Abstract
The scientific community and decision-makers are increasingly concerned about transparency and reproducibility of epidemiologic studies using longitudinal healthcare databases. We explored the extent to which published pharmacoepidemiologic studies using commercially available databases could be reproduced by other investigators. We identified a nonsystematic sample of 38 descriptive or comparative safety/effectiveness cohort studies. Seven studies were excluded from reproduction, five because of violation of fundamental design principles, and two because of grossly inadequate reporting. In the remaining studies, >1,000 patient characteristics and measures of association were reproduced with a high degree of accuracy (median differences between original and reproduction <2% and <0.1). An essential component of transparent and reproducible research with healthcare databases is more complete reporting of study implementation. Once reproducibility is achieved, the conversation can be elevated to assess whether suboptimal design choices led to avoidable bias and whether findings are replicable in other data sources.
Concerns about reproducibility of biomedical science have moved funding agencies, professional research societies, and journal editors to strengthen the transparency of the research process in preclinical, clinical, and population health sciences.1–3 Transparency and reproducibility are intertwined concepts. There is general agreement that transparency can be achieved through a series of such measures as: (1) registration of study protocols before the initiation of research to increase the chance that all study results will become publicly available; (2) reporting guidelines to encourage complete description of all details necessary to reproduce study findings; and (3) making the actual research data available to other researchers to reproduce findings or make additional discoveries.4–7 Funding agencies, such as the National Institutes of Health and the Patient Centered Outcomes Research Institute, have made public statements about the necessity to make research data available for reproduction by independent research groups.8,9
Randomized clinical trials are at the forefront of activities to increase transparency and reproducibility. Regulatory agencies and journal editors require the registration of clinical trial protocols,10 and observational studies are increasingly encouraged to follow suit.4,11,12 Randomized clinical trials have extensive guidelines and standards with regard to design, conduct, and reporting.13,14 After a consortium of pharmaceutical companies in the United States volunteered to make trial data publically available,15 the European Medicines Agency enacted a policy to make clinical reports submitted in support of a marketing authorization application or postauthorization submission publicly available.16
After these developments, the epidemiology research community produced several guidelines for observational studies that encourage completeness of study design reporting. These include elements such as clear description of setting, eligibility criteria, variable definition, measurement, bias, and study size.17,18 Although these guidelines facilitate transparency, Hernán and Wilcox19 explain that direct replication with new data collection, as done in preclinical experiments, is rarely possible for large prospective epidemiologic studies because of their long duration and high costs, making it even more urgent to encourage sharing of original research data.
However, observational studies that make secondary use of existing longitudinal healthcare databases can be reproduced relatively quickly at moderate costs when the source data (e.g., administrative healthcare claims or electronic medical records data) are made available through licensing and data use agreements. There are many examples in the literature of similar studies conducted by different investigators, sometimes using the same source data. In some cases, findings are reproduced, but, in others, findings can also be quite different.20–23 Recognizing the importance of reproducibility, several organizations have created common data models, converting raw source data to fit standardized data table structures to facilitate use of validated code and software tools for rapid, high integrity safety analyses in different data sources.7,24,25
In database studies, reproducibility is the ability of independent investigators to obtain the same findings when applying the same design and operational choices in the same data source. This is in contrast to replicability, which would be the ability to obtain similar findings with application of the same design and choices in different data sources. For healthcare database studies, a near exact reproduction of findings within the same data source should be consistently achievable.
Even with access to relevant healthcare databases and validated software tools to extract and analyze the desired cohorts, the ability to reproduce studies remains dependent on clear reporting of key design, operational, and analytic decisions. Currently, there are no published large-scale examinations of the reproducibility of healthcare database research that quantify this issue and highlight specific areas in which the situation could be improved. Our objective was to attempt reproduction of a select sample of healthcare database studies from the peer-reviewed literature. We look specifically to assess the transparency of information shared with the public and how closely findings could be reproduced.
RESULTS
In total, we reproduced 40 substudies from 31 publications within the Aetion platform and directly tested 6 protocols using Aetion and PROMPT-CM or de novo SAS programming. Of the reproduced studies, 13 included comparative assessments as well as descriptive characteristics for the studied population; all six of the direct test protocols were comparative studies. The directly tested study protocols, a list of studies considered for reproduction, and detailed comparisons of original and reproduced studies are available in the Supplementary Appendix S1.
Difficulty in reproduction of original studies was noted for several reasons: (1) code lists defining outcomes, covariates, and inclusion/exclusion criteria in original studies were not reported (11 of 31; 35.4%); (2) noncode list-based covariate definition unclear, for example calculation of dose or grouping of insurance types (4 of 31; 12.9%); and (3) timing of cohort entry, inclusion criteria, covariate assessment period, or enrollment period unclear (3 of 31; 9.6%).
