Abstract
The aberrant expression level of SARS-CoV-2 cell receptor gene ACE2 was reported in lung adenocarcinoma (LUAD) comorbidity of COVID-19. However, the association of ACE2 expression levels with immunosuppression and metabolic reprogramming in LUAD remains lacking. We investigated the expression level of ACE2, an association of ACE2 expression level with various types of immune signatures, immune ratios, and pathways. We employed a weighted gene co-expression network analysis (WGCNA) R package to identify the gene modules and investigated prognostic roles of hub genes in LUAD. Overexpression of ACE2 level was found in LUAD and ACE2 expression was negatively associated with various types of immune signatures including CD8+ T cells, CD4+ regulatory T cells, NK cells, and T cell activation. Besides, ACE2 upregulation was not only associated with CD8+ T cell/CD4+ regulatory T cell ratios but also linked with downregulation of immune-markers including CD8A, KLRC1, GZMA, GZMB, NKG7, CCL4, and IFNG. Moreover, the ACE2 expression level was found to be associated with the enrichment level of various metabolic pathways and it was also found that the metabolic pathways are directly positively correlated with the increased expression levels of ACE2, indicating that the overexpression of ACE2 is associated with metabolic reprogramming in LUAD. Furthermore, WGCNA based analysis revealed the gene modules in the high-ACE2-expression-level group of LUAD and identified GCLC and SLC7A11 hub genes which are not only highly expressed in lung adenocarcinoma but also correlated with the poor survival prognosis. Our analysis of ACE2 in LUAD tissues suggests that ACE2 is not only a receptor but is also associated with immunosuppression and metabolic reprogramming. This study underlines the clue for understanding the clinical significance of ACE2 in COVID-19 patients with LUAD comorbidity.
Keywords: SARS-CoV-2, ACE2, Lung adenocarcinoma comorbidity, Immunosuppression, Metabolic reprogramming, Immune-markers, Hub genes, Survival prognosis
1. Introduction
Coronavirus disease 2019 (COVID-19) arose from severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Among COVID-19 confirmed cases, patients with any comorbidity yielded poorer clinical outcomes and an increasing number of comorbidities also correlated with poorer clinical outcomes [1]. The mortality rate was high and correlated with general risk factors in COVID-19 patients with cancer comorbidity [2]. Recently, it was shown that lung cancer was the most frequent type of cancer in a COVID-19 cohort [3]. The SARS-CoV-2 induces extensive and aberrant non-effective host immune responses which are linked with crucially fetal severe lung damage through pro-inflammatory cytokines [4]. Clinical evidence indicated that pro-inflammatory cytokines mediated the severe pathogenesis of inflammatory-induced lung injury from different conditions including sepsis, pneumonia, aspiration, and shock [5]. Pro-inflammatory cytokines are also significantly correlated with lung injury in COVID-19 pneumonia [6] and chronic obstructive pulmonary disease (COPD) [7]. In lung injury, the inflammatory process of T cells, neutrophils, and macrophages is crucially driven by the classical proinflammatory cytokines including TNF-α, IFN-γ, IL-1, IL-6, IL-8, IL-18, and IL-32 [8]. Lung cancer patients are estimated with a high mortality rate worldwide [9] and lung adenocarcinoma (LUAD) is one of the major histological subtypes of lung cancer [10]. Angiotensin-converting enzyme 2 (ACE2) is a functional host cell receptor for SARS-related coronavirus (SARS-CoV) [11] and SARS-CoV-2 [12,13]. Recently it was stated that ACE2 expression level is elevated in LUAD [14], potentially demonstrating that LUAD patients are excessively and aberrantly susceptible to SARS-CoV-2 infection. It was found that ACE2 is higher in the lung metastases derived from different cancer subtypes than organ metastases of other sites [15]. It was stated that the protein levels of ACE2 were significantly upregulated in both alveolar tissue and bronchial epithelium of diabetic patients when compared with control, indicating the severity and susceptibility of COVID-19 in patients with diabetes. Zisman et al. revealed the higher expression of ACE2 in failing heart ventricles of patients with idiopathic dilated cardiomyopathy [16]. Lely et al. demonstrated that increased ACE2 expression was found in the glomerular and peritubular capillary endothelia in all primary renal diseases [17]. ACE2 expression level is correlated with immune infiltrations in uterine corpus endometrial carcinoma (UCEC) and kidney renal papillary cell carcinoma (KIRP) [18]. A direct metabolic link was also demonstrated to coronavirus infection and it is necessary to metabolic control in all patients with COVID-19 [19].
In this study, we aim to analyze the expression level of ACE2 in male, female, non-smoker, and smoker of lung adenocarcinoma tissue by computational analysis of gene expression profiling. We compared ACE2 expression levels across the TCGA-LUAD cohort combined with the GTEx database and gene expression omnibus (GEO) datasets. The TCGA-LUAD cohort (level-3 RNA-Seq data) containing 517 tumor samples and 59 adjacent normal samples (https://portal.gdc.cancer.gov/). Also, we analyzed the correlations between ACE2 expression levels and numerous types of immune signatures enrichment levels in LUAD. We identified the Kyoto encyclopedia of genes and genomes (KEGG) [20] pathways which are upregulated in the high expression group of ACE2 and low expression group of ACE2 in lung adenocarcinoma. The correlation of the single-sample gene set enrichment analysis (ssGSEA) score of an individual pathway and the expression level of ACE2 was evaluated. Moreover, we used weighted gene co-expression network analysis (WGCNA) [21] to identify the gene modules (gene ontology) that were differentially enriched between the high-ACE2-expression-level and the low- ACE2-expression-level tumors in LUAD. Finally, hub genes were identified specifically for the gene module and we evaluated the prognostic roles of screened hub genes in lung adenocarcinoma patients.
2. Materials and methods
2.1. Datasets
We used a webserver “gene expression profiling interactive analysis (GEPIA) 2” [22] (http://gepia2.cancer-pku.cn/#index) which contains expression profiling of TCGA-LUAD data with matched TCGA and GTEx [23] normal tissues. The lung adenocarcinoma gene expression profiling datasets were downloaded by searching the NCBI gene expression omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) using the keywords “lung adenocarcinoma”, and “lung cancer”, and identified six gene expression datasets: GSE40791 [24], GSE43458 [25], GSE19804 [26,27], GSE136043 [28], GSE118370 [29], and GSE13213 [30]. Besides, the lung adenocarcinoma TCGA-LUAD cohort was downloaded from the TCGA data portal (https://portal.gdc.cancer.gov/).
2.2. Differential expression of ACE2 in lung adenocarcinoma
We employed GEPIA 2 [22] (http://gepia2.cancer-pku.cn/#index) to identify the differential expression of ACE2 between cancer and normal tissues. GEPIA 2 contains TCGA-LUAD data with matched normal data and the GTEx database [23] of normal tissues. For GEO datasets, the differential expression of ACE2 was screened by using GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r) which is an interactive web tool. GEO2R tool based on the GEOquery and limma R packages from the Bioconductor project (http://www.bioconductor.org/). The thresholds of P-value < 0.05 and |log2FC| or |FC| (fold change) > 0.50 were set to find out significant level.
2.3. Evaluation of the immune signature enrichment levels
We identified the enrichment level of the immune signature in a tumor sample as the single-sample gene-set enrichment analysis (ssGSEA) score [31]. ssGSEA is an extension of GSEA which calculating the separate enrichment scores for each pairing of a sample and gene set. The gene set contains the collection of all marker genes of an immune signature. We included 9 immune signatures: B cell, CD8+ T cells, CD4+ regulatory T cells, NK cells, Tregs, T cell activation, cytolytic activity, MHC class 1, type 1 interferon (IFN), and type 2 IFN. The marker genes set of individual immune signatures are displayed in Supplementary Table S1.
