Graphical abstract
Keywords: Molecular docking, SARS-CoV-2, Aloe vera, COVID-19, Antiviral activity, ADMET properties
Highlights
-
•
A solution has to be found rapidly against COVD-19.
-
•
From a set of 10 compounds of Aloe vera, 3 potential inhibitors of SARS-CoV-2 main protease were identified.
-
•
The binding affinity of ligand-protein interactions and the Lipinski’s rule of five based-on ADME analysis were used to confirm the best candidate.
Abstract
SARS-CoV-2 is the pathogen agent of the new corona virus disease that appeared at the end of 2019 in China. There is, currently, no effective treatment against COVID-19. We report in this study a molecular docking study of ten Aloe vera molecules with the main protease (3CLpro) responsible for the replication of coronaviruses. The outcome of their molecular simulation and ADMET properties reveal three potential inhibitors of the enzyme (ligands 6, 1 and 8) with a clear preference of ligand 6 that has the highest binding energy (−7.9 kcal/mol) and fully obeys the Lipinski’s rule of five.
1. Introduction
A case of unidentified pneumonia was reported, in late December 2019, in Wuhan, Hubei Province, People's Republic of China which became a global pandemic, killing hundreds of thousands of people [1], [2], [3]. Coronavirus disease 2019 (or COVID-19) is an emerging infectious disease caused by a strain of coronavirus called SARS-CoV-2. Its clinical characteristics are very similar to those of viral pneumonia. After analysis on respiratory samples, PRC Centers for Disease Control (CDC) experts declared that the pneumonia, later known as novel coronavirus pneumonia (NCP), was caused by novel coronavirus [4].
In January 2020, the whole-genome sequences of 2019-nCoV from different laboratories and regions have been submitted to GISAID database, which allowed the International Committee on Taxonomy of Viruses (ICTV) and the WHO to permanently name the 2019-nCoV pathogen as Severe Acute Respiratory Syndrome CoronaVirus 2 (SARS-CoV-2) and the causing disease as coronavirus disease 2019 (COVID-2019) [5], [6]. However, SARS-CoV-2 quickly spread to over 150 countries and on March 11th, the World Health Organization (WHO) formally recognized the COVID-19 as a pandemic.
Comparative genomic studies have shown that SARS-CoV-2 belongs to the Beta coronavirus family and is phylogenetically very similar to SARS-CoV-1, which was responsible for an outbreak of acute pneumonia that occurred in November 2002 in Guangdong Province, China.
Also genome analysis of novel coronavirus sequences revealed that the complete genome sequence recognition rates of SARS-CoV and bat SARS coronavirus (SARSr-CoV-RaTG13) were 79.5% and 96% respectively [7]. This implies that the coronavirus might originate from bat. Similar to other viruses, SARS-CoV-2 has many potential natural hosts, intermediate hosts and final hosts. This poses great challenges to prevention and treatment of virus infection and compared with SARS and MERS, this virus has high transmissibility and infectivity, despite of low mortality rate [6]. According to the Centers for Disease Control and Prevention (CDC), the WHO and the U.S. Food and Drug Administration (FDA), despite the biosafety and hygiene measures to limit the large-scale spread of this pandemic, there are currently no medications or vaccines proven to be effective for the treatment or prevention of the SARS-CoV-2.
Furthermore, the prospect of developing a new drug in the short to medium term is not feasible due to many constraints [6].
Some antivirals already used in the treatment of SARS-CoV and MERS-CoV are recommended. These include lopinavir and ritonavir, in combination with nebulized alfa-interferon [8], [9]. Chloroquine, a known antimalarial drug used as an immunomodulant in other coronavirus infections, has been proposed but its use is still controversial in the scientific community [10], [11]. Therefore, it is a potential target for anti-coronaviruses inhibitors screening [12] . Structure-based activity analyses and high-throughput studies have identified potential inhibitors for SARS-CoV and MERS-CoV 3CLpro [13], [14]. This means that an alternative solution to this major public health problem is urgently needed to save lives, and traditional medicine, which has proved its worth around the world when used against several diseases, remains one of the avenues that can be exploited to counter this pandemic.
The role of traditional medicine in the treatment of COVID-19 has recently been reported in the literature [15]. Indeed, medicinal plants are an important source of molecules with various pharmacological properties including antiviral properties that can be used in the search for the solution against COVID-19. Medicinal plants, especially those employed in traditional medicine, have attracted significant attention because they include bioactive compounds that could be used to develop formal drugs against several diseases with no or minimal side-effects [16]. According to WHO, more than 80% of the population in Africa use traditional medicine to solve the primary health problem [17], [18], [19]. Nevertheless, it is not unique to Africa or other developing countries where it is recognized as a traditional medicine. It is also used in the so-called developed or industrialized countries [20], [21].
Aloe vera (L.) Burm.f. considered as a “miraculous plant” or “wonder plant” is a medicinal plant that has been used for more than 3000 years in various cultures [22]. It is one of more than 400 species in the genus Aloe of the family Xanthorrhoeaceae. It is one of the most studied and used medicinal plants worldwide. Its pharmacological properties and phytochemistry are well documented [23], [24]. Since the appearance of COVID-19, there has been some information referring to the use of this plant alone or in combination with others against COVID-19. In addition, in silico (virtual screening) analysis by molecular docking revealed that several secondary metabolites isolated from tropical medicinal plants have the potential to inhibit the main protease of COVID-19 (Mpro), a very promising potential pharmacological target [25].
Very recently, our research group showed in a survey of literature that Aloe vera can be used as potential anti-COVID-19 plant regarding its antiviral activity [26] . This study is designed to identify the potential inhibitors from the set of 10 compounds of Aloe vera by means of molecular docking and ADMET (Absorption, Distribution, Metabolism, Excretion and Toxicity) properties of the inhibitors using SwissADME and preADME server, Korea.
