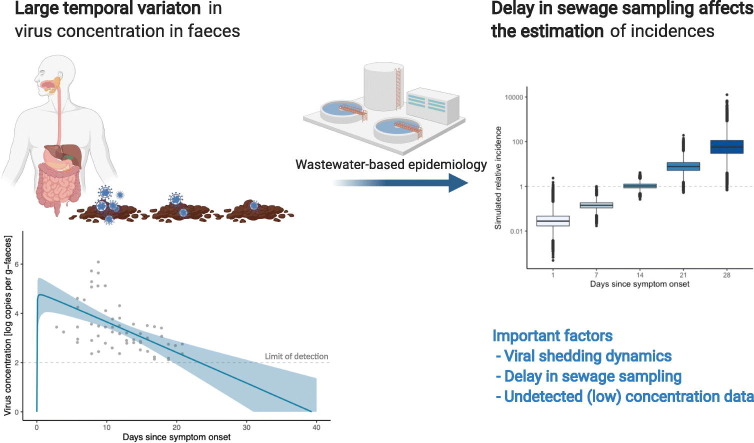
Abstract
Wastewater-based epidemiology (WBE) is one of the most promising approaches to effectively monitor the spread of COVID-19. The virus concentration in faeces and its temporal variations are essential information for WBE. While some clinical studies have reported SARS-CoV-2 concentrations in faeces, the value varies amongst patients and changes over time. The present study aimed to examine how the temporal variations in the concentration of virus in faeces affect the monitoring of disease incidence. We reanalysed the experimental findings of clinical studies to estimate the duration of virus shedding and the faecal virus concentration. Available experimental data as of 23 October 2020 were collected. The viral shedding kinetics was modelled, and the dynamic model was fitted to the collected data by a Bayesian framework. Using posterior distributions, the duration of viral shedding and the concentration of virus copies in faeces over time were computed. We estimated the median concentration of SARS-CoV-2 in faeces as 3.4 (95% CrI: 0.24–6.5) log copies per gram-faeces over the shedding period, and our model implied that the duration of viral shedding was 26.0 days (95% CrI: 21.7–34.9), given the current standard quantification limit (Ct = 40). With simulated incidences, our results also indicated that a one-week delay between symptom onset and wastewater sampling increased the estimation of incidence by a factor of 17.2 (i.e., 101.24 times higher). Our results demonstrated that the temporal variation in virus concentration in faeces affects microbial monitoring systems such as WBE. The present study also implied the need for adjusting the estimates of virus concentration in faeces by incorporating the kinetics of unobserved concentrations. The method used in this study is easily implemented in further simulations; therefore, the results of this study might contribute to enhancing disease surveillance and risk assessments that require quantities of virus to be excreted into the environment.
Keywords: SARS-CoV-2, COVID-19, Viral shedding, Faeces, Wastewater-based epidemiology (WBE), Mathematical modelling
Graphical abstract
1. Introduction
As of 23 October 2020, the novel coronavirus disease 2019 (COVID-19) has spread all over the world. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of this disease. The infection causes general symptoms such as fever, cough, shortness of breath and diarrhoea (Guan et al., 2020), and the disease progression differs according to age (Omori et al., 2020; X.-W. Xu et al., 2020) and clinical history (Wu and McGoogan, 2020). Previous epidemiological studies on COVID-19 have reported that a substantial proportion of infected individuals were even asymptomatic (Lavezzo et al., 2020; Mizumoto et al., 2020). These disease characteristics have caused complications in controlling the transmission of SARS-CoV-2.
To identify hidden chains of infection, more effective ways of monitoring the spread of the disease are required. In this context, wastewater-based epidemiology (WBE) has been attracting attention as a promising potential approach (Larsen and Wigginton, 2020; Lodder and de Roda Husman, 2020). Clinical observations have demonstrated that there is prolonged virus shedding in faeces, ranging from 1 to 33 days (Gupta et al., 2020; Wu et al., 2020; Y. Xu et al., 2020), and therefore wastewater surveillance can enable the monitoring of excreted viruses through which we can capture the presence and possibly the numbers of infected individuals, regardless of their symptoms. As a monitoring system, researchers have successfully reported their detection or quantification results in various countries, such as Australia (Ahmed et al., 2020), Japan (Haramoto et al., 2020), etc. (La Rosa et al., 2020; Medema et al., 2020; Rimoldi et al., 2020). The Netherlands and several municipalities in the United States have already started utilising WBE in practice as a part of their surveillance systems (CDC, 2020; The Dutch National Institute for Public Health and the Environment (RIVM), 2020).