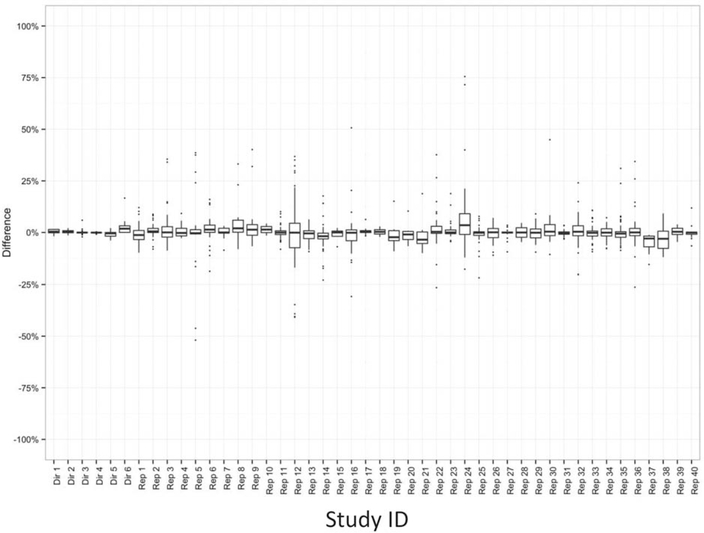
The difference in prevalence of binary characteristics for all reproduced descriptive or comparative studies and protocols are shown in Figure 1. There were 1419 differences (across 983 variables) in the reproduced studies and directly tested protocols. The median absolute difference was 1.7% for reproduced studies and 0.2% for direct testing of protocols. The interquartile ranges for reproduced studies were wider than for directly tested protocols, with outliers where the prevalence differences were >25.0%. For example, in study “RS05,” six outliers with prevalence differences >25.0% reflect differences in prevalence between the original study and reproduction within categories of a comorbidity score. This large difference may be partially attributable to lack of clarity regarding which version of the comorbidity score was implemented for the study and the absence of code lists or algorithms in the publication.
Figure 1.
Differences in binary characteristics for all reproduced (N = 40) and protocols (N = 6). Dir = directly tested protocol; Rep = reproduced study.
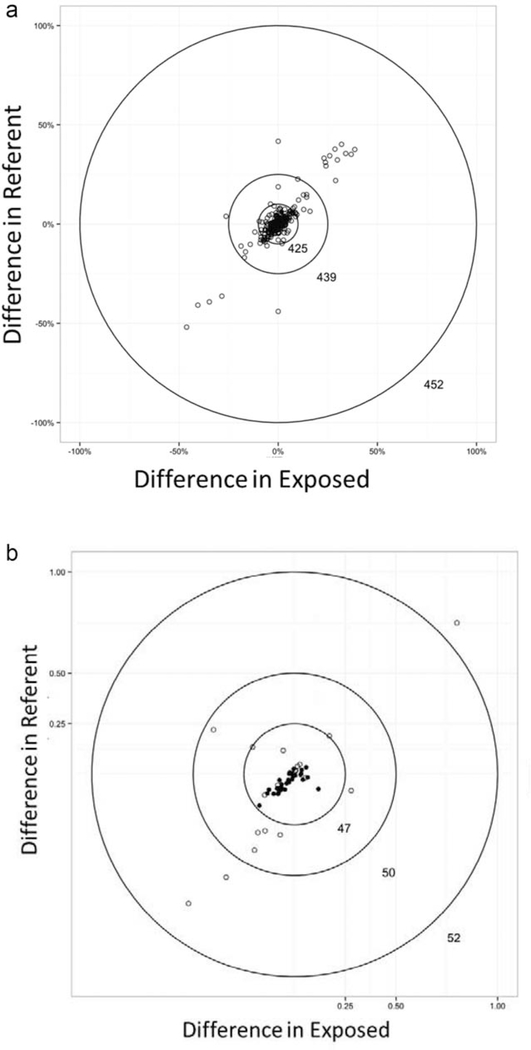
In Figure 2, circle plots A and B show the magnitude of differences in prevalence of binary characteristics between original and reproduced studies or direct tests within each exposure group of comparative studies. The innermost circle has a width of ±10 percentage-point difference in estimated prevalence for exposure and referent groups. This boundary was selected as it is commonly used as a rule of thumb to measure the presence of meaningful confounding. The innermost circle captures 425 of 452 (94.0%) of the measured binary characteristics overall, and 68 of 68 (100%) of the binary characteristics from direct testing of protocols (circle plot A). A small proportion, 4.0% of binary characteristics from reproduced studies, had differences in prevalence >±10% and 2.9% had differences >±25% for both exposure and referent groups. However, the difference in prevalence between original and reproduced studies was of similar magnitude and direction for exposure and referent groups (e.g., differences were nondifferential with respect to exposure). Circle plot B shows the magnitude of standardized differences for continuous characteristics between original and reproduced studies or direct tests within each exposure group of comparative studies. The innermost circle represents a standardized difference of ±0.25. It contains 90% of all continuous characteristics and 100% of continuous characteristics from direct testing. Among the reproduced studies, 4.0% of continuous characteristics had standardized differences >0.5.