2.4. Gene-set enrichment analysis
We conducted gene-set enrichment analysis (GSEA) of the TCGA-LUAD and GEO datasets by using GSEA (R implementation) [[32], [33], [34]]. Our analysis identified the KEGG [20] pathways which are upregulated in the high expression group of ACE2 and low expression group of ACE2 in lung adenocarcinoma (P < 0.05) (expression levels > median versus expression levels < median), respectively. The common pathways in both datasets were selected for calculating the correlations of pathway activities with the expression level of ACE2 in lung adenocarcinoma.
2.5. Correlation of pathway activities with the expression level of ACE2 in lung adenocarcinoma
We used gene sets that are included in the pathway for quantifying the ssGSEA score [31] to identify the activity of a pathway. The marker gene set pathways were displayed in Supplementary Table S2. The Spearman correlation of the ssGSEA score of the pathway and the expression level of ACE2 was used to evaluate the correlation of pathway activities in lung adenocarcinoma.
2.6. Evaluation of ACE2-associated networks in lung adenocarcinoma
We employed WGCNA [21] for identifying the gene modules (gene ontology) that were differentially enriched between the high-ACE2-expression-level and the low- ACE2-expression-level tumors in TCGA-LUAD and GSE40791 (expression levels > median versus expression levels < median), respectively. Hub genes were identified specifically for the gene module by using STRING v11 [35] and NetworkAnalyst [36] tools. The degree of interactions of hub genes with other genes not less than 5 and the minimum required interaction score is 0.40 was set in the STRING v11 tool.
2.7. Survival analysis of hub genes in PrognoScan and GEPIA databases
The prognostic roles of screened hub genes in lung adenocarcinoma patients were analyzed using the PrognoScan database (http://www.abren.net/PrognoScan/) [37] and GEPIA 2 databases [22]. We compared the survival of lung cancer patients classified based on gene expression levels of ACE2 (expression levels > median versus expression levels < median). Kaplan-Meier survival curves were utilized to prove the survival differences, and the log-rank test was used to evaluate the significance of survival differences. Cox P < 0.05 was considered as a significant level.
2.8. Statistical analysis
To evaluate the two variables, we employed Pearson's or Spearman's correlation test. For analyzing the correlations between the expression levels of ACE2 and the enrichment levels (ssGSEA scores) of immune signatures and pathways, we employed Spearman's correlation test because these data were not normally distributed. For analyzing the correlations between the expression levels of ACE2 with the expression levels of a single gene and the ratios of immune signatures, we utilized Pearson's correlation test because these data were normally distributed. For adjusting the multiple tests, the FDR was calculated by the Benjamini and Hochberg method [38]. We used the Student's t-test (P < 0.05 and |FC| > 0.30) for differential expression levels of the immune marker genes between the high expression group of ACE2 (HEA) and low expression group of ACE2 (LEA). For GEO datasets, we used NetworkAnalyst [36] tool for calculating the average expression for the genes having multiple probes.
3. Results
3.1. Over expression of ACE2 in male, female, non-smoker, and smoker of lung adenocarcinoma tissue
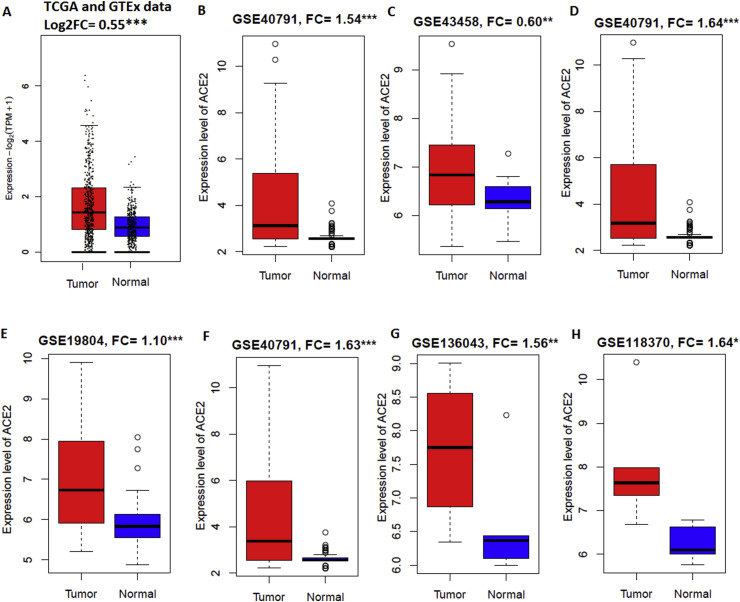
ACE2 is upregulated in various groups of lung adenocarcinoma patients including male (FC = 1.63, P < 0.001), female (FC = 1.10, P < 0.001), non-smoker (FC = 0.60, P < 0.01), and smoker (FC = 1.64, P < 0.001) when compared with respective normal samples (Fig. 1 ). Over expression of ACE2 was found in lung adenocarcinoma of TCGA combined with GTEx data (FC = 0.55, P < 0.001) (Fig. 1A), microarray data (FC = 1.54, P < 0.001) (GSE40791, Fig. 1B), never-smoker (FC = 0.60, P < 0.01) (GSE43458, Fig. 1C), ever or current smoker (FC = 1.64, P < 0.001) (GSE40791, Fig. 1D), female (FC = 1.10, P < 0.001) (GSE19804, Fig. 1E), and male (FC = 1.63, P < 0.001) (GSE40791, Fig. 1F) pateints. In addition, ACE2 is overexpressed (FC = 1.56, P < 0.01) in fresh lung adenocarcinoma tissue specimens (collected immediately after surgical resection) when compared to non-tumor lung tissue (GSE136043, Fig. 1G). In invasive lung adenocarcinoma, elevated expression level of ACE2 was also found when compared with paired normal lung tissues (FC = 1.64, P < 0.05) (GSE118370, Fig. 1H). Upregulation of ACE2 has been demonstrated in LUAD and patients with upregulated ACE2 expression in lung cancer are susceptible to SARS-CoV-2 infection [14,39].
Fig. 1.
ACE2 is upregulated in lung adenocarcinoma patients. A.ACE2 is upregulated in combined TCGA-LUAD and GTEx data. B.ACE2 is upregulated in lung adenocarcinoma of microarray data. C.ACE2 is upregulated when compared to normal lung tissue were paired to the never-smoker lung adenocarcinoma cases. D. Upregulation of ACE2 in ever or current smoking lung adenocarcinoma. E. Genome-wide screening of transcriptional modulation in female lung cancer showed ACE2 is overexpressed. F. Overexpression of ACE2 in male lung adenocarcinoma. G.ACE2 is upregulated in fresh lung adenocarcinoma tissue specimens when compared to non-tumor lung tissue. H.ACE2 is upregulated also in invasive lung adenocarcinoma tissue.*P < 0.05. **P < 0.01, and ***P < 0.001.