One of the most important quantities for WBE is the SARS-CoV-2 concentration in faeces. If the aim of research is to simulate the potential number of cases in a sewershed, the virus concentration in faeces and its time course would affect the simulation. Earlier WBE studies have attempted to simulate the incidence using sewage data (Ahmed et al., 2020; Gonzalez et al., 2020); however, no previous studies have accounted for temporal variations in the virus concentration in faeces, and thus undetectable concentrations that might occur after several weeks from symptom onset have not been considered. In addition, while several clinical studies have reported that there is prolonged SARS-CoV-2 shedding in faeces with binary results (i.e. positive/negative) (Wu et al., 2020) or threshold cycle (Ct) values over time (Han et al., 2020; Y. Xu et al., 2020), quantitative estimates of possible virus excretions have yet to be fully explored.
The present study aims to examine how temporal variations in virus concentrations in faeces affect disease incidence monitoring systems such as WBE by estimating the duration of virus shedding and the virus concentration in faeces. Available time course data of virus concentration in faeces in clinical studies were screened, and subsequently a mathematical model that describes the kinetic viral shedding process was applied to the collected data. The present study also simulated the potential bias for the estimation of incidence with WBE by using an estimated virus shedding time course.
2. Methods
2.1. Data collection
A literature review was conducted to collect available experimental data as of 23 October 2020 using Google Scholar, PubMed and MedRxiv. Seven articles containing the following information were assessed with full text reviews: (a) exact virus copies or Ct values, (b) days from symptom onset or days from hospitalisation and (c) the amount of stool that was quantified by quantitative polymerase chain reaction (qPCR) (Han et al., 2020; Huang et al., 2020; Tan et al., 2020; van Kampen et al., 2020; Wang et al., 2020; Wölfel et al., 2020; Y. Xu et al., 2020). As a result of this review, only patient data reported in Wölfel et al. (2020) were used for further analysis; the other studies were excluded because they reported only Ct values and the experimental details required conversion to virus concentration in faeces [copies/grams (g)-faeces].
The analysed data consist of hospitalised patients involved in a large cluster that occurred in Munich, Germany between 23 and 27 January 2020 (Wölfel et al., 2020). Since it is difficult in retrospective studies to collect stool samples before the onset of symptoms, the data were expressed in the unit of time ‘days after symptom onset’ (Wölfel et al., 2020). The sample size reported in this study was insufficient to stratify the study population into sub-groups and thus exhaustive data were analysed to estimate the temporal variations in virus shedding.
2.2. Model
The time course of virus shedding is modelled as two processes. First, the virus is accumulated in a human host with concentration c 1(t) and then, at a certain point, the virus is shed in concentration c 2(t). According to Teunis et al. (2015), the dynamics can be formulated simply as follows:
| (1) |
where A is the initial concentration of viruses and α and β are the transport rates of viruses. By solving this system, the time course of virus concentration in faeces can be written as
where (see a more detailed biological description in the original study (Teunis et al., 2015)). In the present study, t = 0 was defined as the day of symptom onset, assuming that the accumulation of viruses was complete when the individuals started shedding viruses in their faeces.
One of the properties of this model is that one can analytically calculate important quantities such as peak concentration. Finding the extrema of C(t), the time to peak concentration t peak is written as , and the peak concentration C peak is consequently obtained as .
To define the end of virus shedding, Ct = 40 was used as the threshold (corresponding to approximately 102 copies per g-faeces), following both the original study and the recommended quantification threshold value in current qPCR protocols (Bustin et al., 2009; Wölfel et al., 2020). The duration of virus shedding t total was then computed by seeking the intersection point of C(t) and the threshold value.