Figure 2.
Differences in population characteristics by exposure group for comparative studies. (a) All binary characteristics in reproduced studies (N = 13) or direct test protocols (N = 6). (b) All continuous characteristics in reproduced studies (N = 13) or direct test protocols (N = 6). A. All binary characteristics in reproduced studies (N = 13) or direct test protocols (N = 6). B. All continuous characteristics in reproduced studies (N = 13) or direct test protocols (N = 6). ○ Reproduced comparative study. • Direct test protocol.
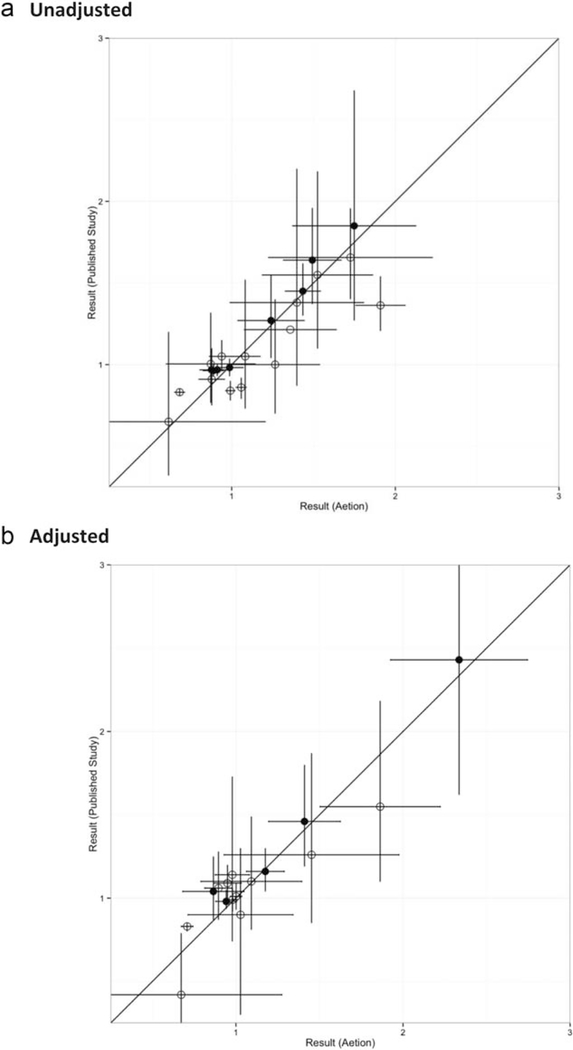
In Figure 3, the y-axis represents the unadjusted or adjusted measure of association from the original study or PROMPT-CM/de novo programmer implementation of a direct test protocol and the x-axis represents the adjusted measure of association from reproduction in Aetion. When the confidence interval crosses the diagonal, the point estimate of the original study is within the confidence limit of the reproduced study and vice versa. The diagonal line angle represents perfect calibration of reproduced adjusted measures of association. We observe that unadjusted and adjusted measures of association are generally well calibrated, with reproduction estimates of similar magnitude as the original estimates (chi-square goodness of fit within tertiles of log hazard ratio; unadjusted: P = 0.46, adjusted: P = 0.82). However, there were a few reproduced studies in which the confidence limits of original and reproduced studies did not overlap. The median difference for unadjusted measures of association for reproduced studies was 0.11 and 0.06, respectively, for directly tested protocols. For adjusted measures of association, the median differences were 0.14 and 0.04, respectively, for reproduced studies and directly tested protocols.
Figure 3.
Measures of association in reproduced comparative studies (N = 13) and protocols (N = 6). (a) Unadjusted. (b) Adjusted. A. All binary characteristics in reproduced studies (N = 13) or direct test protocols (N = 6). B. All continuous characteristics in reproduced studies (N = 13) or direct test protocols (N = 6). ○ Reproduced comparative study. • Direct test protocol.
DISCUSSION
Decision-makers perceive randomized trials as high quality not only because baseline randomization will generally balance confounders, but also because of the stringent requirements for design, implementation, and reporting (either in a published manuscript or via regulatory registration). There is an assumption of a certain standard of conduct when discussing results of an randomized clinical trial that is not yet present for observational database studies, preclinical studies, or other sciences.26 The issue of nonreproducibility of preclinical studies was recently highlighted in a number of science and nature articles, in which separate attempts by pharmaceutical, medical, or other research groups were able to reproduce only 11–45% of published preclinical studies, often after significant efforts by reproducing investigators to communicate with the initial research teams regarding protocol details and procedures.1,2 Similarly, a recent JAMA article found that 35% of published re-analyses of clinical trials had different conclusions than the original findings regarding which patients should be treated.27
Our study is the first large scale exploration of the reproducibility of peer-reviewed publications of database research ever conducted. With access to large healthcare databases that serve as data sources for many published studies, software tools designed to facilitate reproducibility, and an experienced team of pharmacoepidemiologists, we were able to independently reproduce 31 studies and 6 protocols in approximately 4 months (0.6 person-years of effort).