3.2. Association of ACE2 expression level with immune signatures in lung adenocarcinoma
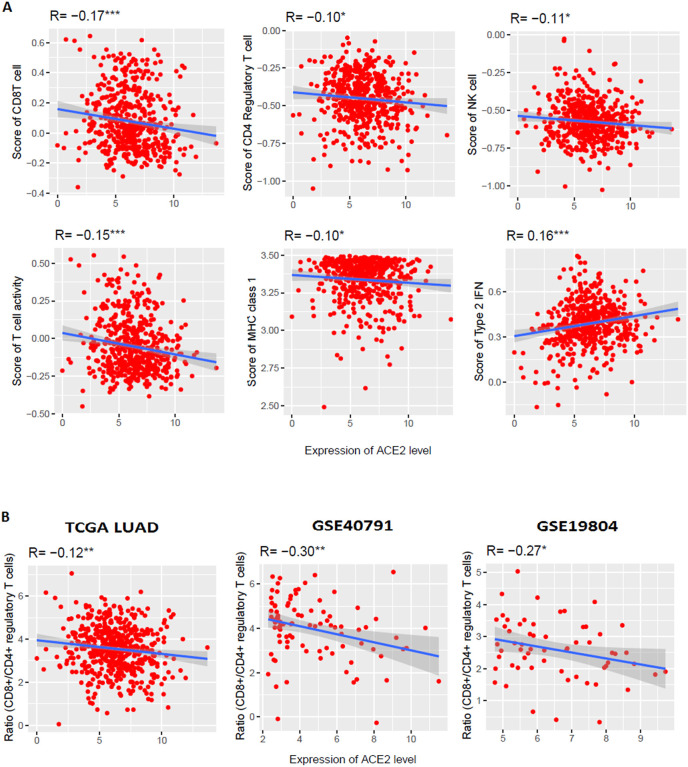
In the TCGA-LUAD cohort, we revealed significant negative correlations of the ACE2 expression levels with the enrichment levels (ssGSEA scores) of CD8+ T cells, CD4+ regulatory T cells, NK cells, T cell activation, and MHC class 1 (Spearman's correlation test, P < 0.05) (Fig. 2 A). Cytolytic activity enrichment score also showed a negative correlation with the expression of ACE2 level (Spearman's correlation test, R = −0.08, P = 0.08). However, type 2 IFN response enrichment level (ssGSEA scores) is positively correlated with the ACE2 expression levels (Spearman's correlation test, P < 0.05) (Fig. 2A). B cell, Tregs, and Type 1 IFN enrichment score is not significantly correlated with the expression level of ACE2. We further investigated the association of ACE2 expression levels with various types of immune signatures in an independent lung adenocarcinoma dataset (GSE40791). In GSE40791 (n = 194), expression level of ACE2 is negatively correlated with the enrichment levels of CD8+ T cells (R = −0.30, P < 0.01), CD4+ regulatory T cells (R = −0.28, P < 0.01), NK cells (R = −0.23, P < 0.05), T cell activation (R = −0.29, P < 0.01), cytolytic activity (R = −0.25, P < 0.05), MHC class 1 (R = −0.17, P < 0.1) (Spearman correlation test) (Supplementary Fig. 1). These results strongly suggest that ACE2 expression level is negatively associated with CD8+ T cells, CD4+ regulatory T cells, NK cells, T cell activation, cytolytic activity, MHC class 1 in lung adenocarcinoma. Lung cancer progression is associated with increased T cell dysfunctions [40]. CD8+ T cells and CD4+ T cells are significantly lower in COVID-19 patients [41]. In lung adenocarcinoma lesions, NK cells were the least abundant immune cell lineage [42]. Strikingly, we found that the ratios of CD8+/CD4+ regulatory T cells had significant negative correlations with the expression level of ACE2 in the TCGA-LUAD cohort and two other GEO datasets (GSE40791 and GSE19804) respectively (Pearson's correlation test, P < 0.05) (Fig. 2B). Altogether, these results suggest that the elevated expression of ACE2 is associated with the immunosuppressive roles in lung adenocarcinoma.
Fig. 2.
The elevated expression level of ACE2 is associated with immune signatures. A. Significant correlation of ACE2 expression levels with immune signatures in lung adenocarcinoma (TCGA data of LUAD). ACE2 expression is negatively correlated with the ssGSEA score of CD8+ T cells, CD4+ regulatory T cell, NK cell, T cell activity, and MHC class 1 and positively correlated with type 2 IFN ssGSEA score (Spearman's correlation test, P < 0.05). B. Significant negative correlation of ACE2 expression levels with CD8+ T cell/CD4+ regulatory T cell ratios in TCGA data of LUAD and two GEO datasets (GSE40791 and GSE19804) (Pearson's correlation test, P < 0.05). The ratio of CD8+ T cell/CD4+ regulatory T cell in a tumor sample is defined as the ratio of average expression levels of their marker genes (log2-transformed for TCGA data). *P < 0.05. **P < 0.01, and ***P < 0.001.
3.3. Identification of differentially expressed immune marker genes between high expression group of ACE2 and low expression group of ACE2 in lung adenocarcinoma
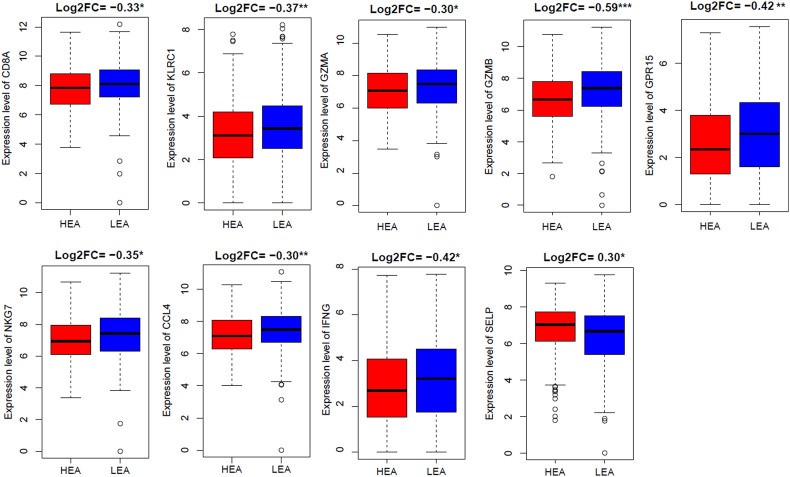
We divided the TCGA-LUAD (n = 517) sample into high expression (expression levels > median) group of ACE2 (HEA) and low expression (expression levels < median) group of ACE2 (LEA) bassed on ACE2 expression level. We chose marker genes (Supplementary Table S1) of significant immune signatures used earlier in this study. Differential analysis of marker genes revealed that CD8A (CD8+ T cells marker), KLRC1 (NK cells marker), GZMA (cytolytic activity and T cell activation marker), GZMB (T cell activation marker), GPR15 (CD4+ regulatory T cells marker), NKG7 (T cell activation marker), CCL4 (T cell activation marker), and IFNG (T cell activation marker) genes are downregulated in HEA (Fig. 3 ). Only type 2 IFN marker SELP is upregulated in LEA (Fig. 3) (Student's t-test, P < 0.05). We validated the expression of these marker genes in an independent GEO dataset (GSE40791) of lung adenocarcinoma. Strikingly, we revealed that the expression of CD8A, KLRC1, GZMA, GZMB, NKG7, CCL4, and IFNG immune marker genes are negatively correlated with the expression level of ACE2 in lung adenocarcinoma of the GSE40791 dataset (Pearson's correlation test, P < 0.05) (Supplementary Fig. S2). In contrast, GPR15 and SELP marker genes had no significant correlation with the expression level of ACE2 in the tumor tissue of GSE40791. These data indicate that the upregulation of ACE2 expression is associated with the downregulation of immune markers including CD8A, KLRC1, GZMA, GZMB, NKG7, CCL4, and IFNG in lung adenocarcinoma tissues. CD8A and GZMA genes were significantly elevated in a cluster that is enriched in lung adenocarcinoma [43]. Increased immunotherapy response is associated with the expression of CCL4 [44]. In the tumor microenvironment of non-small cell lung cancer, decreased IFNG is linked with the stemness of tumor cells [45]. Taken together, the expression level of ACE2 is inversely associated with the anti-tumor immunity in lung adenocarcinoma.
Fig. 3.
Comparative expression levels of the immune marker genes between high expression group of ACE2 (HEA) and low expression group of ACE2 (LEA) in TCGA lung adenocarcinoma. (Student's t-test, *P < 0.05, **P < 0.01, and ***P < 0.001). FC: Fold change.
