Abstract
Objectives
The immunologic profile and opportunistic viral DNA increase were monitored in Italian patients with COVID-19 in order to identify markers of disease severity.
Methods
A total of 104 patients infected with SARS-CoV-2 were evaluated in the study. Of them, 42/104 (40.4%) were hospitalized in an intensive care unit (ICU) and 62/104(59.6%) in a sub-intensive care unit (SICU). Human cytomegalovirus (HCMV) and Epstein-Barr virus (EBV), Parvovirus B19 and Human Herpesvirus 6 virus reactivations were determined by real-time PCR, and lymphocyte subpopulation counts were determined by flow cytometry.
Results
Among opportunistic viruses, only EBV was consistently detected. EBV DNA was observed in 40/42 (95.2%) of the ICU patients and in 51/61 (83.6%) of the SICU patients. Comparing the two groups of patients, the EBV DNA median level among ICU patients was significantly higher than that observed in SICU patients. In parallel, a significant reduction of CD8 T cell and NK count in ICU patients as compared with SICU patients was observed (p < 0.05). In contrast, B cell count was significantly increased in ICU patients (p = 0.0172).
Conclusions
A correlation between reduced CD8+ T cells and NK counts, EBV DNA levels and COVID-19 severity was observed. Other opportunistic viral infections were not observed. The relationship between EBV load and COVID-19 severity should be further evaluated in longitudinal studies.
Keywords: COVID-19, EBV DNA, Lymphocyte subpopulation
Introduction
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2), was first reported in China in December 2019. As of February 21, 2020, more than 7,151,000 cases of COVID-19 have been reported worldwide and there are more than 407,145 deaths. The spread has already taken on pandemic proportions, affecting over 100 countries, including Italy that counts more than 235,270 cases (Ghinai et al., 2020, Remuzzi and Remuzzi, 2020). In February 2020, a large COVID-19 outbreak in Lombardy Italy was observed. The SARS-CoV-2 influence on immunological response, and its relationship with the reactivation of opportunistic viral infection in the clinical course of COVID-19 is still under investigation. Among various pathogens, opportunistic viral infections such as human cytomegalovirus (HCMV) and Epstein-Barr virus (EBV) are particularly relevant in immunocompromised patients (Fishman, 2017, Fishman, 2013) and their reactivation has also been reported in patients in intensive care units (ICU) with no previous immune suppression (Schildermans and De Vlieger, 2020, Limaye and Boeckh, 2010, Frantzeskaki et al., 2015, Ong et al., 2017, Cantan et al., 2019, Coşkun et al., 2017).
On the other hand, the impact of opportunistic viral reactivation in critically ill patients remains to be elucidated.
Finally, the impact of COVID-19 infection on the immune system is not yet well known. In this study, a possible relationship with alterations of the NK and T-cell subpopulations leading to opportunistic virus reactivation were analyzed in patients with COVID-19 having different degrees of severity.
Material and methods
Patient characteristics and comorbidity
Written informed consent was collected. A total of 104 patients infected with SARS-CoV-2 referred to the Fondazione IRCCS Policlinico San Matteo, Pavia between February 2020 and March 2020 were evaluated. Of them, 42/104 (40.4%) were hospitalized in an ICU and 62/104 (59.6%) were hospitalized in a sub-intensive care unit (SICU). The degree of severity of COVID-19 (severe vs mild) was defined using the American Thoracic Society guidelines for community-acquired pneumonia (Metlay et al., 2019). Patient characteristics and comorbidity were described in Table 1 .
Table 1.
Demographic characteristics of the 104 analyzed patients.
| Hospitalization N° 104 |
|||
|---|---|---|---|
| Total | ICUa N° 42 |
SICUb N° 62 |
P-value |
| no (%) | 42 (40.4%) | 62 (59.6%) | |
| Gender | |||
| Male/Female | 36/6 (85.7%/14.3%) | 41/21 (66.1%/33.9%) | 0.0391 |
| Median age (years; IQR) | 61.5 (55−71.25) | 73.5 (57.8−80) | 0.05 |
| Comorbidity | |||
| Heart disease | 19.0% | 33% | 0.25 |
| Hypertension | 40.4% | 48.1% | 0.62 |
| Obesity | 30.9% | 18.5% | 0.27 |
| Lung disease | 4.7% | 14.8% | 0.20 |
| Diabetes | 14.3% | 11.1% | 1.00 |
| Carcinoma | 9.5% | 3.7% | 0.64 |
ICU, Intensive Care Unit.