2. Materials and methods
2.1. Literature review
The literature review was based mainly on the COVID-19 resources that have been made freely available to the scientific community (COVID-19 open research dataset https://pages.semanticscholar.org/coronavirus-research), but also on the usual databases such as Pubmed and Google scholar. The scientific name of the plant (Aloe vera) and COVID-19 were used as keywords for the search. Finally, bibliographical references were made using a bibliographical software “Mendeley”.
2.2. Molecular docking
2.2.1. Protein preparation
The coronavirus encodes more than one dozen protein, among these, the 3C-like protease (3CLpro) is the most studied. The 3CLpro or the COVID-19 virus main protease (Mpro) is a key CoV enzyme which plays a pivotal role in mediating viral replication and transcription, making it an attractive drug target for this virus [27].
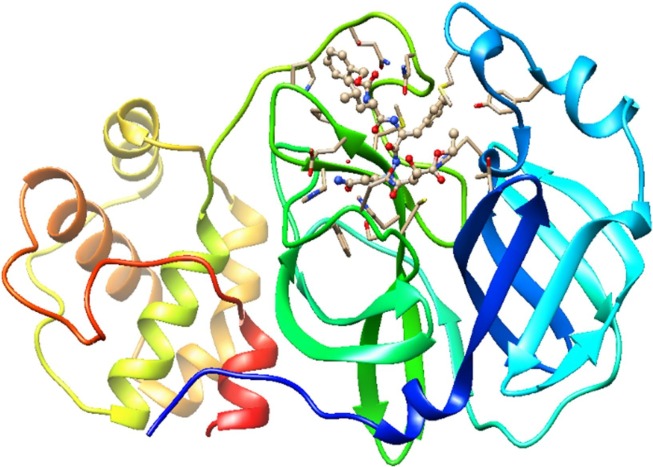
The crystal structures of Mpro (PDB ID: 2GTB and 6LU7) [6], [28] were retrieved from the Protein Data Bank and imported into chimera for visualizing the binding domain of these two complexes and identifying the amino acids in the binding pocket. The hydrogen atoms were added to the protein in order to correct the ionization and tautomeric states of the amino acid residues. Furthermore, the water molecules and complexes bound to receptor molecule were removed before the docking. Incomplete side chains were replaced using Drunbrack rotamer library [29]. In addition, the protein was subjected to energy minimization by applying the AMBER 14SB force field, and AM1-BCC was used for other residues with a maximum number of 200 steps at RMS gradient of 0.02. The optimized protein was saved in pdbqt format and imported to PyRx for molecular docking which was carried out by means of Autodock Vina virtual screening tool [30]. Fig. 1 displayed the complex formed between the COVID-19 Mpro and 2GTB as a potential drug target for 2019-nCoV. According to Xu and co-workers, 2GTB is the main protease found in the CoV associated with the severe acute respiratory syndrome (SARS), and that the main protease in 2019-nCov shares 96% similarity with that in SARS [28].
Fig. 1.
Complex between COVID-19 Mpro and co-crystallized inhibitor 2GTB (PDB).
2.2.2. Generation of ligand dataset
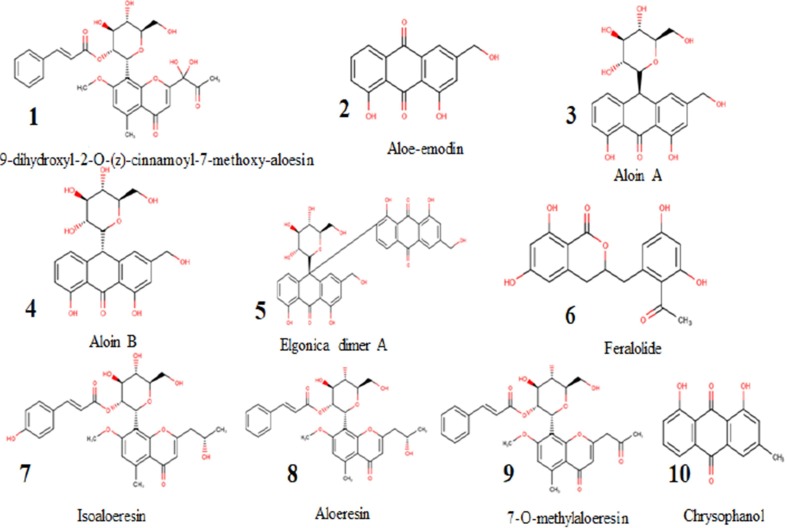
The selected compounds derivatives from various literature resources [31] were drawn using ACD/MarvinSketch (20.9). Fig. 2 shows the 2D structure of the sketched compounds. Furthermore, ligands were imported into ChemDraw to obtain 3D from 2D. The 3D ligands were saved in .sdf format. Ligands optimization was performed by using universal force field (UFF) with conjugate gradients algorithm of 200 Steps, and then analyzed for ADME using the SwissADME database available [32]. The mutagenicity (AMES Test) and carcinogeniticy of the three top compounds were assessed using the preADME server, Korea [33].
Fig. 2.
Structures of compounds 1–10 derived from Aloe vera.