3.4. The expression level of ACE2 is associated with the enrichment of pathways
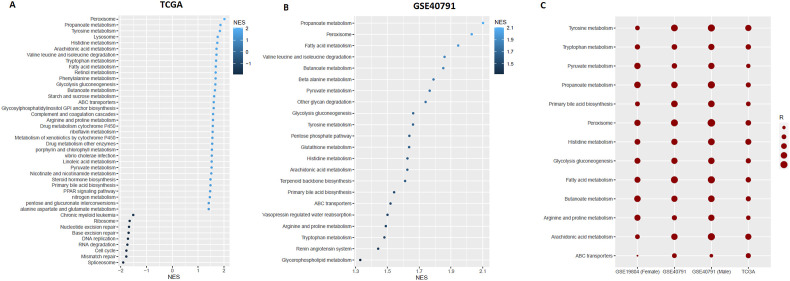
GSEA revealed several KEGG pathways significantly enriched in the high expression group of ACE2 (expression levels > median) and low expression (expression levels < median) group of ACE2 in lung adenocarcinoma (P < 0.05) (Fig. 4 A and B). In TCGA-LUAD and GSE40791 cohorts, the metabolic pathways were highly active in the high expression group of ACE2 (Fig. 4A and B). In the TCGA-LUAD cohort, the active pathways were cell cycle, DNA replication, mismatch repair, spliceosome, base excision repair, RNA degradation, nucleotide excision repair, ribosome, and chronic myeloid leukemia in the low expression group of ACE2 (Fig. 4A) but active pathways were not found in low expression group of ACE2 in GSE40791. The commonly found active pathways in the high expression group of ACE2 were identified in both datasets (Between TCGA-LUAD and GSE40791 cohorts). Interestingly, we identified commonly active 14 pathways in the high expression group of ACE2 including glycolysis gluconeogenesis, fatty acid metabolism, peroxisome, tyrosine metabolism, histidine metabolism, arachidonic acid metabolism, tryptophan metabolism, propanoate metabolism, ABC transporters, arginine, and proline metabolism, valine leucine and isoleucine degradation, butanoate metabolism, primary bile acid biosynthesis, and pyruvate metabolism. This result confirmed the increased metabolic pathway activity in the high expression group of ACE2. Furthermore, we calculated the ssGSEA score of all 14 overlapping pathways across TCGA-LUAD, GSE40791, GSE19804 (Female), and GSE40791 (Male) datasets. Thirteen pathways upregulated in the high expression group of ACE2 are positively correlated (Spearman's correlation test, P < 0.05) with the expression of ACE2 across TCGA-LUAD and GSE40791 data. Also, 12 pathways (except ABC transporters) are also positively correlated with the elevated expression of ACE2 in GSE19804 (Female) and GSE40791 (Male) datasets (Spearman's correlation test, P < 0.05) (Fig. 4C). Dysregulation of energy metabolism and immune evasion are two emerging hallmarks of cancer [46]. Malignant cells reprogram their metabolism and energy production for supporting cell proliferation and survival [47]. Increased glycolytic rate is essential for supporting the cancer cell growth, survival, proliferation, and long-term maintenance [48]. Cancer cells reprogram fatty acid metabolism which is essential for cancer progression, metastasis, and remodeling of the tumor microenvironment [49]. Amino acid metabolism is a crucial target for cancer therapy [50].
Fig. 4.
GSEA (R implementation) based identification of ACE2 expression-specific pathways. A. KEGG pathways enriched in TCGA-LUAD (NES>0 is enriched in high expression group and NES<0 is enriched in low expression group). B. KEGG pathways enriched in GSE40791 (No pathways found in low expression group of ACE2) (P < 0.05). C. Correlation between the pathway enrichment score (ssGSEA) and the expression level of ACE2. upregulated overlapping 13 pathways have direct positive correlations with the expression of ACE2 across the datasets (Spearman's correlation test, P < 0.05).
3.5. ACE2-expression-specific correlated gene signatures are associated with the enrichment of pathways in lung adenocarcinoma
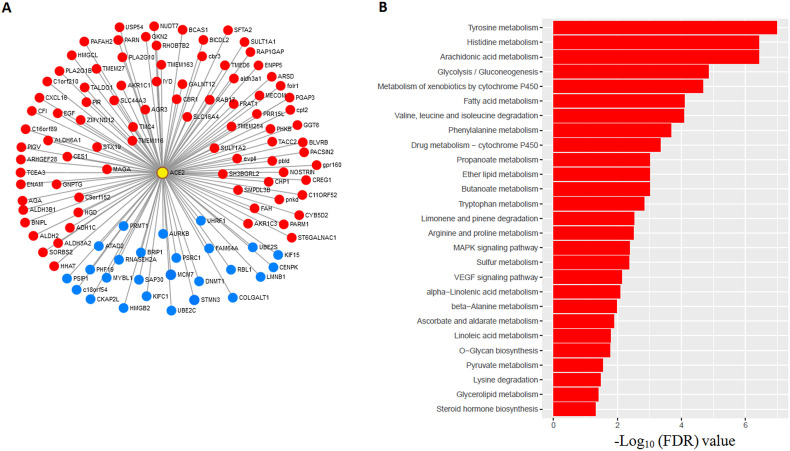
We used the TCGA-LUAD cohort and GSE40791 dataset to identify the correlation (Pearson correlation, R > 0.30, FDR<0.05) between the expression level of ACE2 and all other genes. We found the positively correlated common 87 gene signatures (red indicated in Fig. 5 A) including two transcription factors CREG1 and MECOM between TCGA and GSE40791 (Pearson's correlation test, FDR <0.05, R > 0.3). CREG1 has a potential role in cell proliferation and migration in NSCLC cells [51]. MECOM is significantly amplified in lung adenocarcinoma [10]. Finally, 87 positively correlated genes were inputted into GSEA to identify the pathways. We found 27 KEGG pathways which are mostly associated with metabolism (FDR < 0.05) (Fig. 5B). Interestingly, we got overlapping 11 pathways (fatty acid metabolism, tyrosine metabolism, histidine metabolism, arachidonic acid metabolism, tryptophan metabolism, propanoate metabolism, glycolysis gluconeogenesis, arginine and proline metabolism, valine leucine and isoleucine degradation, butanoate metabolism, and pyruvate metabolism) which are also actively enriched in high expression group ACE2 (Fig. 4A–C). In contrast, we also found negatively correlated common 26 gene signatures (blue indicated in Fig. 5A) including six transcription factors HMGB2, MYBL1, PSIP1, RBL1, SAP30, and UHRF1 between TCGA-LUAD and GSE40791 cohorts (Pearson's correlation test, FDR < 0.05, R < −0.3). In a human lung cancer cell, the HMGB2 gene increased cisplatin sensitivity [52]. Significant pathways were not identified by GSEA for negatively correlated 26 gene signatures. This result further confirmed the association of ACE2 expression level with metabolic pathways in lung adenocarcinoma tissue.
Fig. 5.
Correlated gene signatures with ACE2 and their significant association with pathway enrichment (FDR<0.05) A. 87 (in red) and 26 (in blue) genes showing significant positive and negative expression correlations with ACE2 (yellow circled with red) consistently between TCGA (LUAD) and GSE40791, respectively (Pearson's correlation test, FDR < 0.05, |R| > 0.3). B. Involvement of 87 (in red) positively correlated gene signatures with the enrichment of KEGG pathways. These positively correlated gene signatures were mainly involved with metabolism-associated pathways (FDR<0.05).