There is currently no gold standard defining reproducibility of a database study. In our view, reproducibility of database science is not a binary characteristic, it is a continuum. Studies with detailed and transparent reporting are easier to reproduce than studies that provide little detail on implementation. The absence of comprehensive public reporting on key operational definitions was the most common inhibitor of reproduction of studies by our team of independent investigators. In spite of this, among 31 peer reviewed publications, we were able to reproduce population descriptive measures and measures of association with a high degree of accuracy. The median difference in adjusted measure of effect between the original and reproduction for the studies we examined was reassuringly small and unlikely to have implications for clinical care.
The high accuracy in this reproduction effort can be attributed in part to the efforts of the reproduction team, a group of pharmacoepidemiologists with decades of experience, to make informed guesses regarding variable definitions or other key decisions when these were not clearly specified in the original articles. Intuiting the intent of the original studies was easier in situations in which possible variations for defining a condition were smaller. For example, although there are limited International Classification of Diseases ninth revision – Clinical Modification codes for a condition, such as hypertension, there could be dozens or hundreds of Read codes28 representing potentially relevant clinical concepts in data from the United Kingdom.
Although the median differences between original studies and reproductions were small, the range for a subset of characteristics within some reproduced studies could be quite large compared to the range observed with independent implementation of clearly outlined protocols. The need to make informed guesses contributed to the wide range in differences observed in reproduction, highlighting the need for greater transparency in reporting of database studies to enable complete reproduction of science.
Our study has several limitations. First, our identification of studies for reproduction was nonsystematic and should not be construed as representative of all database science. Future work to quantify reproducibility and transparency of database studies should incorporate a well-defined selection strategy with input from multiple stakeholders. Second, differences in how the Aetion platform and PROMPT-CM or de novo programmers interacted with source data (e.g., Aetion’s data connector vs. conversion of raw data to Sentinel common data model tables) could have contributed to differences in direct testing of protocols, however, our virtually identical results in direct testing alleviate that concern. Third, shifts in guidelines and practice for prescribing or indications for small nonoverlap in years for some of the original studies to the years of data accessible by the reproduction team could have contributed to differences. For more exact reproduction, accessing the exact same years of data for all studies would remove the confounding factor of time. A fourth limitation was that some studies were originally conducted by researchers employed by the data-holding organization and therefore had access to more detailed cost data than general researchers outside the organization. Finally, we did not attempt to reproduce studies that were not cohort studies, required supplemental data linkage, or had obvious flaws in design. This does not mean that the excluded studies were not reproducible.
There are currently a number of guidance documents on the conduct6,29–33 and reporting of database studies.18 However, the lack of incentive for researchers to expend additional effort on open practices remains. We have demonstrated that factors contributing to lack of transparency and reproducibility of peerreviewed publications of database studies can be outlined and quantified. Quantifying the issues paves the way for measuring improvement with implementation of measures designed to raise the bar and incentivize changes in research culture and practice. For example, the Transparency and Openness Committee recently proposed eight universal standards to address the role that journals can play to promote transparency and openness in the sciences.3 The Transparency and Openness Committee standards provide concrete suggestions that journals can adopt to provide positive reinforcement and incentives for open reporting, such as encouraging recognition of researcher’s effort in creating appendices, data, code, and preregistered study protocols through citation of these materials as intellectual contributions as well as issuing badges or letters of recognition from journals for meeting openness standards.3 With the availability of electronic appendices in almost any scientific journal, word limits are no longer a barrier to detailed reporting. When study protocols detailing these decisions are not publically available via preregistration, then this information should be included as online appendices or made available as a citable resource for any peer-reviewed healthcare database study. Currently, there are no standards for contents of e-appendices, however, as the field moves toward greater transparency of reporting, there may be a need for guidelines regarding the contents of appendices detailing operational decisions.
Full reproducibility in healthcare database studies occurs when independent investigators are able to apply the same design and analytic choices to the same source data, and are able to obtain the same analytic population and estimated measures of association (or at least a near exact reproduction). The scientific community as a whole would benefit and the credibility of healthcare database studies would increase if greater effort were directed toward ensuring that public reporting for database studies contained sufficient detail to allow full reproduction. Without reproduction there can be no replication and without replication there can be little trust in our research output.
