biotechnol. J. 2020;15(5):1900225. doi: 10.1002/biot.v15.510.1002/biot.201900225. [DOI] [PubMed] [Google Scholar]
- Li N., et al. ‘Molecular diagnosis of COVID - 19: Current situation and trend in China. (Review) 2020;11:1–13. doi: 10.3892/etm.2020.9142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S.B., Yeo T., Lee W.D., Bhagat A.A.S., Tan S.J., Tan D.S.W., Lim W.-T., Lim C.T. Addressing cellular heterogeneity in tumor and circulation for refined prognostication. Proc. Natl. Acad. Sci. USA. 2019;116(36):17957–17962. doi: 10.1073/pnas.1907904116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q., Wen D., Wu J., Liu L., Wu W., Fang X., Kong J. Microfluidic Immunoassays for Sensitive and Simultaneous Detection of IgG/IgM/Antigen of SARS-CoV-2 within 15 min. Anal. Chem. 2020;92(14):9454–9458. doi: 10.1021/acs.analchem.0c01635. [DOI] [PubMed] [Google Scholar]
- Makler A., Asghar W. Exosomal biomarkers for cancer diagnosis and patient monitoring. Expert Rev. Mol. Diagnost. 2020;20(4):387–400. doi: 10.1080/14737159.2020.1731308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinski J., Tadimety A., Burklund A., Zhang J.X.J. Scalable Signature-Based Molecular Diagnostics Through On-chip Biomarker Profiling Coupled with Machine Learning. Ann. Biomed. Eng. 2020;48(10):2377–2399. doi: 10.1007/s10439-020-02593-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagrath S., Sequist L.V., Maheswaran S., Bell D.W., Irimia D., Ulkus L., Smith M.R., Kwak E.L., Digumarthy S., Muzikansky A., Ryan P., Balis U.J., Tompkins R.G., Haber D.A., Toner M. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450(7173):1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasseri B., Soleimani N., Rabiee N., Kalbasi A., Karimi M., Hamblin M.R. Point-of-care microfluidic devices for pathogen detection. Biosens. Bioelectron. 2018;117:112–128. doi: 10.1016/j.bios.2018.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen S., Lo T.-W., Fouladdel S., Zeinali M., Keller E., Azizi E., Ramnath N., Nagrath S. Simultaneous Single Cell Gene Expression and EGFR Mutation Analysis of Circulating Tumor Cells Reveals Distinct Phenotypes in NSCLC. Adv. Biosys. 2020;4(8):2000110. doi: 10.1002/adbi.v4.810.1002/adbi.202000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H., Zhang H., Matei I.R., Costa-Silva B., Hoshino A., Rodrigues G., Psaila B., Kaplan R.N., Bromberg J.F., Kang Y., Bissell M.J., Cox T.R., Giaccia A.J., Erler J.T., Hiratsuka S., Ghajar C.M., Lyden D. Pre-metastatic niches: organ-specific homes for metastases. Nat. Rev. Cancer. 2017;17(5):302–317. doi: 10.1038/nrc.2017.6. [DOI] [PubMed] [Google Scholar]
- Pekin D., Skhiri Y., Baret J.-C., Le Corre D., Mazutis L., Ben Salem C., Millot F., El Harrak A., Hutchison J.B., Larson J.W., Link D.R., Laurent-Puig P., Griffiths A.D., Taly V. Quantitative and sensitive detection of rare mutations using droplet-based microfluidics. Lab Chip. 2011;11(13):2156. doi: 10.1039/c1lc20128j. [DOI] [PubMed] [Google Scholar]
- Punnoose E.A., Lackner M.R. Challenges and opportunities in the use of CTCs for companion diagnostic development’., Recent results in cancer research. Fortschritte der Krebsforschung. Progres dans les recherches sur le cancer, Germany. 2012;195:241–253. doi: 10.1007/978-3-642-28160-0_22. [DOI] [PubMed] [Google Scholar]