3.6. Identification of ACE2 expression specific interaction networks in lung adenocarcinoma
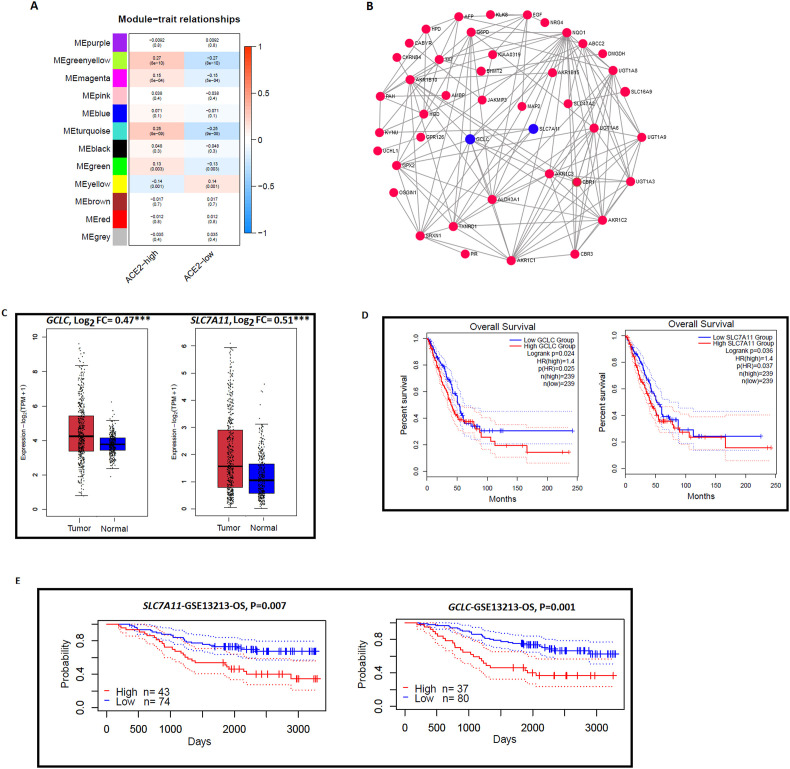
We used the TCGA-LUAD cohort and GSE40791 dataset to identify the ACE2 expression specific interaction networks modules. In the TCGA-LUAD cohort, WGCNA identified four gene modules (green-yellow, turquoise, magenta, and green) that were more highly enriched in the high-ACE2-expression-level than in the low- ACE2-expression-level tumors (Fig. 6 A). On the other hand, only one gene module (indicated yellow) was found to be more highly enriched in the low-ACE2-expression-level tumors (Fig. 6A). The GO terms in the highly enriched green-yellow module are oxidoreductase activity, small molecule metabolic process, and alcohol dehydrogenase (NADP+) activity. In addition, we also identified gene modules in GSE40791 (Supplementary Fig. S3 A). WGCNA identified only one gene modules (pink) that was more highly enriched in the high-ACE2-expression-level than in the low- ACE2-expression-level tumors and three gene modules (turquoise, green, and brown) that were found to be more highly enriched in the low-ACE2-expression-level tumors (Supplementary Fig. S3 A). The GO terms in the highly enriched pink modules are carboxylic acid metabolic process, oxoacid metabolic process, oxidation-reduction process, organic acid metabolic process, small molecule metabolic process, oxidoreductase activity, monocarboxylic acid metabolic process, and catalytic activity. Interestingly, two GO terms oxidoreductase activity and small molecule metabolic process are commonly found in the highly enriched module of high-ACE2-expression-level tumor of both datasets (TCGA-LUAD and GSE40791). In contrast, no commonly enriched GO term was found in the module of low-ACE2-expression-level tumor of both datasets (TCGA-LUAD and GSE40791). It indicates that ACE2 expression is associated with a metabolic process in lung adenocarcinoma.
Fig. 6.
Module coexpression networks of ACE2 in lung adenocarcinoma. A. WGCNA based identification of gene modules (gene ontology) more highly enriched in high-ACE2-expression-level and low-ACE2-expression-level in TCGALUAD cohorts. B. A subnetwork of the green-yellow gene module active in the high-ACE2-expression-level group of TCGA-LUAD. GCLC and SLC7A11 (indicated blue color) hub genes are associated with survival prognosis. C.GCLC and SLC7A11 hub genes are significantly highly expressed in GTEx combined with TCGA-LUAD cohorts. D. Kaplan-Meier curves shows that the elevated expression of GCLC and SLC7A11 hub genes are associated with a worse overall survival (OS) prognosis in TCGA-LUAD cohorts by GEPIA 2 (log-rank test, P < 0.05). E. Kaplan-Meier curves also show that GCLC and SLC7A11 hub genes are associated with shorter survival prognosis (OS) in GSE13213 dataset by Prognoscan (log-rank test, P < 0.05).
From the green-yellow gene module (Fig. 6A) of the TCGA-LUAD cohort, we identified 24 hub genes (Supplementary Table S3). In addition, we also identified 37 hub genes (Supplementary Table S4) in the pink module (Supplementary Fig. S3 A) of GSE40791. Interestingly, we found common 14 hub genes (NQO1, EGF, CBR1, CBR3, AKR1B10, AKR1C1, AKR1C2, AKR1C3, ALDH3A1, G6PD, GCLC, GPX2, SLC7A11, and SRXN1) between TCGA-LUAD and GSE40791 datasets. These hub genes were mainly involved in metabolism-related pathways (KEGG, FDR<0.001) identified by GSEA, including arachidonic acid metabolism (AKR1C3, AKR1C1, CBR1, and CBR3), metabolism of xenobiotics by cytochrome P450 (AKR1C1, AKR1C2, AKR1C3, and ALDH3A1), glutathione metabolism (G6PD, GCLC, and GPX2), and steroid hormone biosynthesis (AKR1C1, AKR1C2, and AKR1C3). A subnetwork (Fig. 6B) from the green-yellow gene module identified 14 hub genes including GCLC and SLC7A11 (indicated blue) which are not only associated with higher expression in lung adenocarcinoma but also linked with the survival prognosis in LUAD patients (Fig. 6C–E). Interestingly, we found that GCLC and SLC7A11 expression levels are higher in TCGA-LUAD when compared with TCGA and GTEx combined normal lung tissue (Fig. 6C). Furthermore, overexpression of GCLC (FC = 0.96, P < 0.001) and SLC7A11 (FC = 2.44, P < 0.001) were also found in lung adenocarcinoma tissues of GSE40791 dataset (two-sided Student's t-test) (Supplementary Fig. S3 B). Survival analysis using GEPIA 2 revealed that GCLC and SLC7A11 hub genes are associated with shorter overall survival (OS) prognosis in the TCGA-LUAD cohort (Fig. 6D). Moreover, PrognoScan revealed the elevated expression of GCLC and SLC7A11 had a negative correlation with survival prognosis (OS) in an independent GSE13213 dataset (Fig. 6E). In lung adenocarcinoma, high expression of GCLC is a potential predictor of treatment failure through cisplatin resistance [53]. Overexpression of SLC7A11 was found in lung cancer cells for satisfying the metabolic requirements of cancer cell growth and survival [54].