Our reproduction effort demonstrates that when reporting of details of study implementation are sufficiently transparent, healthcare database studies can be reproduced with great accuracy; however, there is great variability in the degree to which recently published healthcare database studies are reproducible. We contrasted the degree of variability in population characteristics and estimates for measure of association from independent implementation of clearly specified protocols with the variability observed using all publically available information to reproduce published database studies. In our investigation, the greatest driver of variability between the original and the reproduction was the lack of detailed code lists describing how original researchers defined their target population and its characteristics. Reporting details of the algorithms used to define the analytic population and its characteristics is a necessary component for transparency and reproducibility of healthcare database science that has been recognized in recent guidelines for database studies, such as REporting of studies Conducted using Observational Routinely-collected Data (RECORD).34 However, other inhibitors of reproduction included the lack of explicit reporting of key design choices. For example, the exact washout period for new users, the covariate assessment period, or the follow-up period. Which of these periods included the index date? Was a stockpiling algorithm used to account for dispensations with overlapping day’s supply? Was there allowance for an induction period before ascertaining outcomes? What were the reasons for censoring? Are inpatient outcomes defined as occurring on the date of admission, discharge, or somewhere in between? If an as-treated analysis was reported, how was adding or switching to other medications handled? These are all operational questions that researchers must make decisions about in programming code when making use of the longitudinal data streams available in large healthcare databases. However, these decisions frequently remain behind the scenes, as public reporting often lacks the degree of detail necessary for reproduction.
These key operational questions are the “gears and levers” that investigators adjust when using semiautomated software solutions, such as the Aetion platform or the US Food and Drug Administration (FDA) PROMPT tools to customize and implement database studies. Although key operational decisions cannot and should not be automated, newly available software tools can implement researcher’s choices with validated code and further facilitate transparency and reproducibility of studies via creation of reports with detailed description of those decisions for online appendices.
Our large scale reproduction demonstrates the need for a joint effort by researchers, journal editors, regulators, healthcare technology assessment organizations, and other stakeholders to continue working on creative solutions for facilitating transparency and reproducibility of database studies. Once complete documentation and full reproducibility of specific analyses has been achieved, the next step for database science in healthcare is replication and elevation of the conversation to seek understanding of whether different causal contrasts led to different results or whether suboptimal design and analytic choices led to avoidable bias.
METHODS
Identification of studies
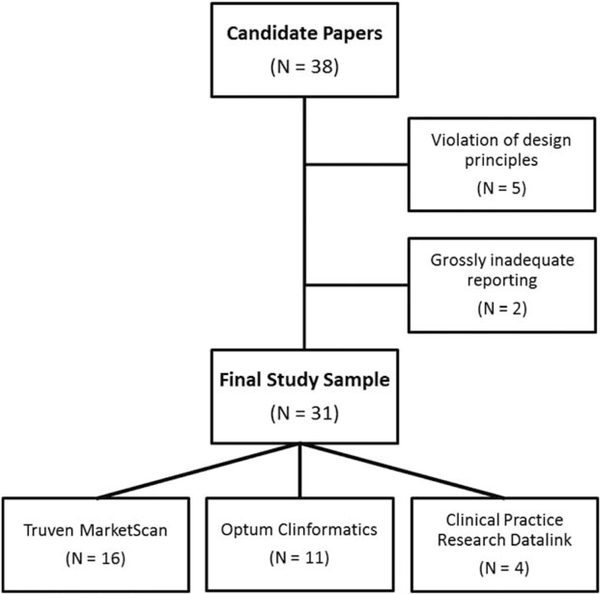
We used a nonsystematic snowball strategy to identify 38 healthcare database studies published in the peer-reviewed literature between 2008 and 2014 and available through PubMed (Figure 4). Suitable candidate publications for this evaluation of reproducibility included cohort studies conducted in databases licensed by Brigham and Women’s Hospital or Boehringer Ingelheim, the two groups that conducted this study. These databases included the MarketScan claims database from Truven (United States), the United Healthcare claims data from the Clinformatics DataMart by Optum (United States), and the Clinical Practice Research Datalink primary care electronic medical record databases (England).
Table 1.
Metrics to quantify reproducibility
| Measure | Reproduction of published study | Direct test of protocol |
|---|---|---|
| Difference | Oprevalence − Aprevalence | Dprevalence − Aprevalence |
| Absolute difference | abs(Ohazard ratio − Ahazard ratio) | abs(Dhazard ratio − Ahazard ratio) |
| Standardized difference |
Metrics to quantify reproducibility
A, Aetion reproduction; D, direct test protocol (Prospective Routine Observational Monitoring Program Tool – Cohort Matching [PROMPT-CM] or independent programming); O, original study.
We only attempted to reproduce cohort studies that used a study design compatible with the FDA Prospective Routine Observational Monitoring Program Tool – Cohort Matching (PROMPT-CM)35 for active drug safety surveillance. This tool allows semiautomated cohort identification, creation of baseline covariates, and follow-up for outcomes within the Sentinel Program’s distributed data network.36 We did not attempt to reproduce studies that required access to supplemental linked data or chart reviews. Small nonoverlap in years of data available to the original investigators and the reproduction team was allowed if there were no new drugs in class entering or exiting the market and no factors likely to influence the descriptive characteristics of the studied population in the nonoverlapping time were identified. We excluded five articles implemented in a way that violated fundamental design principles, such as inclusion of immortal time or inconsistent temporality (e.g., defining exposure after start of follow-up). Another two articles were excluded from reproduction because only adjusted results were reported without sufficient detail on key operational parameters necessary to implement the study. In the 31 articles remaining, we allowed reproduction of multiple substudies per article (e.g., a study among prevalent users and a study among incident users or studies among patients <65 or ≥65 years of age).