Sub-Intensive Care Unit, In bold is the significant value.
Characterization of serological status for HCMV, EBV and parvovirus B19
Antiviral capsid antigen (anti-VCA), anti-EBNA, anti−HCMV and anti-parvovirus B19 specific IgG were quantified from serum samples using a chemiluminescent assay according to manufacturers’ instructions (Liaison, Diasorin, Saluggia, Italy). Positive results were defined as IgG> 20U/mL for VCA and EBNA IgG, IgG>14 U/mL for CMV IgG and Index>1.1 for Parvovirus B19 IgG.
Viral quantification in COVID-19 patients
Nasal swabs (NS) or bronchoalveolar lavage (BAL) and whole blood were collected during the hospitalization from all patients, and 200 μL of clinical specimens were extracted using the QIAsymphony® instrument with QIAsymphony® DSP Virus/Pathogen Midi Kit (QIAGEN, Hilden, Germany). Specific real-time reverse transcriptase-polymerase chain reaction (RT-PCR) targeting RNA-dependent RNA polymerase and E genes were used to detect the presence of SARS-CoV-2 according to the WHO guidelines (Anon, 2020) and Corman et al. protocols (Corman et al., 2020). The CMV DNA and EBV DNA load were determined by home-made real-time PCR in CMV and EBV seropositive subjects using primers and probes and Applied Biosystems Instruments (ThermoFisher, Waltham, Massachusetts, United States) as reported (Baldanti et al., 2008, Furione et al., 2012). At the same time, Parvovirus B19 and Human Herpesvirus 6 were analyzed in a fraction of patients (Corcoran et al., 2010, Watzinger et al., 2004).
Lymphocyte subpopulations count and characterization
Blood samples were stained with CYTO-STAT tetraCHROME CD45-FITC/CD4-RD1/CD8-ECD/CD3-PC5 and CYTO-STAT tetraCHROME CD45-FITC/CD56-RD1/CD19-ECD/CD3-PC5, all from Beckman Coulter, Milan, Italy).The percentage of CD4+ and CD8 + T lymphocytes and CD56 + NK and CD19 + B cells was determined by Navios Flow Cytometer System (Beckman Coulter). Using FlowCountFluorospheres, CD4+, CD8 + T cell subsets and CD56 + NK and CD19 + B cells were expressed as absolute counts (cells/μl). Gating strategy was set up on CD45+ and side scatter (SSC).
Statistical analysis
Descriptive data were reported or considered as absolute and relative frequencies, median and interquartile range (IQR) based on the type of variable distribution. For qualitative variables, Fisher’s test was used, while the Mann-Whitney test was used for quantitative variables in order to perform comparison between groups. Spearman’s test was used for the correlation analysis. All tests were two-tailed. A p-value <0.05 was considered statistically significant. Analyses were performed using the GraphPad Prism 5 (GraphPad Software, CA, USA).
Results
Clinical characteristics
Male COVID-19 patients were more frequent in both units (p = 0.039). The most common symptoms detected in patients on admission or during hospitalization in ICU and SICU, respectively, were fever (80.9% and 100%; p = 0.02), interstitial lung disease (100% and 55.6%; p < 0.001), dyspnea (100% and 37%; p < 0.001), and diarrhea (2.3 and 3.7%; p > 0.05). Overall, all the patients except one were positive for EBV IgG (99%), while 100/104 subjects (96.2%) were CMV seropositive at time of hospitalization. Finally, 72/104 (69.2%) were positive for Parvovirus B19 IgG.