4. Discussion
Several previous studies have identified the aberrant expression of ACE2 in lung adenocarcinoma based on gene expression profiling data [14,39]. Zhang et al. observed the higher expression of ACE2 in LUAD and LUSC and also predicting prognosis in these two common lung cancer types [55]. Samad et al. revealed that ACE2 is highly expressed in lung adenocarcinoma (LUAD) and lung squamous carcinoma (LUSC) and this is associated with poor lung cancer outcomes [56]. We analyzed the expression level of ACE2 in male, female, non-smoker, and smoker of lung adenocarcinoma tissue. Our analysis revealed that elevated expression level of ACE2 was found in lung adenocarcinoma of male, female, non-smoker, and smoker patients (Fig. 1). In addition, ACE2 is upregulated in fresh lung adenocarcinoma tissue specimens when compared to non-tumor lung tissue (Fig. 1G), and also overexpressed in invasive lung adenocarcinoma tissue (Fig. 1H). Lung cancer patients are more susceptible to SARS-CoV-2 infection than normal individuals [3]. Since ACE2 is upregulated in lung cancer patients, Qi Kong et al. verified the susceptibility of lung cancer patients in each age stage, subtype, and pathological stage to SARS-CoV-2 infection [14]. Zhang et al. demonstrated that the expression of SARS-CoV-2 entry gene ACE2 reveals the susceptibility of COVID-19 in lung cancer patients [55]. Altogether, it is rational that male, female, non-smoker, and smoker patients of lung adenocarcinoma are susceptible to the risk of SARS-CoV-2 infection.
Our results revealed that ACE2 expression level is negatively correlated with the ssGSEA score of CD8+ T cells, CD4+ regulatory T cell, NK cell, T cell activity (Fig. 2A and Supplementary Fig. S1). In lung tissues, CD8+ T cells and NK cells are significantly inversely correlated with the expression levels of SARS-CoV-2 entry receptor ACE2 [57], indicating the increasing sensitivity of SARS-CoV-2 infection. It was stated that the chronic presence of lung tumor is associated with CD8+ T cell dysfunctions, sensitizes them to programmed cell death, and poor clinical responses in immunotherapeutic trials [58]. A significant decrease of both CD8+ T cells and CD4+ regulatory T cell were observed in the onset of SARS and both of the immune cells rapidly loss during the acute phase of SARS [59]. In lung cancer, the potentiality of NK cells is reduced that ultimately limited the protective role of NK cells against tumors in the early stage of lung cancer [60]. Deficiencies of NK cells are associated with increased susceptibility to viruses [61] and human NK cells are essential to clear virally infected cells [62]. A decreased amount of CD8+ T cells, CD4+ regulatory T cells, and NK cells in lung adenocarcinoma indicated the increased susceptibility of patients to the SARS-CoV-2 infection. Besides, we found a significant negative correlation of ACE2 expression levels with CD8+ T cell/CD4+ regulatory T cell ratios (Fig. 2B). Moreover, CD8A, KLRC1, GZMA, GZMB, NKG7, CCL4, and IFNG immune markers have an inverse association with the expression level of ACE2 in lung adenocarcinoma tissues (Fig. 3 and Supplementary Fig. S2). Taken together, our results underline that the expression of ACE2 is associated with viral spread and immunopathology in lung adenocarcinoma.
Cancer-associated metabolic reprogramming is linked with gene expression, cellular differentiation, and tumor microenvironment [63]. Since ACE2 expression is directly correlated with dysregulation of the metabolic pathways (Fig. 4A–C), higher expression of ACE2 has profound effects on tumorigenesis of lung adenocarcinoma through metabolic reprogramming. WGCNA based GO identification in green-yellow module further proves the association of ACE2 with metabolic process in lung adenocarcinoma (Fig. 6A). For replication and spreading, the virus induces host metabolic pathways including glycolysis, fatty acid synthesis, and/or glutaminolysis [64]. Metabolic control of glucose and lipid levels are key factors in COVID-19 patients [19]. Altogether, the high expression level of ACE2 is not only associated with tumorigenesis but also linked with SARS-CoV-2 replication and spreading through inducing the metabolic pathways in lung adenocarcinoma.
This study has identified the role of ACE2 in lung adenocarcinoma diagnosis and prognosis and may provide therapeutic targets for lung adenocarcinoma. On the other hand, this study also proved that lung adenocarcinoma patients are more susceptible to SARS-CoV-2 infections than normal populations. This study has some drawbacks. Our generated results from the bioinformatics analysis need to be verified by clinical and experimental conditions. However, to translate these findings into clinical application, further experimental and clinical validation would be warranted.
5. Conclusions
Overexpression of ACE2 level was negatively associated with various types of immune signatures and immune ratios. The expression level of ACE2 was found to be associated with the enrichment level of various metabolic pathways and it was also found that the metabolic pathways are directly positively correlated with the increased expression levels of ACE2, indicating that the overexpression of ACE2 is associated with metabolic reprogramming in LUAD. Our findings may provide new insights into the roles of ACE2 in immunosuppression and metabolic reprogramming of LUAD tissues and the clinical significance of ACE2 in COVID-19 patients with LUAD comorbidities. Future studies warranted for the inhibition of ACE2 in LUAD tissues as a therapeutic approach should be carefully evaluated.
6. Availability of data and materials
The lung adenocarcinoma TCGA-LUAD cohort was downloaded from the TCGA data portal (https://portal.gdc.cancer.gov/). In addition, the lung adenocarcinoma gene expression profiling datasets were downloaded from the NCBI gene expression omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/).
Author contributions
MNU conceived the research, designed analysis strategies, performed data analyses, and wrote the manuscript. RA proof-read the whole manuscript to eliminate grammatical errors. ML and ZA performed data analyses and helped for preparing the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cbi.2021.109370.
Abbreviations
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- ACE2
Angiotensin-converting enzyme 2
- LUAD
lung adenocarcinoma
- COVID-19
Coronavirus disease 2019
- WGCNA
Weighted gene co-expression network analysis
- GTEx
Genotype-tissue expression
- TCGA
The cancer genome atlas
- GEO
Gene expression omnibus
- KEGG
Kyoto encyclopedia of genes and genomes
- ssGSEA
Single-sample gene set enrichment analysis
- SARS-CoV
SARS-related coronavirus
- GEPIA
Expression profiling interactive analysis
- GSEA
Gene-set enrichment analysis
- NK
Natural killer