For the purpose of this study, we considered all publicly available information for each study, including the article itself, appendices, additional online materials published on the journal website, or materials explicitly referenced in the article and available online.
Reproduction of studies
Historically, reproducing a study conducted in a healthcare database involved independent double programming. This resource-heavy and time-consuming strategy would not have been viable for the dozens of studies that we planned to replicate. Therefore, we used the Aetion Evidence Platform (Aetion, New York, NY); specifically the Aetion Safety, Aetion Effectiveness, and Aetion Population Description applications. Common to these applications is a cohort selector with a semantic interface and a library of stored definitions for exposures, outcomes, and confounder variables based on user specifications. Methodological details concerning cohort identification, including a Consolidated Standards of Reporting Trials patient flow diagram, and statistical analyses are provided via an automated reporting tool.
Identification of the relevant analytic population and all statistical analyses used in reproduction applied the methods, as described in the original publications. When the description of study implementation was insufficiently detailed, the reproduction team made educated guesses about the intent of the original investigators and adapted code lists/algorithms from other articles studying similar conditions.
Direct reproduction of protocols: Aetion analytics vs. accepted standard approaches
In addition to replicating studies in the peer-reviewed literature, we developed six protocols and directly examined reproducibility by executing the protocols via the Aetion platform and either the Sentinel Program’s extensively validated, open source PROMPT-CM35 tool or independent programming with SAS software (SAS Institute, Cary, NC). We chose the PROMPT-CM modules as a current standard because these preprogrammed SAS macros have been comprehensively tested in a controlled data environment and are currently in use by the FDA for active drug safety surveillance.35,37 Detailed information about the modular programs, including program code, can be found at www.mini-sentinel.org. The PROMPT-CM tool is designed to run on source data that has been converted to table structures specified in the Sentinel common data model. We directly tested four protocols with Aetion and PROMPT-CM using United Healthcare as source data; these protocols were based on previously published analyses.38–40 The remaining two protocols tested comparability of study findings using Aetion and de novo SAS programming with Medicare Claims Synthetic Public Use Files41 as source data and were based on examples in the user manual.42
We conducted direct testing of study protocols in addition to reproduction of published database studies for two reasons: (1) to provide a baseline of how closely one can expect results to align when investigating teams independently implement the same protocol with different software tools (either Aetion PROMPT-CM or de novo SAS programming); (2) to verify that results from Aetion are not specific to the platform (a proprietary software tool). We chose to use the Aetion platform to carry out the large scale reproduction of healthcare database studies because it required less person-time to implement per study than either PROMPT-CM or de novo SAS programming.
Metrics to quantify reproducibility
In order to quantify the reproducibility of studies, we focused on two categories: (1) reproduction of the patient cohort and (2) reproduction of the statistical analysis. For the former, we compared population descriptive measures between our reproduction and the original published study; for the latter, we compared measures of association. Others have used metrics, such as a kappa statistic, in which original and reproduced studies are defined as concordant if they are on the same side of null, as well as a binary metric in which concordant results are defined as having a ratio of reproduction to original estimates between 0.5 and 1.5.43 However, these metrics may be overly generous in classifying reproductions as concordant with original studies. Requiring only that estimates are on the same side of null or have magnitude within 50% leaves considerable room for meaningful differences in both the operational aspect of how the estimate was obtained and how such an estimate might be interpreted clinically. We chose to use metrics that focus on absolute differences and calibration in order to provide a more nuanced picture of the degree to which studies are reproducible.
Population descriptive measures
We computed the difference in prevalence of binary characteristics as well as standardized differences for continuous characteristics reported in the original and reproduced studies (Table 1). Differences closer to 0.0 reflect a closer match between original and reproduced studies. We also calculated differences and standardized differences for population characteristics in direct testing of protocols using PROMPT-CM35 or de novo programming with SAS (SAS Institute, version 9.3) and Aetion. For comparative studies (as opposed to descriptive studies), these differences were computed within exposure group.
Measures of association
We computed absolute differences for crude and adjusted relative measures of association (e.g., hazard ratio) for comparative studies. We produced calibration plots for original and reproduced measures of occurrence and association and report a Chi-Square goodness of fit statistic.
Supplementary Material
Figure 4.
Consolidated Standards of Reporting Trials diagram.
Study Highlights
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
The scientific community and decision-makers are increasingly concerned about transparency and reproducibility of biomedical science.
WHAT QUESTION DID THIS STUDY ADDRESS?
Recent high profile efforts to reproduce preclinical and clinical studies have drawn attention to this issue; however, there has not yet been a large-scale effort to evaluate reproducibility of healthcare database studies.
WHAT THIS STUDY ADDS TO OUR KNOWLEDGE
With sufficient transparency in reporting, healthcare database studies can be reproduced with great accuracy; however, there is great variability in the degree to which recently published healthcare database studies are reproducible. The reproduction team made informed guesses in >50% of reproduced studies, highlighting the need for greater transparency in reporting.