2.2.3. Molecular docking studies
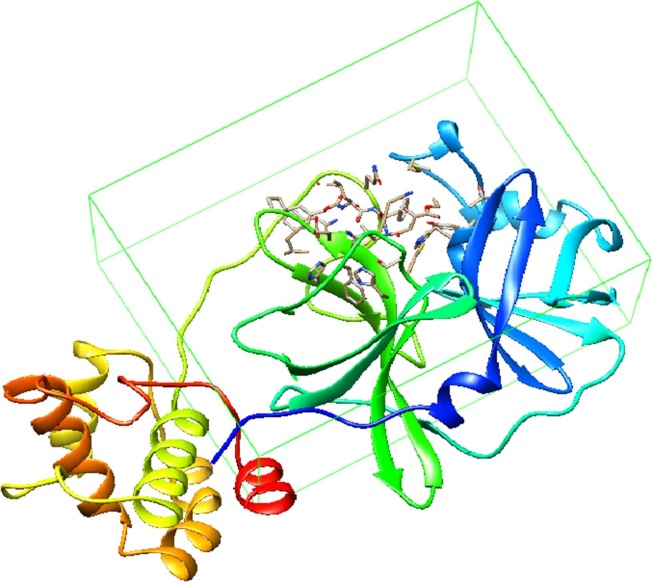
A primary objective in molecular docking was the ability to estimate the scoring function and evaluate protein-ligand interactions in order to predict the binding affinity and activity of the ligand molecule [34]. Autodock Vina and PyRx were used to generate the bioactive binding poses of ligands dataset in the active site of COVID-19 Mpro. Protein coordinates from the bound ligand of 2GTB were used to define the active site as shown in Fig. 3 .
Fig. 3.
Active site of 2GTB.
Discovery Studio 2020 Client software was used to model non-bonded polar and hydrophobic contacts in the inhibitor site of COVID-19 3CLpro/Mpro (PDB ID: 2GTB). Hit molecules which showed the expected interactions with the critical amino acids present in the active site of the protein, may show potent antagonist properties towards COVID-19 3CLpro/Mpro protease.
3. Results
The binding affinities and the main interactions between COVID-19 Mpro and some Aloe vera metabolite compounds were obtained by using Autodock Vina. Several compounds from Aloe vera have been reported to show antiviral bioactivities. We investigated ten compounds from this plant as potential inhibitors of the COVID-19 Mpro. Since Lopinavir and Nelfinavir are commonly used to treat human immunodeficiency virus (HIV)/acquired immunodeficiency syndrome patients that can represent potential treatment options [35], they were used as drug standards for comparison.
The binding affinities or Vina fitness score obtained from docking 2GTB with the native ligand (reference ligand), nelfinavir, lopinavir, and ten Aloe vera compounds are given in Table 1 .
Table 1.
Binding affinity (kcal/mol) of 2GTB and Ligands.
| Receptor PDB ID | Ligands | Binding Affinity (ΔG en Kcal/mol) |
|---|---|---|
| 2GTB | Lopinavir | −8.4 |
| Nelfinavir | −8.1 | |
| 6 | −7.9 | |
| 1 | −7.7 | |
| 8 | −7.7 | |
| Ref. Ligand | −7.4 | |
| 7 | −7.3 | |
| 3 | −7.1 | |
| 5 | −7.1 | |
| 9 | −7.0 | |
| 10 | −6.8 | |
| 2 | −6.7 | |
| 4 | −6.7 |
The binding affinity values range from 7.9 to 6.7 kcal/mol. Free enthalpies reveal a strong competition in the ligands 6, 1 and 8; 7, 3, 5 and 9; 10, 2 and 4. However, there is no doubt that the ligand 6 exhibits the higher binding affinity (7.9 kcal/mol), slightly higher to ligands 1 and 8 which have exactly the same binding energy (7.7 kcal/mol). The poorest ligands obtained in this study are ligands 2 and 4 that exhibit lower energy (6.7 kcal/mol). This complex has the lowest complexation energy (-6.7 kcal/mol). Overall, values of binding affinity of ligands suggest that three ligands (6, 1 and 8) obtained from the set of 10 compounds of Aloe vera are the best ligands that can be used as potential inhibitors of the COVID-19 Mpro. It is interesting to compare the binding site affinity of the best docked pose of ligands with that of the cocrystalised ligand. The docked binding energy of the co-crystalised ligand is placed in the fourth position, a bit lower than those of the three best ligands. The binding energies of all compounds are somewhat close to that of the reference ligand, except for compounds 10, 2 and 4. Further, the binding site of compounds is compared with one of the co-crystalised ligands used in this study (Discussion section).