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Guan W.-J., Liang W.-H., Zhao Y., Liang H.-R., Chen Z.-S., Li Y.-M., Liu X.-Q., Chen R.-C., Tang C.-L., Wang T., Ou C.-Q., Li L., Chen P.-Y., Sang L., Wang W., Li J.-F., Li C.-C., Ou L.-M., Cheng B., Xiong S., Ni Z.-Y., Xiang J., Hu Y., Liu L., Shan H., Lei C.-L., Peng Y.-X., Wei L., Liu Y., Hu Y.-H., Peng P., Wang J.-M., Liu J.-Y., Chen Z., Li G., Zheng Z.-J., Qiu S.-Q., Luo J., Ye C.-J., Zhu S.-Y., Cheng L.-L., Ye F., Li S.-Y., Zheng J.-P., Zhang N.-F., Zhong N.-S., He J.-X. China medical treatment expert group for COVID-19, comorbidity and its impact on 1590 patients with COVID-19 in China: a nationwide analysis. Eur. Respir. J. 2020;55 doi: 10.1183/13993003.00547-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuderer N.M., Choueiri T.K., Shah D.P., Shyr Y., Rubinstein S.M., Rivera D.R., Shete S., Hsu C.-Y., Desai A., de Lima Lopes G., Grivas P., Painter C.A., Peters S., Thompson M.A., Bakouny Z., Batist G., Bekaii-Saab T., Bilen M.A., Bouganim N., Larroya M.B., Castellano D., Del Prete S.A., Doroshow D.B., Egan P.C., Elkrief A., Farmakiotis D., Flora D., Galsky M.D., Glover M.J., Griffiths E.A., Gulati A.P., Gupta S., Hafez N., Halfdanarson T.R., Hawley J.E., Hsu E., Kasi A., Khaki A.R., Lemmon C.A., Lewis C., Logan B., Masters T., McKay R.R., Mesa R.A., Morgans A.K., Mulcahy M.F., Panagiotou O.A., Peddi P., Pennell N.A., Reynolds K., Rosen L.R., Rosovsky R., Salazar M., Schmidt A., Shah S.A., Shaya J.A., Steinharter J., Stockerl-Goldstein K.E., Subbiah S., Vinh D.C., Wehbe F.H., Weissmann L.B., Wu J.T.-Y., Wulff-Burchfield E., Xie Z., Yeh A., Yu P.P., Zhou A.Y., Zubiri L., Mishra S., Lyman G.H., Rini B.I., Warner J.L. COVID-19 and Cancer Consortium, Clinical impact of COVID-19 on patients with cancer (CCC19): a cohort study. Lancet. 2020;395 doi: 10.1016/S0140-6736(20)31187-9. 1907–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liang W., Guan W., Chen R., Wang W., Li J., Xu K., Li C., Ai Q., Lu W., Liang H., Li S., He J. Cancer patients in SARS-CoV-2 infection: a nationwide analysis in China. Lancet Oncol. 2020;21:335–337. doi: 10.1016/S1470-2045(20)30096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Addeo A., Obeid M., Friedlaender A. COVID-19 and lung cancer: risks, mechanisms and treatment interactions. J Immunother Cancer. 2020;8 doi: 10.1136/jitc-2020-000892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodman R.B., Pugin J., Lee J.S., Matthay M.A. Cytokine-mediated inflammation in acute lung injury. Cytokine Growth Factor Rev. 2003;14:523–535. doi: 10.1016/s1359-6101(03)00059-5. [DOI] [PubMed] [Google Scholar]
- 6.Chen L.-D., Zhang Z.-Y., Wei X.-J., Cai Y.-Q., Yao W.-Z., Wang M.-H., Huang Q.-F., Zhang X.-B. Association between cytokine profiles and lung injury in COVID-19 pneumonia. Respir. Res. 2020;21:201. doi: 10.1186/s12931-020-01465-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du Y., Yang C., Li B., Wu X., Lv Y., Jin H., Cao Y., Sun J., Luo Q., Gong W., Zhang H., Liu B., Wu J.-F., Dong J. Association of pro-inflammatory cytokines, cortisol and depression in patients with chronic obstructive pulmonary disease. Psychoneuroendocrinology. 2014;46:141–152. doi: 10.1016/j.psyneuen.2014.04.020. [DOI] [PubMed] [Google Scholar]
- 8.Atamas S.P., Chapoval S.P., Keegan A.D. Cytokines in chronic respiratory diseases. F1000 Biol Rep. 2013;5 doi: 10.3410/B5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong Q., Xiang Z., Wu Y., Gu Y., Guo J., Geng F. Analysis of the susceptibility of lung cancer patients to SARS-CoV-2 infection. Mol. Canc. 2020;19:80. doi: 10.1186/s12943-020-01209-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Subbarayan K., Ulagappan K., Wickenhauser C., Seliger B. Expression and clinical significance of SARS-CoV-2 human targets in neoplastic and non-neoplastic lung tissues. Curr. Cancer Drug Targets. 2020 doi: 10.2174/1568009620666201207145019. [DOI] [PubMed] [Google Scholar]
- 16.Zisman L.S., Keller R.S., Weaver B., Lin Q., Speth R., Bristow M.R., Canver C.C. Increased angiotensin-(1-7)-forming activity in failing human heart ventricles: evidence for upregulation of the angiotensin-converting enzyme Homologue ACE2. Circulation. 2003;108:1707–1712. doi: 10.1161/01.CIR.0000094734.67990.99. [DOI] [PubMed] [Google Scholar]
- 17.Lely A.T., Hamming I., van Goor H., Navis G.J. Renal ACE2 expression in human kidney disease. J. Pathol. 2004;204:587–593. doi: 10.1002/path.1670. [DOI] [PubMed] [Google Scholar]
- 18.Yang J., Li H., Hu S., Zhou Y. ACE2 correlated with immune infiltration serves as a prognostic biomarker in endometrial carcinoma and renal papillary cell carcinoma: implication for COVID-19. Aging (N Y) 2020;12:6518–6535. doi: 10.18632/aging.103100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bornstein S.R., Dalan R., Hopkins D., Mingrone G., Boehm B.O. Endocrine and metabolic link to coronavirus infection. Nat. Rev. Endocrinol. 2020:1–2. doi: 10.1038/s41574-020-0353-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45 doi: 10.1093/nar/gkx247. W98–W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.GTEx Consortium The genotype-tissue expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y., Foreman O., Wigle D.A., Kosari F., Vasmatzis G., Salisbury J.L., van Deursen J., Galardy P.J. USP44 regulates centrosome positioning to prevent aneuploidy and suppress tumorigenesis. J. Clin. Invest. 2012;122:4362–4374. doi: 10.1172/JCI63084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kabbout M., Garcia M.M., Fujimoto J., Liu D.D., Woods D., Chow C.-W., Mendoza G., Momin A.A., James B.P., Solis L., Behrens C., Lee J.J., Wistuba I.I., Kadara H. ETS2 mediated tumor suppressive function and MET oncogene inhibition in human non-small cell lung cancer. Clin. Canc. Res. 2013;19:3383–3395. doi: 10.1158/1078-0432.CCR-13-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu T.-P., Tsai M.-H., Lee J.-M., Hsu C.-P., Chen P.-C., Lin C.-W., Shih J.-Y., Yang P.-C., Hsiao C.K., Lai L.-C., Chuang E.Y. Identification of a novel biomarker, SEMA5A, for non-small cell lung carcinoma in nonsmoking women. Cancer Epidemiol. Biomark. Prev. 2010;19:2590–2597. doi: 10.1158/1055-9965.EPI-10-0332. [DOI] [PubMed] [Google Scholar]
- 27.Lu T.-P., Hsiao C.K., Lai L.-C., Tsai M.-H., Hsu C.-P., Lee J.-M., Chuang E.Y. Identification of regulatory SNPs associated with genetic modifications in lung adenocarcinoma. BMC Res. Notes. 2015;8:92. doi: 10.1186/s13104-015-1053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang N., Zou C., Zhu Y., Luo Y., Chen L., Lei Y., Tang K., Sun Y., Zhang W., Li S., He Q., Zhou J., Chen Y., Luo J., Jiang W., Ke Z. HIF-1ɑ-regulated miR-1275 maintains stem cell-like phenotypes and promotes the progression of LUAD by simultaneously activating Wnt/β-catenin and Notch signaling. Theranostics. 2020;10:2553–2570. doi: 10.7150/thno.41120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu L., Lu C., Huang Y., Zhou J., Wang X., Liu C., Chen J., Le H. SPINK1 promotes cell growth and metastasis of lung adenocarcinoma and acts as a novel prognostic biomarker. BMB Rep. 2018;51:648–653. doi: 10.5483/BMBRep.2018.51.12.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomida S., Takeuchi T., Shimada Y., Arima C., Matsuo K., Mitsudomi T., Yatabe Y., Takahashi T. Relapse-related molecular signature in lung adenocarcinomas identifies patients with dismal prognosis. J. Clin. Oncol. 2009;27:2793–2799. doi: 10.1200/JCO.2008.19.7053. [DOI] [PubMed] [Google Scholar]
- 31.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mootha V.K., Lindgren C.M., Eriksson K.-F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., Houstis N., Daly M.J., Patterson N., Mesirov J.P., Golub T.R., Tamayo P., Spiegelman B., Lander E.S., Hirschhorn J.N., Altshuler D., Groop L.C. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 34.Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: an R Package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., Jensen L.J., von Mering C. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia J., Gill E.E., Hancock R.E.W. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat. Protoc. 2015;10:823–844. doi: 10.1038/nprot.2015.052. [DOI] [PubMed] [Google Scholar]