Viral detection and quantification
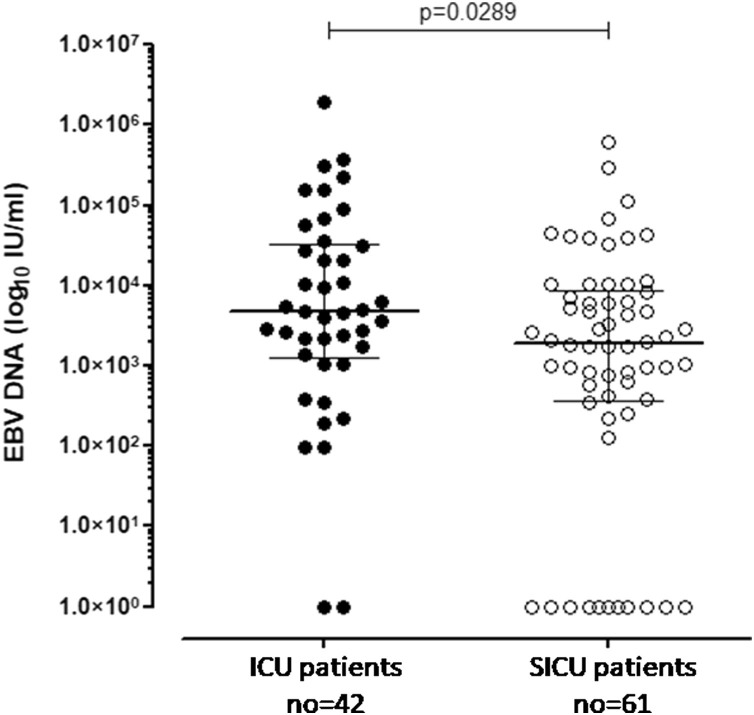
CMV, Parvovirus B19 and HHV-640 DNA were negative in all patients analyzed. EBV positive DNA was observed in 91/103 EBV seropositive patients (88.3%). Of them, 40/42 patients (95.2%) were admitted to the ICU and 51/61 patients (83.6%) were admitted to the SICU. In the remaining 2/42 patients (4.8%) and 10/61 patients (16.4%) in the ICU and SICU, respectively, no EBV DNAemia events were reported (p = 0.1161). The EBV DNA median level among ICU patients [4709 (IQR 1284–32075) IU/mL] was significantly higher than that observed in SICU patients [median 1890 (IQR 370.1–8820) IU/mL; p = 0.0289) as shown in Figure 1 .
Figure 1.
EBV DNA was measured in 42 COVID19-positive patients in ICU (black dots) and in 62 COVID19-positive patients in SICU (white dots). Median and IQR are shown and p-value was given in the graph.
Lymphocyte subpopulation counts
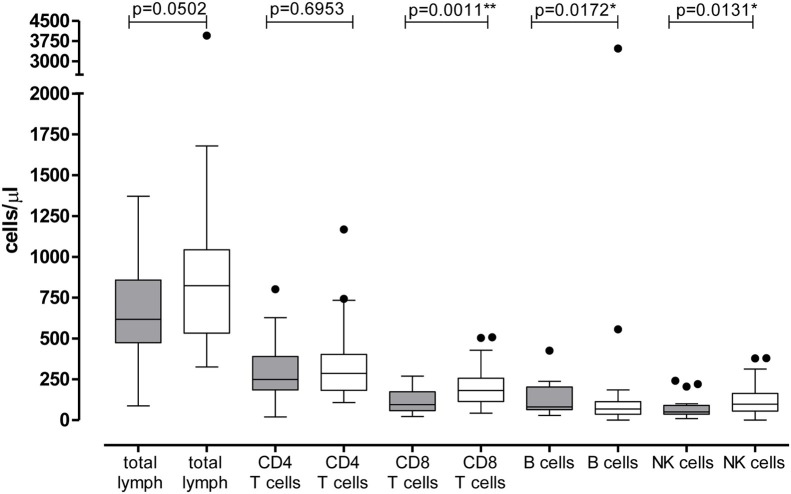
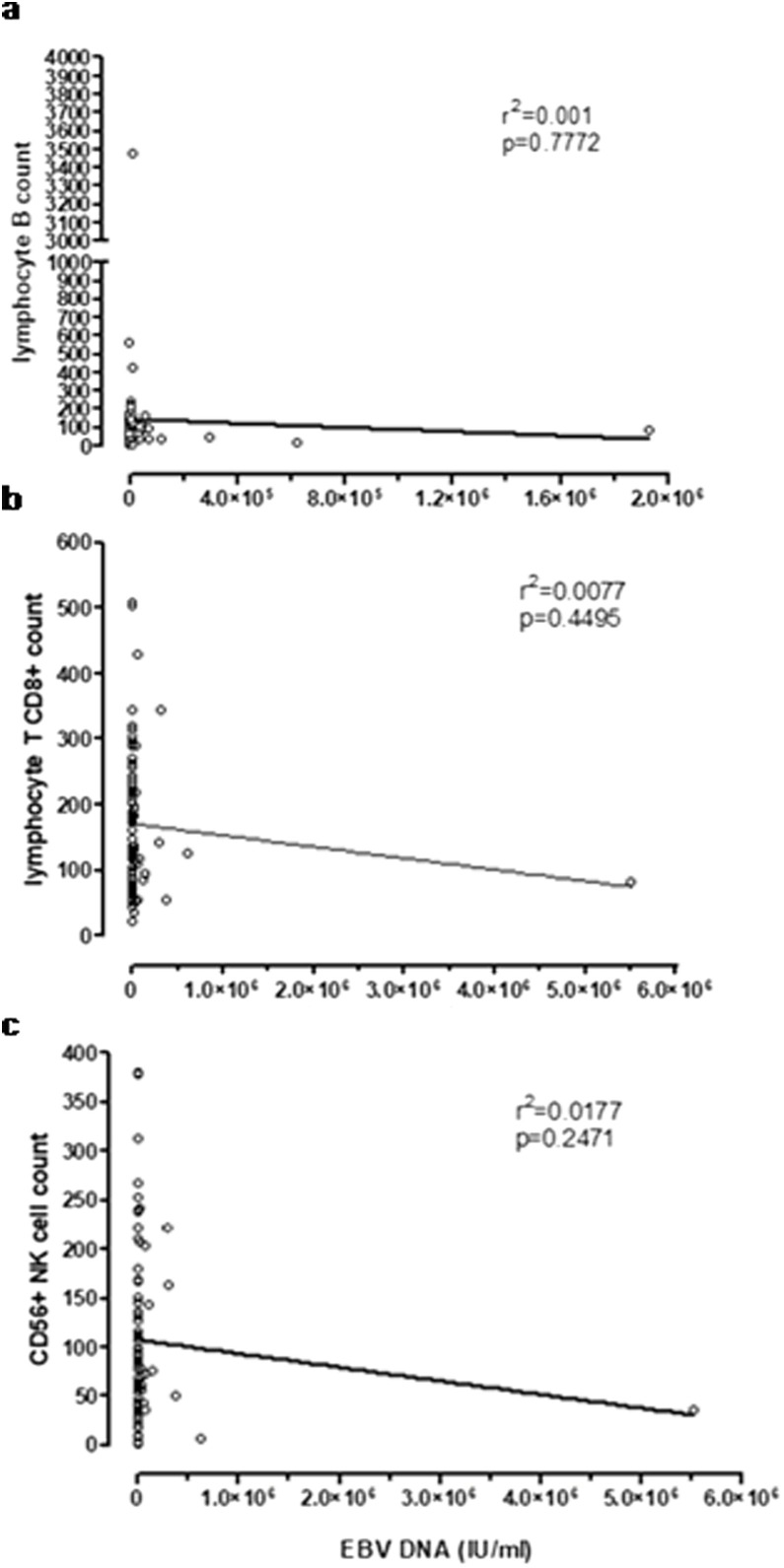
Lymphocyte subpopulation counts were available for 31/42 ICU patients (73.8%) and for 55/62 SICU patients (88.7%). A slight reduction of the total lymphocyte count was observed in ICU patients as compared with SICU patients [median 617.5 (IQR 