2.3. Model fit
To jointly estimate the parameters (C 0, α, β), a hierarchical Bayesian framework was used to account for individual variations and provide for the uncertainty of the estimates. Here, the expected value is denoted as u = ln (C(t)) with natural log scale. By assuming that the log-transformed concentration is normally distributed, the likelihood is simply
where y is the observed data (i.e. the log-scaled virus concentration data) and ϵ is the standard deviation. The variation in the observed data in a hierarchical model framework is described as
with mean vector μ = (μ α, μ β, μ C0, μ ϵ) and standard deviation vector σ = (σ α, σ β, σ C0, σ ϵ).
The hyperparameters in this framework were specified positive non-informative prior probability distributions for parameters μ and σ. Using the Hamiltonian Monte Carlo (HMC) algorithm, we obtained estimates and 95% credible intervals (CrI) by sampling the posterior distributions. All of the computations above were implemented in R-4.0.0 with a package {rstan}-2.21.2 (R Core Team, 2020; Stan Development Team, 2020).
2.4. Simulation of relative incidence
Suppose that the aim here is to calculate the potential number of infected individuals per day using WBE surveillance. If observational data on the average number of SARS-CoV-2 ribonucleic acid (RNA) copies in sewage per day and complete information on the symptom onset of all of the infected individuals are available, the crude daily incidence with the average daily water volume and the average amount of faeces per person per day is simulated as:
where is the average virus concentration in faeces.
The crucial point here is that the virus concentration is the variable that is dependent on the time between sewage sampling and symptom onset. Thus, by denoting the concentration in faeces at day τ from symptom onset as , the sensitivity of the expected incidence to the virus concentration in faeces was examined in terms of the relative incidence defined as
where is the baseline concentration defined by the median of the (quantified) concentration reported in Wolfel et al. (Wölfel et al., 2020). For simplicity, the analysis assumes that all of the individuals had the same symptom onset (τ days before sampling) and the dates were simulated as τ = 1, 7, 14, 21, 28 by changing and fixing the other conditions.
3. Results
3.1. Time course of viral shedding
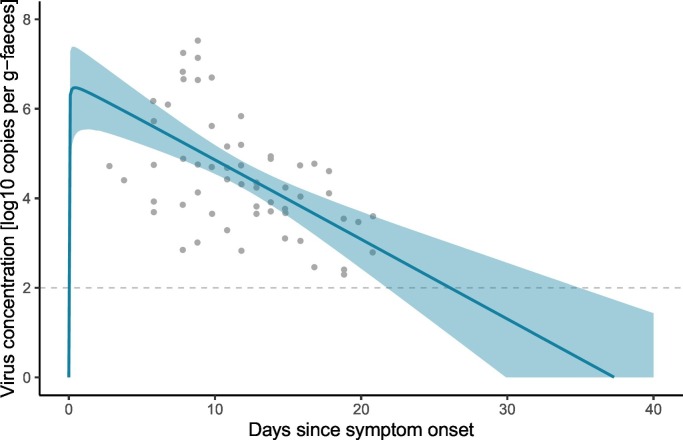
The estimated curve for the time course of virus shedding is shown in Fig. 1 . The results of model fitting indicate that the virus concentration in faeces rapidly increases after symptom onset, and that prolonged shedding occurs until the concentration becomes lower than the quantification limits. The time to peak concentration t peak was estimated to be 0.34 (95% CrI: 0.20–1.9) days. If the end of virus shedding is defined as the time point at which the virus concentration becomes lower than the quantification limit of qPCR (in this analysis, two log(copies)), the duration of virus shedding was estimated to be 26.0 days (95% CrI: 21.7–34.9) days.
Fig. 1.