HOW THIS MIGHT CHANGE CLINICAL PHARMACOLOGY AND THERAPEUTICS
Detailed reporting of key design choices and codes used to characterize the analytic population are a necessary component for reproducibility of healthcare database studies. Barriers to reproducibility can be outlined and quantified, paving the way for measuring improvement with implementation of measures designed to incentivize changes in research culture and practice.
ACKNOWLEDGMENTS
Dr. Wang was supported by grant number R00HS022193 from the Agency for Healthcare Research and Quality. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Agency for Healthcare Research and Quality.
Footnotes
Additional Supporting Information may be found in the online version of this article.
CONFLICT OF INTEREST/DISCLOSURES
The authors of this manuscript have longstanding interest in developing software to facilitate transparent and reproducible database research. Several were involved in development of both the Aetion platform and the FDA’s PROMPT tools. Dr. Wang was a key developer of the open source PROMPT-CM tool for the FDA’s Sentinel Program and a consultant to Aetion, a software company. Dr. Wang’s effort on this project was supported by her R00 award from AHRQ; she has received less than $5,500 in consulting fees per year in the last two years as a consultant to Aetion. Dr. Rassen is Chief Scientific Officer of Aetion and was involved with development of software tools for the FDA’s Sentinel Initiative. Ms. Patrick is employed by Aetion and Ms. Garry is a consultant to Aetion. Dr. Verpillat and Dr. Bartels are employed by Boehringer Ingelheim.
References
- 1.Begley CG & Ellis LM Drug development: raise standards for preclinical cancer research. Nature 483, 531–533 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Kaiser J The cancer test. Science 348, 1411–1413 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Nosek BA et al. Scientific standards. Promoting an open research culture. Science 348, 1422–1425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.European Network of Centres for Pharmacoepidemiology and Pharmacovigilance (ENCePP). <http://www.encepp.eu/> (2014). Accessed 13 December 2014.
- 5.PCORI Methodology Committee. (eds. Hickam D, Totten A, Berg A, Rader K, Goodman S & Newhouse R) The PCORI Methodology Report. 2013. <http://www.pcori.org/assets/2013/11/PCORI-Methodology-Report.pdf>. Accessed 13 December 2014.
- 6.Dreyer NA et al. GRACE principles: recognizing high-quality observational studies of comparative effectiveness. Am. J. Manag. Care 16, 467–471 (2010). [PubMed] [Google Scholar]
- 7.Curtis LH et al. Design considerations, architecture, and use of the Mini-Sentinel distributed data system. Pharmacoepidemiol. Drug Saf 21 Suppl 1, 23–31 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Collins FS & Tabak LA Policy: NIH plans to enhance reproducibility. Nature 505, 612–613 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodman SK & Krumholz HM Open science: PCORI’s efforts to make study results and data more widely available. <http://www.pcori.org/blog/open-science-pcoris-efforts-make-study-results-anddata-more-widely-available> (2015).
- 10.Laine C et al. Clinical trial registration-looking back and moving ahead. N. Engl. J. Med . 356, 2734–2736 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Lash TL & Vandenbroucke JP Should preregistration of epidemiologic study protocols become compulsory? Reflections and a counterproposal. Epidemiology 23, 184–188 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Williams RJ, Tse T, Harlan WR & Zarin DA Registration of observational studies: is it time? CMAJ 182, 1638–1642 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.ICH. The International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use. <http://www.ich.org/> (2014). Accessed 13 December 2014.
- 14.Altman DG et al. The revised CONSORT statement for reporting randomized trials: explanation and elaboration. Ann. Intern. Med . 134, 663–694 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Strom BL, Buyse M, Hughes J & Knoppers BM Data sharing, year 1-access to data from industry-sponsored clinical trials. N. Engl. J. Med 371, 2052–2054 (2014). [DOI] [PubMed] [Google Scholar]
- 16.European Medicines Agency. European Medicines Agency policy on publication of clinical data for medicinal products for human use. <http://www.ema.europa.eu/docs/en_GB/document_library/Other/2014/10/WC500174796.pdf> (2014). Accessed 20 December 2014.
- 17.von Elm E et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Int. J. Surg 12, 1495–1499 (2014). [DOI] [PubMed] [Google Scholar]
- 18.RECORD. REporting of studies Conducted using Observational Routinely-collected Data (RECORD). <http://www.record-statement.org/> (2014).