473.8–858) cells/μl vs 823 (IQR 533–1044) cells/μl; p = 0.0502]. In detail, CD4 T cell count was not statistically different between the two groups of patients [median 249 (IQR 185.3–393.3) cells/μl vs 286 (IQR 182–403) cells/μl, respectively; p = 0.6953]. On the other hand, we observed a significant reduction of CD8+ T cell count in ICU patients as compared with SICU patients [median 95 (IQR 58–174) cells/μl vs 181 (115–257) cells/μl; p = 0.0011]. Similarly, a lower median NK count was reported in ICU patients as compared with the group of SICU patients [50 (IQR 36–90) cells/μl vs 98 (IQR 56–163) cells/μl; p = 0.0131]. In addition, comparing the two groups of patients, B cell count was significantly increased in ICU patients [median 81 (IQR 65–203) cells/μl vs 69 (36–113) cells/μl, respectively; p = 0.0172) (Figure 2 ). No linear correlation between EBV load and CD8 T cells, B cells or NK cells was observed (Figure 3 ).
Figure 2.
Lymphocyte subpopulation counts were compared in the 42 patients of the Intensive Care Unit (grey bars) and in the 62 patients of the Infectious Disease Department (white bars). The p-value was measured by the Mann Whitney test and was given for each group of comparison; *p < 0.05; **p < 0.01.
Figure 3.