Fitted model to the observed virus concentration in faeces. Grey plots represent observed data, the blue curve shows the posterior median and the shaded zones are the 95% credible intervals. The dashed line indicates the quantification limit threshold of qPCR (2 log copies/g-faeces). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2. Expected virus concentration
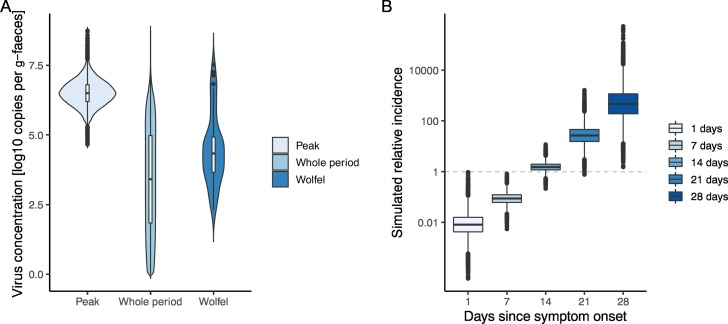
With HMC posterior samples, the possible range of virus concentrations in faeces during the shedding period and at peak concentration was estimated. Those estimates with uncertainty are shown in Fig. 2(A). If the quantitative data from Wölfel et al. to calculate the expected virus concentration is used (Wölfel et al., 2020), the median was estimated to be 4.3 (95% percentile: 2.4–7.2) [log copies per g-faeces]; however, the estimates in the present study suggest that the median of concentration over the whole shedding period was 3.4 (95% CrI: 0.24–6.5) [log copies per g-faeces]. For peak concentration C peak, the estimate was 6.5 (95% CrI: 5.6–7.4) [log copies per g-faeces].
Fig. 2.
Estimated virus concentration in faeces (A) and simulated relative incidence caused by changing the delay between wastewater sampling and symptom onset (B). (A) From the left of the panel, violin plots of estimated concentrations in faeces on the day of peak shedding, of the entire shedding period and of raw quantified experimental data reported by Wölfel et al. are visualised (Wölfel et al., 2020). (B) Relative incidences for different sewage samples are plotted, in which we assumed that all of the individuals had the same symptom onset (τ days before sampling, as shown on the x-axis). By definition, the relative incidence equals 1 when the quantitative data in Wölfel et al. are used for the virus concentration in faeces (Wölfel et al., 2020).
3.3. Simulated relative incidence
To assess the potential impact of the timing of sewage sampling for WBE, the relative incidence was simulated, by changing the delay between sewage sampling and symptom onset. Fig. 2(B) illustrates the results of simulated relative incidences in which the value equals 1 when Wölfel et al.'s quantified data are used for the virus concentration in faeces (Wölfel et al., 2020). The results indicate that the (log-converted) expected incidence increased linearly with longer intervals, and a one-week delay increased the estimated incidence by a factor of 17.2 (i.e., 101.24 times higher).
4. Discussion
In this study, the time course of SARS-CoV-2 shedding in faeces by applying a simple dynamic model was estimated. Results from the analysis show that there is a large temporal variation in virus concentration in faeces (Fig. 1), resulting in potential bias when simulating the number of infected cases with the data on virus concentration in sewage (Fig. 2). The model describes the kinetics of the accumulation and excretion processes (Eq. (1)) and thereby accounts for unobserved concentrations that were under the detection limits of real-time PCR.
The duration of virus shedding in faeces was estimated to be 26.0 days (95% CrI: 21.7–34.9) days from symptom onset. This result is consistent with previous clinical reports (Gupta et al., 2020; Wu et al., 2020). While earlier studies have suggested possible durations with only binary (positive/negative) results (Wu et al., 2020), the present analysis provides a credible range of shedding duration with uncertainty. Furthermore, the proposed method is easily applicable to other data when it becomes available, or even when the threshold values are changed. Several clinical studies have reported Ct values over time, but the amount of stool in RNA extractions was not recorded (Han et al., 2020; Huang et al., 2020; van Kampen et al., 2020; Wang et al., 2020). If such information is available, that evidence would be synthesized with the proposed method and induce more valuable implications for both the natural history of COVID-19 infection and environmental surveillance.