- Raimondi M.T., Donnaloja F., Barzaghini B., Bocconi A., Conci C., Parodi V., Jacchetti E., Carelli S. Bioengineering tools to speed up the discovery and preclinical testing of vaccines for SARS-CoV-2 and therapeutic agents for COVID-19. Theranostics. 2020;10(16):7034–7052. doi: 10.7150/thno.47406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboud J., Xu G., Garrett A., Adriko M., Yang Z., Tukahebwa E.M., Rowell C., Cooper J.M. Paper-based microfluidics for DNA diagnostics of malaria in low resource underserved rural communities. Proc. Natl. Acad. Sci. USA. 2019;116(11):4834–4842. doi: 10.1073/pnas.1812296116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riethdorf S., Fritsche H., Müller V., Rau T., Schindlbeck C., Rack B., Janni W., Coith C., Beck K., Jänicke F., Jackson S., Gornet T., Cristofanilli M., Pantel K. Detection of Circulating Tumor Cells in Peripheral Blood of Patients with Metastatic Breast Cancer: A Validation Study of the CellSearch System. Clin. Cancer Res. 2007;13(3):920–928. doi: 10.1158/1078-0432.CCR-06-1695. [DOI] [PubMed] [Google Scholar]
- Riordon J., Sovilj D., Sanner S., Sinton D., Young E.W.K. Deep Learning with Microfluidics for Biotechnology. Trends Biotechnol. 2019;37(3):310–324. doi: 10.1016/j.tibtech.2018.08.005. [DOI] [PubMed] [Google Scholar]
- Salman A., Carney H., Bateson S., Ali Z. Shunting microfluidic PCR device for rapid bacterial detection. Talanta. 2020;207:120303. doi: 10.1016/j.talanta.2019.120303. [DOI] [PubMed] [Google Scholar]
- Schwarzenbach H., Hoon D.S.B., Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer. 2011;11(6):426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- Singh D.K., Ahrens C.C., Li W., Vanapalli S.A. Label-free, high-throughput holographic screening and enumeration of tumor cells in blood. Lab Chip. 2017;17(17):2920–2932. doi: 10.1039/c7lc00149e. [DOI] [PubMed] [Google Scholar]
- Sinkala E., Sollier-Christen E., Renier C., Rosàs-Canyelles E., Che J., Heirich K., Duncombe T.A., Vlassakis J., Yamauchi K.A., Huang H., Jeffrey S.S., Herr A.E. Profiling protein expression in circulating tumour cells using microfluidic western blotting. Nat. Commun. 2017;8(1) doi: 10.1038/ncomms14622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Shi Q., Wei W. Single cell proteomics in biomedicine: High-dimensional data acquisition, visualization, and analysis. Proteomics. 2017;17(3-4):1600267. doi: 10.1002/pmic.v17.3-410.1002/pmic.201600267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F.u., Ganguli A., Nguyen J., Brisbin R., Shanmugam K., Hirschberg D.L., Wheeler M.B., Bashir R., Nash D.M., Cunningham B.T. Smartphone-based multiplex 30-minute nucleic acid test of live virus from nasal swab extract. Lab Chip. 2020;20(9):1621–1627. doi: 10.1039/d0lc00304b. [DOI] [PubMed] [Google Scholar]
- Sun Z., Shi K.e., Yang S., Liu J., Zhou Q., Wang G., Song J., Li Z., Zhang Z., Yuan W. Effect of exosomal miRNA on cancer biology and clinical applications. Mol. Cancer. 2018;17(1) doi: 10.1186/s12943-018-0897-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S.J., Yobas L., Lee G.Y.H., Ong C.N., Lim C.T. Microdevice for the isolation and enumeration of cancer cells from blood. Biomed. Microdevices. 2009;11(4):883–892. doi: 10.1007/s10544-009-9305-9. [DOI] [PubMed] [Google Scholar]
- Tay A., Pavesi A., Yazdi S.R., Lim C.T., Warkiani M.E. Advances in microfluidics in combating infectious diseases. Biotechnol. Adv. 2016;34(4):404–421. doi: 10.1016/j.biotechadv.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The malERA Consultative Group on Diagnoses, 2011. A research agenda for malaria eradication: Diagnoses and diagnostics. PLoS Med., 8, 1. http://doi.org/10.1371/journal.pmed.1000396. [DOI] [PMC free article] [PubMed]