EBV DNA load was correlated to B cell count (a), CD8 + T cell count (b) and NK count (c); r (Remuzzi and Remuzzi, 2020) and p value were calculated using Spearman’s correlation test and results were given in each graph.
Discussion
The strict relation between immunosuppression and reactivation of latent viruses and an increased prevalence of opportunistic viral infections is well known, especially in transplanted patients and in ICU patients (Fishman, 2013, Schildermans and De Vlieger, 2020, Limaye and Boeckh, 2010, Frantzeskaki et al., 2015, Ong et al., 2017, Cantan et al., 2019, Coşkun et al., 2017).
The impact of COVID-19 infection on the immune system is still largely unknown. We observed a significant loss of NK and CD8+ T cells in COVID-19 patients, which paralleled the severity of the infection. Normally, in healthy subjects lymphocyte CD3+ counts range from 690 to 2540 cells/μl, lymphocyte CD4+ and CD8+ counts range from 410 to 1590 and from 190 to 1140 cells/μl, respectively; moreover, in healthy subjects, we reported that CD19+ B cell counts and CD56 + NK cell counts range from 163 to 288 cells/μl and from 151 to 296 cells/μl.
Surprisingly, this NK and CD8+ T cell reduction was associated with presence of EBV DNA only. Despite that the large majority of patients were seropositive for both CMV and B19, no evidence of circulating CMV and B19 DNA was observed. In this study, we analyzed the correlation between COVID-19 infection and immunological and virological factors, classifying patients according to the severity of the COVID-19 disease. Overall, a higher proportion of interstitial lung disease and dyspnea was observed among ICU patients as compared to SICU patients (p < 0.001). Interestingly, positive EBV DNA was observed in approximately 87% of patients. The rate of positive EBV DNA observed was higher in COVID-19 patients with severe infection (ICU patients) as compared to patients with mild symptoms (SICU patients). Moreover, the median EBV DNA levels in patients with severe COVID-19 was significantly higher than in patients with mild COVID-19 disease, supporting the evidence of a higher immune impairment in the previous patients. However, no linear correlation between EBV DNA load and lymphocyte counts was observed. In line with these results, a reduced total lymphocyte count in patients referred to the ICU was observed, reporting a significant reduction in terms of both CD8+ and NK cells as compared to SICU patients, while no significant reduction of CD4+ T cell counts was observed. Recent studies underlined a marked lymphopenia in a large fraction of COVID-19 patients (Huang et al., 2020, Tan et al., 2020) with a reduction of total T lymphocytes, CD4+ T cells and CD8+ T cells and a more profound reduction in severe cases (Chen et al., 2020).
Of note, the higher increase of B cell count observed in patients with severe symptoms as compared to patients with milder symptoms was in line with results obtained by Chen and colleagues (Chen et al., 2020). However, our data was in contrast with Wang and colleagues’ observation, reporting a decrease of B cells in patients with severe COVID-19 disease (Wang et al., 2020).
Since B lymphocytes are the site of latency for EBV, an increase of B cells in patients with severe symptoms could be related to the increased EBV DNA levels as occurring in patients with PTLD (post-transplant lymphoproliferative disorder). On the other hand, the increase of B cell counts during COVID-19 could reflect the antibody response to SARS-CoV-2. Finally, the role of increased levels of EBV DNA on B cell functions, with a potential impact on antibody production, is unknown. Thus, we can only speculate that the presence of detectable EBV DNA may play a role in the severity of COVID-19 disease on the basis of the higher frequency of EBV DNA positive patients and the higher EBV DNA levels.
Interestingly, among all opportunistic viruses, reactivation in COVID-19 patients seems to be restricted to EBV, an event somewhat corroborating a role in the severity of the disease. In fact, despite the low rate of CD8+ T cells and NK cells in patients with severe symptoms, CMV reactivation, for example, was never observed. Usually, detectable CMV DNA in whole blood is more frequent than EBV DNA in immunocompromised patients with reduced lymphocyte counts.
In conclusion, our data support the possible correlation between lymphopenia and EBV load with COVID-19 disease severity. In order to corroborate our hypothesis, monitoring of the immunological profile and opportunistic virus reactivation over time is warranted in order to better understand the role in the pathogenesis of the disease or its relationship with the severity of symptoms.
Funding
This work was supported by the Ministero della Salute, Fondazione IRCCS Policlinico San Matteo Ricerca Corrente (grant 80207).