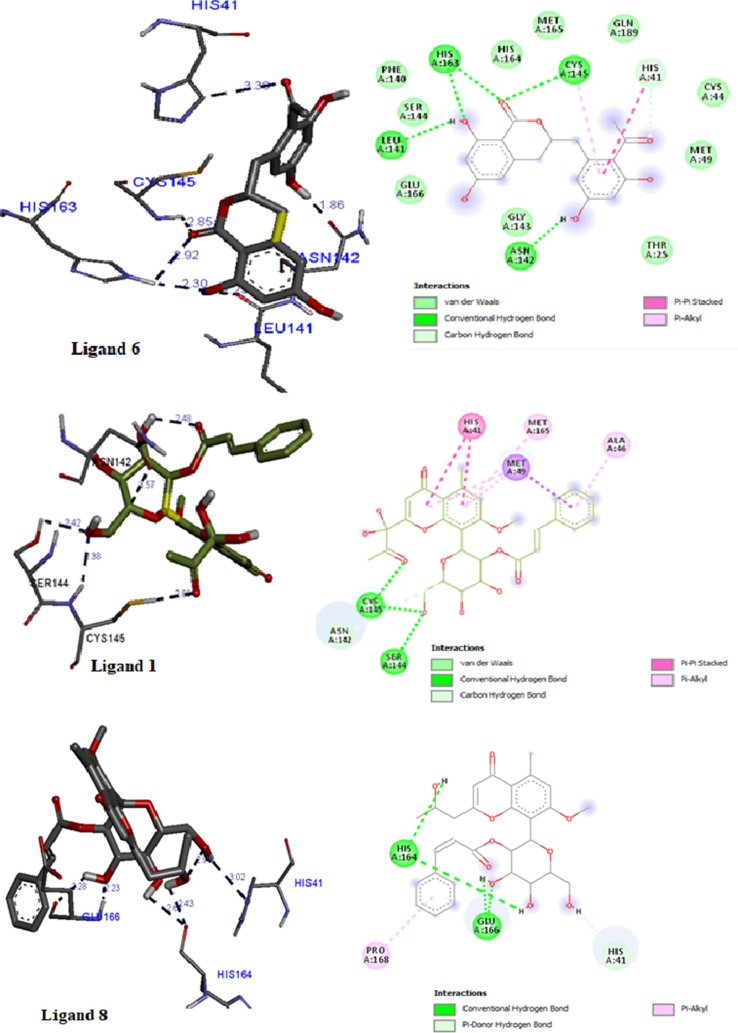
As showed in Fig. 4 a, the complexes between ligands and the receptor are mainly stabilized by hydrogen bonds involving O—H and C O groups where the ligand can act simultaneously as donor and acceptor [36]. However, other interactions like dispersion forces [37], π-π interactions [38], and hydrophobic interactions [39] contribute to the stability of the complexes (Fig. 4 b). H-bond distances and angles between the protein target and the best ligands along with the involved groups in the H-bonds are gathered in Table 2 , and Table 3 shows that all compounds from Aloe vera form hydrogen-bonds with the key amino acid residues of COVID-19 Mpro.
Fig. 4.
(a) (left). H-bonding interactions between the best ligands with COVID-19 Mpro protein target; (b) (right) All types of interactions between the best ligands with COVID-19 Mpro.
Table 2.
Hydrogen-bonds parameters (distances and angles) derived from docking of COVID-19 Mpro with the three best ligands.
| Ligand | AA Group | Ligand Group | δ (Å) | θ(°) |
|---|---|---|---|---|
| 1 | CYS145 | O C2 | 2.09 | 160 |
| HIS163 | H—O | 1.89 | 163 | |
| CYS145 | H—O | 1.77 | 160 | |
| 6 | HIS163 | H—O | 2.30 | 163 |
| LEU141 | H—O | 2.29 | 135 | |
| ASN142 | H—O | 1.86 | 153 | |
| CYS145 | O C2 | 2.24 | 158 | |
| CYS145 | H—O | 1.77 | 160 | |
| 8 | GLU166 | H—O | 2.23 | 155 |
| CYS145 | O C2 | 2.60 | 165 | |
| GLU166 | H—O | 2.28 | 140 | |
| HIS164 | H—O | 2.43 | 155 | |
Table 3.
Amino acids residues involved in H-bonds in both ligands and receptors.
| Inhibitor | Amino acids with H-bonds |
|---|---|
| 2GTB (PDB) | LYS5, ALA7, THR25, HIS41, MET49, TYR54, VAL125, TYR126, GLY127, PHE140, LEU141, ASN142, GLY143, SER144, CYS145, HIS163, HIS164, MET165, GLU166, LEU167, PRO168, HIS172, ASP187, ARG188, GLN189, GLN192, ALA198, LYS236, TYR237, GLN273 |
| Lopinavir | ARG188, GLU166, GLN189 |
| Nelfinavir | GLN189, GLU166, GLN192 |
| 1 | CYS145, HIS163, CYS145 |
| 2 | GLU166, ARG188 |
| 3 | GLU166, SER144, LEU141, HIS164, GLN168, CYS145, ASN142 |
| 4 | GLU166, LEU141, CYS145, ASN142, HIS41, GLN189, HIS164, |
| 5 | THR 26, HIS164, HIS163, LEU141, THR26 |
| 6 | HIS163, CYS145, LEU141, ASN142, CYS145 |
| 7 | GLU166, THR190, GLN189, ASN142, PHE140 |
| 8 | CYS145, GLU166, HIS164, GLU166 |
| 9 | GLN189, HIS141, GLY143, |
| 10 | ASN142, HIS41 |
Modelling of contact areas as illustrated in Fig. 4 shows lipophilic and hydrogen-bonding interactions involving typical hydrophobic and polar amino acid residues, which is crucial for the inhibition of COVID-19 Mpro [35].
3.1. Prediction of drug-likeness descriptors and toxicity
The potential inhibitors or the best docked ligands can be selected based on their binding energy. Nevertheless, ADMET (Absorption, Distribution, Metabolism, Excretion and Toxicity) properties are key steps and appropriate methodologies employing in drug development and discovery processes [40]. Physicochemical property is an important parameter of a molecule that influences efficacy, safety or metabolism which could be predicted by using Lipinski’s rule of five (RO5) that is: molecular mass < 500; Hydrogen-bond donors (HBD) < 5; Hydrogen-bond acceptors (HBA) < 10; and Log P < 5 [41].
The results obtained from ADME analysis with SwissADME software (Table 4 ) reveal that among the best three ligands predicted above, only the ligand 6 fully obeyed the Lipinski’s rule of five. It is worthy to remind here that this ligand is the best one, having the highest binding energy. The ligand 1 only meets one criterion (log P) while the ligand 8 meets two criteria (log P and HBD). Another ligand that meets all criteria is the ligand 2. However, it exhibits the lowest binding energy.
Table 4.