- Théry C., Amigorena S., Raposo G., Clayton A. Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids. Curr. Protoc. Cell Biol. 2006;30(1) doi: 10.1002/0471143030.2006.30.issue-110.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- Umer M., Vaidyanathan R., Nguyen N.-T., Shiddiky M.J.A. Circulating tumor microemboli: Progress in molecular understanding and enrichment technologies. Biotechnol. Adv. 2018;36(4):1367–1389. doi: 10.1016/j.biotechadv.2018.05.002. [DOI] [PubMed] [Google Scholar]
- Vaidyanathan R., Soon R.H., Zhang P., Jiang K., Lim C.T. Cancer Diagnosis: From Tumor to Liquid Biopsy and Beyond. Lab Chip. 2018 doi: 10.1039/C8LC00684A. [DOI] [PubMed] [Google Scholar]
- Vasioukhin V., Anker P., Maurice P., Lyautey J., Lederrey C., Stroun M. Point mutations of the N-ras gene in the blood plasma DNA of patients with myelodysplastic syndrome or acute myelogenous leukaemia. Br. J. Haematol. 1994;86(4):774–779. doi: 10.1111/j.1365-2141.1994.tb04828.x. [DOI] [PubMed] [Google Scholar]
- Warkiani M.E., Tay A.K.P., Khoo B.L., Xiaofeng X.u., Han J., Lim C.T. Malaria detection using inertial microfluidics. Lab Chip. 2015;15(4):1101–1109. doi: 10.1039/c4lc01058b. [DOI] [PubMed] [Google Scholar]
- Yao Y., Chen X.i., Zhang X., Liu Q.i., Zhu J., Zhao W., Liu S., Sui G. Rapid Detection of Influenza Virus Subtypes Based on an Integrated Centrifugal Disc. ACS Sens. 2020;5(5):1354–1362. doi: 10.1021/acssensors.9b02595. [DOI] [PubMed] [Google Scholar]
- Yeh Y.-T., Gulino K., Zhang YuHe, Sabestien A., Chou T.-W., Zhou B., Lin Z., Albert I., Lu H., Swaminathan V., Ghedin E., Terrones M. A rapid and label-free platform for virus capture and identification from clinical samples. Proc. Natl. Acad. Sci. USA. 2020;117(2):895–901. doi: 10.1073/pnas.1910113117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo J.C., Kenry, Zhao Z., Zhang P., Wang Z., Lim C.T. Label-free extraction of extracellular vesicles using centrifugal microfluidics. Biomicrofluidics. 2018;12(2):024103. doi: 10.1063/1.5019983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi A., Yoshioka Y., Yamamoto Y., Ishikawa M., Ikeda S.-I., Kato T., Kiyono T., Takeshita F., Kajiyama H., Kikkawa F., Ochiya T. Malignant extracellular vesicles carrying MMP1 mRNA facilitate peritoneal dissemination in ovarian cancer. Nat. Commun. 2017;8(1) doi: 10.1038/ncomms14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Zhou X., He M., Shang Y., Tetlow A.L., Godwin A.K., Zeng Y. Ultrasensitive detection of circulating exosomes with a 3D-nanopatterned microfluidic chip. Nat. Biomed. Eng. 2019;3(6):438–451. doi: 10.1038/s41551-019-0356-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Zhou X., Zeng Y. Multiplexed immunophenotyping of circulating exosomes on nano-engineered ExoProfile chip towards early diagnosis of cancer. Chem. Sci. 2019;10(21):5495–5504. doi: 10.1039/c9sc00961b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Jiang H.-R. A review on continuous-flow microfluidic PCR in droplets: Advances, challenges and future. Analytica Chimica Acta. 2016;914:7–16. doi: 10.1016/j.aca.2016.02.006. [DOI] [PubMed] [Google Scholar]
- Zhuang J., Yin J., Lv S., Wang B., Mu Y. Advanced “lab-on-a-chip” to detect viruses – Current challenges and future perspectives. Biosens. Bioelectron. 2020;163:112291. doi: 10.1016/j.bios.2020.112291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilionis R., Nainys J., Veres A., Savova V., Zemmour D., Klein A.M., Mazutis L. Single-cell barcoding and sequencing using droplet microfluidics. Nat. Protoc. 2017;12(1):44–73. doi: 10.1038/nprot.2016.154. [DOI] [PubMed] [Google Scholar]