Conflict of interest
The authors declare that they have no competing interests.
Author contributions
Stefania Paolucci and Irene Cassaniti: data analysis and interpretation, drafted the article; Federica Novazzi, Loretta Fiorina and GiudittaComolli: sample processing. Antonio Piralla: software and data analysis.Raffaele Bruno, Renato Maserati, Roberto Gulminetti, Stefano Novati and Francesco Mojoli: patient enrollment. Fausto Baldanti: critical revision of the article and final approval of the version to be published. All authors contributed toward data analysis, drafting and revising the paper, gave final approval of the version to be published and agree to be accountable for all aspects of the work.
Acknowledgements
We thank Daniela Sartori for careful preparation of the manuscript.
Contributor Information
for the San Matteo Pavia COVID-19 Task Force:
R Bruno, M Mondelli, E Brunetti, A Di Matteo, E Seminari, L Maiocchi, V Zuccaro, L Pagnucco, B Mariani, S Ludovisi, R Lissandrin, A Parisi, P Sacchi, SFA Patruno, G Michelone, R Gulminetti, D Zanaboni, S Novati, R Maserati, P Orsolini, M Vecchia, M Sciarra, E Asperges, M Colaneri, A Di Filippo, M Sambo, S Biscarini, M Lupi, S Roda, TC Pieri, I Gallazzi, M Sachs, P Valsecchi, S Perlini, C Alfano, M Bonzano, F Briganti, G Crescenzi, AG Falchi, R Guarnone, B Guglielmana, E Maggi, I Martino, P Pettenazza, S Pioli di Marco, F Quaglia, A Sabena, F Salinaro, F Speciale, I Zunino, M De Lorenzo, G Secco, L Dimitry, G Cappa, I Maisak, B Chiodi, M Sciarrini, B Barcella, F Resta, L Moroni, G Vezzoni, L Scattaglia, E Boscolo, C Zattera, MF Tassi, V Capozza, D Vignaroli, M Bazzini, G Iotti, F Mojoli, M Belliato, L Perotti, S Mongodi, G Tavazzi, G Marseglia, A Licari, I Brambilla, D Barbarini, A Bruno, P Cambieri, G Campanini, C. Cavanna, G Comolli, M Corbella, R Daturi, M Furione, B Mariani, P Marone, R Maserati, E Monzillo, S Paolucci, M Parea, E Percivalle, A Piralla, F Rovida, A Sarasini, M Zavattoni, G Adzasehoun, M Ardizzone, L Bellotti, V Brunco, E Cabano, G Casali, L Capella, D Devitis, L Dossena, G Frisco, G Garbagnoli, F Gardellini, A Girello, A Guerrizio, V Landini, C Lucchelli, V Maliardi, P Piemontese, S Pezzaia, M Premoli, C Rebuffa, C Zanello, J Bagnarino, F Bergami, A Bonetti, G Caneva, I Cassaniti, A Corcione, R Di Martino, A Di Napoli, A Ferrari, G Ferrari, L Fiorina, A Gallone, F Giardina, A Girardi, A Mercato, C Merla, F Novazzi, G Ratano, B Rossi, G Saveriaempillai, IM Sciabica, M Tallarita, E Vecchio Nepita, J Vitali, A Cerino, S Varchetta, B Oliviero, S Mantovani, D Mele, M Calvi, M Tizzoni, C Nicora, A Triarico, V Petronella, C Marena, A Muzzi, P Lago, S Cutti, V Novelli, F Comandatore, G BatistiBiffignandi, S Gaiarsa, M Rettani, C Bandi, and A Ferrari
References
- https://www.who.int/docs/default-source/coronaviruse/protocol-v2-1.pdf.