- 19.Hernan MA & Wilcox AJ Epidemiology, data sharing, and the challenge of scientific replication. Epidemiology 20, 167–168 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Meier CR, Schlienger RG, Kraenzlin ME, Schlegel B & Jick H HMG-CoA reductase inhibitors and the risk of fractures. JAMA 283, 3205–3210 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Smeeth L, Douglas I, Hall AJ, Hubbard R & Evans S Effect of statins on a wide range of health outcomes: a cohort study validated by comparison with randomized trials. Br. J. Clin. Pharmacol 67, 99– 109 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suissa S & Azoulay L Metformin and the risk of cancer: time-related biases in observational studies. Diabetes Care 35, 2665–2673 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schneeweiss S, Huybrechts KF & Gagne JJ Interpreting the quality of health care database studies on the comparative effectiveness of oral anticoagulants in routine care. J. Comp. Eff. Res 3, 33–41 (2013). [Google Scholar]
- 24.Overhage JM, Ryan PB, Reich CG, Hartzema AG & Stang PE Validation of a common data model for active safety surveillance research. J. Am. Med. Inform. Assoc . 19, 54–60 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Curtis LH, Brown J & Platt R Four health data networks illustrate the potential for a shared national multipurpose big-data network. Health Aff. (Millwood) 33, 1178–1186 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Open Science Collaboration. Psychology. Estimating the reproducibility of psychological science. Science 349, aac4716 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Ebrahim S et al. Reanalyses of randomized clinical trial data. JAMA 312, 1024–1032 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Robinson D, Schulz E, Brown P & Price C Updating the Read Codes: user-interactive maintenance of a dynamic clinical vocabulary. J. Am. Med. Inform. Assoc . 4, 465–472 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.ISPE. Guidelines for good pharmacoepidemiology practices (GPP). Pharmacoepidemiol. Drug Saf 17, 200–208 (2008). [DOI] [PubMed] [Google Scholar]
- 30.US Department of Health and Human Services Food and Drug Administration. Guidance for industry and FDA staff best practices for conducting and reporting pharmacoepidemiologic safety studies using electronic healthcare data sets. <http://www.rees-france.com/en/article.php3?id_article5506> (2013).
- 31.European Network of Centres for Pharmacoepidemiology and Pharmacovigilance. ENCePP Guide on Methodological Standards in Pharmacoepidemiology. <http://www.encepp.eu/standards_and_guidances/methodologicalGuide.shtml> (2014).
- 32.Johnson ML, Crown W, Martin BC, Dormuth CR & Siebert U Good research practices for comparative effectiveness research: analytic methods to improve causal inference from nonrandomized studies of treatment effects using secondary data sources: the ISPOR Good Research Practices for Retrospective Database Analysis Task Force Report--Part III. Value Health 12, 1062–1073 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Berger ML, Mamdani M, Atkins D & Johnson ML Good research practices for comparative effectiveness research: defining, reporting and interpreting nonrandomized studies of treatment effects using secondary data sources: the ISPOR Good Research Practices for Retrospective Database Analysis Task Force Report—Part I. Value Health 12, 1044–1052 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Langan SM et al. Setting the RECORD straight: developing a guideline for the REporting of studies Conducted using Observational Routinely collected Data. Clin. Epidemiol 5, 29–31 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gagne JJ, Wang SV, Rassen JA & Schneeweiss S A modular, prospective, semi-automated drug safety monitoring system for use in a distributed data environment. Pharmacoepidemiol. Drug Saf 23, 619–627 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gagne JJ et al. Active safety monitoring of newly marketed medications in a distributed data network: application of a semi-automated monitoring system. Clin. Pharmacol. Ther . 92, 80–86 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toh S, Baker MA, Brown JS, Kornegay C, Platt R & Mini-Sentinel Investigators. Rapid assessment of cardiovascular risk among users of smoking cessation drugs within the US Food and Drug Administration’s Mini-Sentinel program. JAMA Intern. Med 173, 817–819 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Toh S et al. Comparative risk for angioedema associated with the use of drugs that target the renin-angiotensin-aldosterone system. Arch. Intern. Med 172, 1582–1589 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Solomon DH et al. Relationship between selective cyclooxygenase-2 inhibitors and acute myocardial infarction in older adults. Circulation 109, 2068–2073 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Schneeweiss S, Rassen JA, Glynn RJ, Avorn J, Mogun H & Brookhart MA High-dimensional propensity score adjustment in studies of treatment effects using health care claims data. Epidemiology 20, 512–522 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.CMS 2008–2010 Data Entrepreneurs’ Synthetic Public Use File (DE-SynPUF). <https://www.cms.gov/Research-Statistics-Data-and-Systems/Downloadable-Public-Use-Files/SynPUFs/DE_Syn_PUF.html> (2014).
- 42.Centers for Medicare and Medicaid Services (CMS) Linkable 2008–2010 Medicare Data Entrepreneurs’ Synthetic Public Use File (DE-SynPUF). <https://www.cms.gov/Research-Statistics-Data-and-Systems/Downloadable-Public-Use-Files/SynPUFs/Downloads/SynPUF_Codebook.pdf> (2013).
- 43.Van Le H, Beach KJ, Powell G, Pattishall E, Ryan P & Mera RM Performance of a semi-automated approach for risk estimation using a common data model for longitudinal healthcare databases. Stat. Methods Med. Res 22, 97–112 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.