- 37.Mizuno H., Kitada K., Nakai K., Sarai A. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med. Genom. 2009;2:18. doi: 10.1186/1755-8794-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 39.Chai P., Yu J., Ge S., Jia R., Fan X. Genetic alteration, RNA expression, and DNA methylation profiling of coronavirus disease 2019 (COVID-19) receptor ACE2 in malignancies: a pan-cancer analysis. J. Hematol. Oncol. 2020;13:43. doi: 10.1186/s13045-020-00883-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thommen D.S., Schreiner J., Müller P., Herzig P., Roller A., Belousov A., Umana P., Pisa P., Klein C., Bacac M., Fischer O.S., Moersig W., Savic Prince S., Levitsky V., Karanikas V., Lardinois D., Zippelius A. Progression of lung cancer is associated with increased dysfunction of T cells defined by coexpression of multiple inhibitory receptors. Cancer Immunol Res. 2015;3:1344–1355. doi: 10.1158/2326-6066.CIR-15-0097. [DOI] [PubMed] [Google Scholar]
- 41.Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., Chen L., Li M., Liu Y., Wang G., Yuan Z., Feng Z., Zhang Y., Wu Y., Chen Y. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front. Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lavin Y., Kobayashi S., Leader A., David Amir E.-A., Elefant N., Bigenwald C., Remark R., Sweeney R., Becker C.D., Levine J.H., Meinhof K., Chow A., Kim-Shulze S., Wolf A., Medaglia C., Li H., Rytlewski J.A., Emerson R.O., Solovyov A., Greenbaum B.D., Sanders C., Vignali M., Beasley M.B., Flores R., Gnjatic S., Pe’er D., Rahman A., Amit I., Merad M. Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell. 2017;169:750–765. doi: 10.1016/j.cell.2017.04.014. e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Givechian K.B., Garner C., Benz S., Song B., Rabizadeh S., Soon-Shiong P. An immunogenic NSCLC microenvironment is associated with favorable survival in lung adenocarcinoma. Oncotarget. 2019;10:1840–1849. doi: 10.18632/oncotarget.26748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blons H., Garinet S., Laurent-Puig P., Oudart J.-B. Molecular markers and prediction of response to immunotherapy in non-small cell lung cancer, an update. J. Thorac. Dis. 2019;11:S25–S36. doi: 10.21037/jtd.2018.12.48. S36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song M., Ping Y., Zhang K., Yang L., Li F., Zhang C., Cheng S., Yue D., Maimela N.R., Qu J., Liu S., Sun T., Li Z., Xia J., Zhang B., Wang L., Zhang Y. Low-dose IFNγ induces tumor cell stemness in tumor microenvironment of non-small cell lung cancer. Can. Res. 2019;79:3737–3748. doi: 10.1158/0008-5472.CAN-19-0596. [DOI] [PubMed] [Google Scholar]
- 46.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 47.Vanhove K., Graulus G.-J., Mesotten L., Thomeer M., Derveaux E., Noben J.-P., Guedens W., Adriaensens P. The metabolic landscape of lung cancer: new insights in a disturbed glucose metabolism. Front Oncol. 2019;9:1215. doi: 10.3389/fonc.2019.01215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liberti M.V., Locasale J.W. The warburg effect: how does it benefit cancer cells? Trends Biochem. Sci. 2016;41:211–218. doi: 10.1016/j.tibs.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koundouros N., Poulogiannis G. Reprogramming of fatty acid metabolism in cancer. Br. J. Canc. 2020;122:4–22. doi: 10.1038/s41416-019-0650-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lukey M.J., Katt W.P., Cerione R.A. Targeting amino acid metabolism for cancer therapy. Drug Discov. Today. 2017;22:796–804. doi: 10.1016/j.drudis.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clark D.J., Mei Y., Sun S., Zhang H., Yang A.J., Mao L. Glycoproteomic approach identifies KRAS as a positive regulator of CREG1 in non-small cell lung cancer cells. Theranostics. 2016;6:65–77. doi: 10.7150/thno.12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arioka H., Nishio K., Ishida T., Fukumoto H., Fukuoka K., Nomoto T., Kurokawa H., Yokote H., Abe S., Saijo N. Enhancement of cisplatin sensitivity in high mobility group 2 cDNA-transfected human lung cancer cells. Jpn. J. Canc. Res. 1999;90:108–115. doi: 10.1111/j.1349-7006.1999.tb00673.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hiyama N., Ando T., Maemura K., Sakatani T., Amano Y., Watanabe K., Kage H., Yatomi Y., Nagase T., Nakajima J., Takai D. Glutamate-cysteine ligase catalytic subunit is associated with cisplatin resistance in lung adenocarcinoma. Jpn. J. Clin. Oncol. 2018;48:303–307. doi: 10.1093/jjco/hyy013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ji X., Qian J., Rahman S.M.J., Siska P.J., Zou Y., Harris B.K., Hoeksema M.D., Trenary I.A., Heidi C., Eisenberg R., Rathmell J.C., Young J.D., Massion P.P. xCT (SLC7A11)-mediated metabolic reprogramming promotes non-small cell lung cancer progression. Oncogene. 2018;37:5007–5019. doi: 10.1038/s41388-018-0307-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang H., Quek K., Chen R., Chen J., Chen B. Expression of the SARS-CoV-2 receptor ACE2 reveals the susceptibility of COVID-19 in non-small cell lung cancer. J. Canc. 2020;11:5289–5292. doi: 10.7150/jca.49462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Samad A., Jafar T., Rafi J.H. Identification of angiotensin-converting enzyme 2 (ACE2) protein as the potential biomarker in SARS-CoV-2 infection-related lung cancer using computational analyses. Genomics. 2020;112:4912–4923. doi: 10.1016/j.ygeno.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duijf P.H.G. Baseline pulmonary levels of CD8+ T cells and NK cells inversely correlate with expression of the SARS-CoV-2 entry receptor ACE2. BioRxiv. 2020 doi: 10.1101/2020.05.04.075291. [DOI] [Google Scholar]
- 58.Prado-Garcia H., Romero-Garcia S., Aguilar-Cazares D., Meneses-Flores M., Lopez-Gonzalez J.S. Tumor-induced CD8+ T-cell dysfunction in lung cancer patients. Clin. Dev. Immunol. 2012:741741. doi: 10.1155/2012/741741. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li T., Qiu Z., Han Y., Wang Z., Fan H., Lu W., Xie J., Ma X., Wang A. Rapid loss of both CD4+ and CD8+ T lymphocyte subsets during the acute phase of severe acute respiratory syndrome. Chin. Med. J. 2003;116:985–987. [PubMed] [Google Scholar]
- 60.Aktaş O.N., Öztürk A.B., Erman B., Erus S., Tanju S., Dilege Ş. Role of natural killer cells in lung cancer. J. Canc. Res. Clin. Oncol. 2018;144:997–1003. doi: 10.1007/s00432-018-2635-3. [DOI] [PubMed] [Google Scholar]
- 61.Orange J.S. Human natural killer cell deficiencies and susceptibility to infection. Microb. Infect. 2002;4:1545–1558. doi: 10.1016/s1286-4579(02)00038-2. [DOI] [PubMed] [Google Scholar]
- 62.van Erp E.A., van Kampen M.R., van Kasteren P.B., de Wit J. Viral infection of human natural killer cells. Viruses. 2019;11 doi: 10.3390/v11030243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pavlova N.N., Thompson C.B. The emerging hallmarks OF cancer metabolism. Cell Metabol. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sanchez E.L., Lagunoff M. Viral activation of cellular metabolism. Virology. 2015:479–480. doi: 10.1016/j.virol.2015.02.038. 609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The lung adenocarcinoma TCGA-LUAD cohort was downloaded from the TCGA data portal (https://portal.gdc.cancer.gov/). In addition, the lung adenocarcinoma gene expression profiling datasets were downloaded from the NCBI gene expression omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/).