Lipinski parameters for dataset from SwissADME.
| Code | Compound name | Molecular weight (Da) | Log P | HBD | HBA | Violation | yes/no | Solubility | Log S (Mol/L) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 9-dihydroxyl-2-O-(z)-cinnamoyl-7-methoxy-Aloesin | 570.54 | 2.55 | 5 | 12 | 2 | no | Moderately soluble | −4.01 |
| 2 | Aloe-emodin | 270.40 | 1.95 | 3 | 5 | 0 | yes | Soluble | −3.43 |
| 3 | Aloin A | 418.89 | 2.20 | 7 | 9 | 1 | yes | Soluble | −2.95 |
| 4 | Aloin B | 418.39 | 1.40 | 7 | 9 | 1 | yes | Soluble | −2.95 |
| 5 | Elgonica dimer A | 686.61 | 2.79 | 10 | 134 | 3 | no | Poorly soluble | −6.57 |
| 6 | Feralolide | 344.3 | 1.80 | 4 | 7 | 0 | yes | Moderately soluble | −5.48 |
| 7 | IsoAloeresin | 556.56 | 2.70 | 5 | 11 | 2 | no | Moderately soluble | −5.06 |
| 8 | Aloeresin | 538.56 | 3.25 | 3 | 10 | 1 | yes | Moderately soluble | −4.30 |
| 9 | 7-O-methylAloeresin | 554.60 | 1.40 | 4 | 11 | 2 | no | Moderately soluble | −5.59 |
| 10 | Chrysophanol | 268.26 | 2.50 | 2 | 4 | 0 | yes | Moderately soluble | −4.11 |
Turning next to the potential toxicity of the three hits, we have predicted their mutanogenicity and carcinogenicity, as well as their hERG (the human ether-à-go-go-related gene) inhibition, BBB (the blood-brain barrier), P-gp substrate (P-glycoprotein), the inhibition constant and Cytochrome P450 inhibition (Drug-drug interaction) as metabolic indicators including CYPs: 1A2, 2C19, 2C9, 2D6 and 3A4. These parameters are gathered in Table 5 .
Table 5.
Pharmacokinetics and toxicity properties of the three potential inhibitors.
| Ligand | Ames_test | Carcino_Rat | BBB permeant | hERG | P-gp S. | Inhibition Constant (nM) | 1A2 | 2C19 | 2C9 | 2D6 | 3A4 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 6 | Mutagen | Positive | No | Low risk | No | 3.19 | No | No | No | No | No |
| 1 | Non-mutagen | Negative | No | Ambiguous | Yes | 22.53 | No | No | No | No | No |
| 8 | Non-mutagen | Negative | No | High risk | Yes | 27.80 | No | No | No | No | Yes |
The investigated pharmacokinetics and toxicity properties of the three molecules reveal that they are potential drugs candidates. The pharmacokinetic evaluation relating to inhibition of Cytochrome P450 and substrate of P-glycoprotein show that these molecules with highest binding affinity are found to be non-inhibitors of all CYPs except CYP3A4 for ligand 8. Interestingly, the inhibition constant values are in agreement with the binding affinities of the three compounds. Ligand 6 shows least inhibition constant value (3.19 nM). At this stage, one might remember that there are several tools for predicting ADMET properties and the results of two ADMET servers are not always the same. For instance, one of the widely used servers of ADMET predictions is vNN-ADMET [42]. ADMET properties were performed using this latter server for ligand 6 and the results provide no prediction for Ames mutagenicity in addition to some discrepancy where ligand 6 is found to be an inhibitor of CYP3A4.
4. Discussion
Herbal medicines and purified natural products provide a rich resource for novel antiviral drug development. Identification of the antiviral mechanisms from these natural agents has shed light on where they interact with the viral life cycle, such as viral entry, replication, assembly, and release, as well as on the targeting of virus–host-specific interactions. In this communication, we summarize the antiviral activity of Aloe vera and perform the molecular docking of some of its compounds (ten ligands) with the COVID-19 main protease to identify the potential inhibitors of the COVID-19 protein, as well as the ADME analysis.
Several proteins that play a key role in COVID-19 viral infection and are considered as potential pharmacological targets have recently been described. These include: 3CLpro (Coronavirus main protein); PLpro (papain-like protease); RdRp (RNA-dependent RNA polymerase); S protein (viral spike glycoprotein); TMPRSS2 (transmembrane protease serine 2); ACE2 (angiotensin-converting enzyme 2); and AT2 (angiotensin AT2 receptor) [43]. 3CLpro/Mpro (PDB ID: 2GTB) is currently recognized as one of the most studied pharmacological targets in the research and development of anti COVID-19 drugs, particularly in terms of both number of patents and number of potential drug candidates [6]. It is a proteolytic enzyme necessary for the cleavage of viral polyprotein into several functionally active protein units. Its choice as a pharmacological target in this study is justified by the fact that its active site is perfectly conserved and would not be affected by mutations. In addition, the high level of homology with 3CLpro/Mpro SARS-CoV-1 (~96%) allows the use of clinically approved anti-SARS-CoV-1 drugs as positive controls in the virtual screening of isolated secondary metabolites of A. vera; for validation in anti-COVID-19 clinical research in this health emergency [40].