- Baldanti F., Gatti M., Furione M., Paolucci S., Tinelli C., Comoli P. Kinetics of Epstein-Barr virus DNA load in different blood compartments of pediatric recipients of T-cell-depleted HLA-haploidentical stem cell transplantation. J ClinMicrobiol. 2008;46:3672–3677. doi: 10.1128/JCM.00913-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantan B., Luyt C.E., Martin-Loeches I. Influenza Infections and Emergent Viral Infections in Intensive Care Unit. SeminRespirCrit Care Med. 2019;40:488–497. doi: 10.1055/s-0039-1693497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H. Clinical and immunologic features in severe and moderate Coronavirus Disease 2019. J Clin Invest. 2020 doi: 10.1172/JCI137244. pii: 137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran C., Hardie D., Yeats J., Smuts H. Genetic variants of human parvovirus B19 in South Africa: cocirculation of three genotypes and identification of a novel subtype of genotype 1. J ClinMicrobiol. 2010;48:137–142. doi: 10.1128/JCM.00610-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coşkun O., Yazici E., Şahiner F., Karakaş A., Kiliç S., Tekin M. Cytomegalovirus and Epstein-Barr virus reactivation in the intensive care unit. Med KlinIntensivmedNotfmed. 2017;112(3):239–245. doi: 10.1007/s00063-016-0198-0. [DOI] [PubMed] [Google Scholar]
- Fishman J.A. Overview: cytomegalovirus and the herpesviruses in transplantation. Am J Transplant. 2013;13(Suppl 3):1–8. doi: 10.1111/ajt.12002. [DOI] [PubMed] [Google Scholar]
- Fishman J.A. Infection in Organ Transplantation. Am J Transplant. 2017;17:856–879. doi: 10.1111/ajt.14208. [DOI] [PubMed] [Google Scholar]
- Frantzeskaki F.G., Karampi E.S., Kottaridi C., Alepaki M., Routsi C., Tzanela M. Cytomegalovirus reactivation in a general, non-immunosuppressed intensive care unit population: incidence, risk factors, associations with organ dysfunction, and inflammatory biomarkers. J Crit Care. 2015;30:276–281. doi: 10.1016/j.jcrc.2014.10.002. [DOI] [PubMed] [Google Scholar]
- Furione M., Rognoni V., Cabano E., Baldanti F. Kinetics of human cytomegalovirus (HCMV) DNAemia in transplanted patients expressed in international units as determined with the Abbott RealTime CMV assay and an in-house assay. J ClinVirol. 2012;55:317–322. doi: 10.1016/j.jcv.2012.08.017. [DOI] [PubMed] [Google Scholar]
- Ghinai I., McPherson T.D., Hunter J.C., Kirking H.L., Christiansen D., Joshi K. First known person-to-person transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the USA. Lancet. 2020;395(10230):1137–1144. doi: 10.1016/S0140-6736(20)30607-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China. Lancet. 2020;15(395):497–506. doi: 10.1016/S0140-6736(20)30183-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limaye A.P., Boeckh M. CMV in critically ill patients: pathogen or bystander? Rev Med Virol. 2010;20:372–379. doi: 10.1002/rmv.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metlay J.P., Waterer G.W., Long A.C., Anzueto A., Brozek J., Crothers K. Diagnosis and treatment of adults with community-acquired pneumonia: an official clinical practice guideline of the American Thoracic Society and Infectious Disease Society of America. Am J RespirCrit Care Med. 2019;200(7):e45–e67. doi: 10.1164/rccm.201908-1581ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong D.S.Y., Bonten M.J.M., Spitoni C., VerduynLunel F.M., Frencken J.F., Horn J. Epidemiology of Multiple Herpes Viremia in Previously Immunocompetent Patients with Septic Shock.Molecular Diagnosis and Risk Stratification of Sepsis Consortium. Clin Infect Dis. 2017;64:1204–1210. doi: 10.1093/cid/cix120. [DOI] [PubMed] [Google Scholar]
- Remuzzi A., Remuzzi G. COVID-19 and Italy: what next? Lancet. 2020;395(10231):1225–1228. doi: 10.1016/S0140-6736(20)30627-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schildermans J., De Vlieger G. Cytomegalovirus: A Troll in the ICU? Overview of the Literature and Perspectives for the Future. Front Med (Lausanne) 2020;7:188. doi: 10.3389/fmed.2020.00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Wang Q., Zhang D., Ding J., Huang Q., Tang Y.Q. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Transduct Target Ther. 2020;5(33) doi: 10.1038/s41392-020-0148-4. eCollection 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Nie J., Wang H., Zhao Q., Xiong Y., Deng L. Characteristics of peripheral lymphocyte subset alteration in COVID-19 pneumonia. J Infect Dis. 2020 doi: 10.1093/infdis/jiaa150. pii: jiaa150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watzinger F., Suda M., Preuner S., Baumgartinger R., Ebner K., Baskova L. Real-time quantitative PCR assays for detection and monitoring of pathogenic human viruses in immunosuppressed pediatric patients. J ClinMicrobiol. 2004;42:5189–5198. doi: 10.1128/JCM.42.11.5189-5198.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]