Thus, it should be noted that the best anti COVID-19 drug candidate is the molecule that can specifically bind to one of the above-mentioned pharmacological targets to form a stable complex. Thermodynamically, this is a compound with the highest possible binding energy expressed in terms of free enthalpy (ΔG) in absolute terms. On the other hand, this molecule should fully obey the Lipinski’s rule of five. From ADME properties, the best candidate is the molecule that fully obeys the Lipinski’s rule of five. The results of virtual screening of 10 Aloe vera compounds based on the docking scores, hydrogen bonding interactions and the Lipinski’s rule of five suggest that three molecules (6, 1 and 8) are potential inhibitors of the protease 3CLPro, an enzyme that plays a key role in post-translational protein regulation, particularly the cleavage of viral polyprotein into functional protein units. The ligand 6 is the best drug candidate among the three hits since in addition to fully obey the Lipinski’s rule of five, it exhibits the highest binding energy. It should be mentioned that these three top compounds have already each shown biological activity, even though they are not yet approved by the U.S. Food and Drug Administration (FDA) as drugs. Kurizaki and co-workers reported the antioxidant, anticancer and anti-fungal activities of feralolide (ligand 6) [44], while ligand 1 exhibits anticancer and anti-inflammatory activities [45], and ligand 8 is reported to have an anti-inflammatory effect [46]. In fact, previous study dealing with virtual screening and repurposing of FDA approved drugs against COVID-19 main protease, showed that the temozolomide, an anticancer drug, can be used for COVID-19 treatment [47]. With regards to the anti-inflammatory effect, Fidan and Aydoğdu formulated a hypothesis according to which Montelukast that is a leukotriene inhibitor with an anti-inflammatory effect could be used against COVID-19 [48]. A recent study validated its binding with the Mpro and the docking score was assessed to −6.5 kcal/mol [49].
Compared with Lopinavir and Nelfinavir which are protease inhibitors recommended for the treatment of SARS and MERS, the docking affinity scores of the three top compounds are very close to those of two anti-HIV drugs. Finally, another interesting facet is to compare the ligand binding site residues of compounds with those of the native ligand. The amino acids residues that form hydrogen bonds for all compounds as well as the co-crystalised ligand are displayed in Table 4. It can be seen that all amino acids residues involving in the hydrogen bonding interaction in ligands 6, 1 and 8 in the Mpro’s pocket are present in the co-crystalised binding site. The binding site of the co-crystalised ligand is better represented by compound 3 which is in the top five and by compound 4 albeit having the lowest binding affinity.
To this end, Aloe vera can be considered as a high-potential anti-COVID-19 plant drug candidate for the management of this disease in the world. Indeed, several experimental studies have shown that the Aloe vera plant is endowed with formidable virucidal properties with a broad spectrum of action. From the point of view of toxicity, the innocuousness of the extracts of this plant has been proven experimentally both in vitro and in vivo. All this scientific evidence is thus generating a growing interest in the urgent galenic formulation of Aloe vera as an essential drug for the management of COVID-19. For example, Aloe vera contains virucidal secondary metabolites (anthraquinones) which, like Lopinavir (an antiretroviral), may act alone or in synergy with SARS-CoV-2 protease 3CLpro.
In addition to intrinsic antiviral properties, it is noteworthy that Aloe vera is also endowed with anti-inflammatory and immuno-modulatory properties. To this effect, it is not excluded that a phyto-drug based on Aloe vera extracts can attenuate in the patient the expression of pro-inflammatory factors and receptors likely to induce acute respiratory distress which is the main cause of mortality associated with COVID-19 while strengthening the immune system. As combination therapies based on viral protease inhibitors are the best therapeutic option, Aloe vera and its major secondary metabolites may play an important role in the management of COVID-19.
As most of in silico investigations concern the available FDA approved drugs as effective inhibitors of SARS-CoV-2, our results could pave the way for clinical research on anti COVID-19 herbal medicine. Indeed, in addition to its secondary metabolites endowed with virucidal properties, Aloe vera contains Zinc (40.8 ppm) [50]. Its use as an herbal medicine brings this trace element to the patient. This chemical element, although indispensable as an enzymatic co-factor, a slight increase in its intracellular concentration inhibits the replication of retroviruses including SARS-CoV-1 [43] important in the management of COVID-19.
5. Conclusion
In this great health emergency that the world is facing, a solution has to be found rapidly to save lives around the globe. While number of scientists are searching curable molecules using synthesis technics, another way is to promote traditional medicine for searching lead compounds (hits) from plants. Knowing that many phytocompounds are known, we can refer to the computational chemistry and bioinformatics resources to help in the study of different activities of medicinal plants such as Aloe vera.
This study was conducted in order to pinpoint the best drug candidates from the set of 10 compounds of Aloe vera using molecular docking and ADMET properties. The reactivity of the COVID-19 main protease (Mpro) with 10 isolated Aloe vera compounds showed that the most stable complex is obtained with feralolide or ligand 6 (−7.9 kcal/mol), followed by 9-dihydroxyl-2-O-(z)-cinnamoyl-7-methoxy-Aloesin or ligand 1 (−7.7 kcal/mol) and Aloeresin or ligand 8 (−7.7 kcal/mol). Finally, the Lipinski’s rule of five based-on ADME analysis confirms the ligand 6 to be the best drug candidate.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Peeri N.C., Shrestha N., Rhaman M.S., Zaki R., Tan Z., Bibi S., Baghbanzadeh M., Aghamohammadi N., Zhang W., Hague U. The SARS, MERS and novel coronavirus (COVID-19) epidemics, the newest and biggest global health threats: what lessons have we learned? Int. J. Epidemiol. 2020;20:1–10. doi: 10.1093/ije/dyaa033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Di W., Tiantian W., Qun L., Zhicong Y. The SARS-CoV-2 outbreak: what we know. Int. J. Infect. Dis. 2020;94:44–48. doi: 10.1016/j.ijid.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng J. SARS-CoV-2: an emerging coronavirus that causes a global threat. Int. J. Biol. Sci. 2020;16:1678–1685. doi: 10.7150/ijbs.45053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet (London, England) 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shu Y., McCauley J.J.E. GISAID: Global initiative on sharing all influenza data–from vision to reality. Euro. Surveill. 2017;22 doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu C., Zhou Q., Li Y., Garner L.V., Watkins S.P., Carter L.J., Smoot J., Gregg A.C., Daniels A.D., Jervey S., Albaiu D. Research and development on therapeutic agents and vaccines for COVID-19 and related human Coronavirus diseases. ACS Cent. Sc. 2020;25:315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.N. Chen, M. Zhou, X. Dong, J. Qu, F. Gong, Y. Han, Y. Qiu, J. Wang, et al., Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 395 (2020) 507–513. [DOI] [PMC free article] [PubMed]
- 8.Cao B., Wang Y., Wen D. A trial of lopinavir-ritonavir in adults hospitalized with severe COVID-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sarma P., Prajapat M., Medhi B. Therapeutic options for the treatment of 2019-novel coronavirus: an evidence-based approach. Indian J. Pharmacol. 2020;52:1–5. doi: 10.4103/ijp.IJP_119_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.C. Andrea, I. Giulia, I. Mariachiara, G. Antonino, E. Sharon, A systematic review on the efficacy and safety of chloroquine for the treatment of COVID-19, (2020) https://doi.org/10.1016/j.jcrc.2020.03.005.
- 11.Chong Y.P., Song J.Y., Seo Y.B., Choi J.P., Shin H.S. Antiviral treatment guidelines for Middle East Respiratory Syndrome. Infection Chemother. 2015;47:212–222. doi: 10.3947/ic.2015.47.3.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Needle D., Lountos G.T., Waugh D.S. Structures of the Middle East respiratory syndrome 274 coronavirus 3C-like protease reveal insights into substrate specificity. Acta Crystallogr. D Biol. 2015;275:1102–1111. doi: 10.1107/S1399004715003521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar V., Tan K.P., Wang Y.M., et al. Identification, synthesis and evaluation of SARS-CoV and MERS-CoV 3C-like protease inhibitors. Bioorg. Med. Chem. 2016;24:3035–3042. doi: 10.1016/j.bmc.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pillaiyar T., Manickam M., Namasivayam V., et al. An overview of severe acute respiratory syndrome–coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J. Med. Chem. 2016;59:6595–6628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020 doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qamar M.T., Maryam A., Muneer I., et al. Computational screening of medicinal plant phytochemicals to discover potent pan-serotype inhibitors against dengue virus. Sci. Rep. 2019;9:1–16. doi: 10.1038/s41598-018-38450-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ipona E.N., Inkoto L.C., Bongo G.N., Mulenga C.M., Ilinga B.L., Shetonde O.M., Mbala B.M., Tshilanda D.D., Mvingu B.K., Kayembe J.S., Mpiana P.T., Ngbolua K.N. Ethno-Botanical survey and ecological study of medicinal plants traditionally used against erectile dysfunction in Democratic Republic of the Congo. Biosci. Bioeng. 2019;4:85–91. [Google Scholar]
- 18.Tshilanda D.D., Onyamboko D.N.V., Babady B.P., Ngbolua K.N., Tshibangu D.S.T., Dibwe E.F., Mpiana P.T. Anti-sickling Activity of ursolic acid isolated from the leaves of Ocimum gratissimum L. (Lamiaceae) Nat. Prod. Bioprospect. 2015;5:215–221. doi: 10.1007/s13659-015-0070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mpiana P.T., Mudogo V., Tshibangu D.S.T., Kitwa E.K., Kanangila A.B., Lumbu J.B.S., Ngbolua K.N., Atibu E.K., Kakule M.K. Antisickling activity of anthocyanins from Bombax pentadrum, Ficus capensis and Ziziphus mucronata: Photodegradation effect. J. Ethnopharmacol. 2008;120:413–418. doi: 10.1016/j.jep.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 20.Kroll D.J. Concerns and needs for research in herbal supplement pharmacotherapy and safety. J. Herbal Pharmather. 2001;1:3–23. [Google Scholar]
- 21.K.N. Ngbolua, P.T. Mpiana, V. Mudogo, Pharmacopée traditionnelle et lutte contre la drépanocytose: Méthodes de sélection et d’évaluation de l’activité des plantes médicinales. Editions Universitaires Européennes, Riga : Latvia. ISBN : 978-613-9-51486-1, (2019).
- 22.Olamide E.A., Abatan M.O. Phytochemical and acute toxicity of ethanolic extract of Enantia chlorantha (oliv) stem bark in albino rats. Interdiscip. Toxicol. 2013;6:145–151. doi: 10.2478/intox-2013-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukherjee P.K., Nema N.K., Maity N., Mukherjee K., Harwansh R.K. Phytochemical and therapeutic profile of Aloe vera. J. Nat. Remedies. 2014;14:1–26. [Google Scholar]
- 24.Singh S., Sharma P.K., Kumar N., Dudhe R. Biological activities of Aloe vera. Int. J. Pharm. Technol. 2010;2:259–580. [Google Scholar]
- 25.Sharrif M.M., Sandeep K.V. Aloe vera their chemicals composition and applications: A review. Int. J. Biol. Med. Res. 2011;2:466–471. [Google Scholar]
- 26.Mpiana P.T., Ngbolua K.N., Tshibangu D.S.T., Kilembe J.T., Gbolo B.Z., Mwanangombo D.T., Inkoto C.L., Lengbiye E.M., Mbadiko C.L., Matondo A., Bongo G.N., Tshilanda D.D. Aloe vera (L.) Burm. f. as a potential anti COVID-19 plant: A minireview of its antiviral activity. Eur. J. Med. Plants. 2020;31:86–93. [Google Scholar]
- 27.Yang L., Wen K.S., Ruan X., Zhao Y.X., Wei F., Wang Q. Response of plant secondary metabolites to environmental factors. Molecules. 2018;23(2018):1–26. doi: 10.3390/molecules23040762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Z. Xu, C. Peng, Y. Shi, Z. Zhu, K. Mu, X. Wang, Nelfinavir was predicted to be a potential inhibitor of 2019-nCoV main protease by an integrative approach combining homology modelling, molecular docking and binding free energy calculation (2020).
- 29.Shapovalov M.S., Dunbrack R.L. A smoothed backbone-dependent rotamer library for proteins derived from adaptive kernel density estimates and regressions. Structure. 2011;19:844–858. doi: 10.1016/j.str.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim J.H., Yoon J.-Y., Seo Y.Y., Choi S.-K., Kwon S.J., Cho I.S., Jeong M.H., Kim Y.H., Choi G.S. Tyrosinase inhibitory components from Aloe vera and their antiviral activity. J. of Enzyme Inhib. Med. Chem. 2017;32:78–83. doi: 10.1080/14756366.2016.1235568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daina A., Olivier M., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, druglikeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee S.K., Chang G.S., Lee I.H., Chung J.E., Sung K.Y., No K.T. The preADME: PC-based program for batch prediction of ADME properties. EuroQSAR. 2004;9:5–10. [Google Scholar]
- 34.Verdonk M.L., Cole J.C., Hartshon J.M., Murray W.C., Taylor R.D. Improved protein-ligand docking using gold. Proteins. 2003;52:609–623. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- 35.S. Khaerunnisa, K. Hendra, A. Rizki, S. Suhartati, S. Soetjipto, Potential inhibitor of COVID-19 Main Protease (Mpro) from several medicinal plant compounds by molecular docking study, (2020) doi:10.20944/preprints202003.0226.v1.
- 36.Matondo A., Mukeba C.T., Muzomwe M., Nsimba B.M., Tsalu P.V. Unravelling syn- and anti- orientation in the regioselectivity of carbonyl groups of 5-fluorouracil an anticancer drug toward proton donors. Chem. Phys. Lett. 2018;712:196–207. [Google Scholar]
- 37.Trujillo C., Sanchez-Sanz G. A Study of π−π stacking interactions and aromaticity in polycyclic aromatic hydrocarbon/nucleobase complexes. ChemPhysChem. 2016;17:395–405. doi: 10.1002/cphc.201501019. [DOI] [PubMed] [Google Scholar]
- 38.Kasende O.E., Matondo A., Muya J.T., Scheiner S. Interactions between temozolomide and guanine and its S and Se-substituted analogues. Int. J. Quant. Chem. 2017;117:157–169. [Google Scholar]
- 39.Muya J.T., Mwanangombo D.T., Tsalu P.V., Mpiana P.T., Tshibangu D.S.T., Hoeil C. Conceptual DFT study of the chemical reactivity of four natural products with anti-sickling activity. SN Appl. Sci. 2019;1:1457–1474. [Google Scholar]
- 40.Ye Z., Yang Y., Li X., Cao D., Ouyang D. An Integrated Transfer Learning and Multitask Learning Approach for Pharmacokinetic Parameter Prediction. Mol. Pharm. 2018;16:533–541. doi: 10.1021/acs.molpharmaceut.8b00816. [DOI] [PubMed] [Google Scholar]
- 41.Lagorce D., Douguet D., Miteva M.A., Villoutreix B.O. Computational analysis of calculated physicochemical and ADMET properties of protein-protein interaction inhibitors. Sci. Rep. 2017;7:46277. doi: 10.1038/srep46277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schyman P., Liu R., Desai V., Wallqsvit A. vNN web server for ADMET predictions. Front. Pharmacol. 2017;8:889–902. doi: 10.3389/fphar.2017.00889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin L.T., Hsu W.H., Lin C.C. Antiviral natural products and herbal medicines. J. Trad. Complement Med. 2014;4:24–35. doi: 10.4103/2225-4110.124335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karuzaki A., Watanabe T., Devkota H.P. Chemical constituent from the flowers of Aloe arborescens. Nat. Prod. Commun. 2019:1–4. [Google Scholar]
- 45.Abd-Alla H.I., Shaaban M., Shaaban K.A., Abu-Gabal N.S., Shalaby N.M.M., Laatsch H. New bioactive compounds from Aloe hijazensis. Nat. Prod. Res. 2009;23:1035–1049. doi: 10.1080/14786410802242851. [DOI] [PubMed] [Google Scholar]
- 46.Speranza G., Morelli C.F., Tubaro A., Altinier G., Duri L., Manitto P. Planta Med. 2005;71:79–81. doi: 10.1055/s-2005-837756. [DOI] [PubMed] [Google Scholar]
- 47.Kandeel M., Al-Nazawi M. Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease. Life Sc. 2020;257 doi: 10.1016/j.lfs.2020.117627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fidan C., Aydoğdu A. As a potential treatment of COVID-19: Montelukast. Med. Hypothesis. 2020 doi: 10.1016/j.mehy.2020.109828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huynh T., Wang H., Luan B. In silico exploration of the molecular mechanism of clinically oriented drugs for possible inhibiting SARS-CoV-2’s main protease. J. Chem. Phys. Lett. 2020;11:4413–4420. doi: 10.1021/acs.jpclett.0c00994. [DOI] [PubMed] [Google Scholar]
- 50.te Velthuis A.J.W., van den Worm S.H.E., Sims A.C., Baric R.S., Snijder E.J., van Hemert M.J. Zn2+ inhibits Coronavirus and arterivirus RNA polymerase activity in vitro and zinc ionophores block the replication of these viruses in cell culture. Plos Pathogens. 2010;6:1–10. doi: 10.1371/journal.ppat.1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]