In the context of WBE, the findings on the peak and median concentrations indicated that there might be bias if simulations that are based on faecal virus concentration data use only quantified experimental results by truncating unobserved concentrations. There are large variations in concentration depending on the timing of taking faecal samples (Fig. 1, Fig. 2(A)); a one-week delay increases the estimation of incidence by a factor of 17.2 (Fig. 2(B)). Previous studies have attempted to simulate the potential incidence with the quantified virus concentration in sewage (Ahmed et al., 2020; Gonzalez et al., 2020); however, since there is a strong dependency between the observed concentration in sewage and the sampling delay from symptom onset, the interpretation of quantities averaged over time must be carefully specified. Ideally, epidemiological data such as case series based on symptom onset should be incorporated to simulate incidences more precisely.
There are several limitations in this study. First, the analysed data are a single cohort that consists of diagnosed cases in Germany. The disease progression might differ by age, race or medical history and, consequently, those factors might affect the estimates. Since the sample size was not sufficient to stratify the data into sub-groups, additional data are needed for further analysis. An implication for ongoing clinical studies is that the quantitative description about stool samplings (e.g., the amount of swabbed stool) and the clear definition of disease progressions would be beneficial, especially for synthesizing available evidence. Second, the modelling method assumed that the virus shedding in faeces starts from symptom onset. If the peak had occurred before symptom onset, it would not be captured with this analysis. While earlier clinical studies have indicated that the peak in faeces might be around symptom onset (Han et al., 2020; Y. Xu et al., 2020) and in throat swabs (He et al., 2020), there was no publicly available data that contain viral loads from the day of infection to symptom onset. To obtain a conclusive estimate of the peak timing, observational data such as (prospective) periodic stool sampling or human challenge studies are needed.
Despite the abovementioned limitations, the present analysis would be beneficial for surveillance systems in different sectors. Findings of the present study indicate that the temporal variation in virus concentrations affects microbial monitoring systems such as WBE and repeated testing in hospitals. Especially for the estimation of incidence, the virus concentrations in faeces must be adjusted by incorporating the kinetics of unobserved concentrations (i.e. concentrations lower than the quantification limits). The method used in this study is easily implemented in other simulations and therefore the results of this study might contribute to enhancing disease surveillance and risk assessments that require data on the quantities of viruses that have been excreted into the environment.
Funding
This work was supported by JSPS KAKENHI (Grant Number 20J00793), JST-Mirai Program (Grant Number JPMJMI18DB), and Grants-in-Aid for Crossdepartmental Young Researcher Grants of Hokkaido University, Japan.
CRediT authorship contribution statement
Fuminari Miura: Conceptualization, Methodology, Data curation, Formal analysis, Writing - Original draft, Masaaki Kitajima: Writing - Review & editing, Funding acquisition, Ryosuke Omori: Methodology, Writing - Original draft, Funding acquisition.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Acknowledgment
The authors would like to thank Enago for the English language editing.
Editor: Damia Barcelo
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728:138764. doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., Wittwer C.T. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- CDC National Wastewater Surveillance System (NWSS) 2020. https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/wastewater-surveillance.html URL. (WWW Document)
- Gonzalez R., Curtis K., Bivins A., Bibby K., Weir M.H., Yetka K., Thompson H., Keeling D., Mitchell J., Gonzalez D. COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res. 2020;186:116296. doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.-J., Ni Z.-Y., Hu Y., Liang W.-H., Ou C.-Q., He J.-X., Liu L., Shan H., Lei C.-L., Hui D.S.C., Du B., Li L.-J., Zeng G., Yuen K.-Y., Chen R.-C., Tang C.-L., Wang T., Chen P.-Y., Xiang J., Li S.-Y., Wang J.-L., Liang Z.-J., Peng Y.-X., Wei L., Liu Y., Hu Y.-H., Peng P., Wang J.-M., Liu J.-Y., Chen Z., Li G., Zheng Z.-J., Qiu S.-Q., Luo J., Ye C.-J., Zhu S.-Y., Zhong N.-S., China Medical Treatment Expert Group for Covid-19 Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Parker J., Smits S., Underwood J., Dolwani S. Persistent viral shedding of SARS-CoV-2 in faeces - a rapid review. Color. Dis. 2020;22:611–620. doi: 10.1111/codi.15138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M.S., Seong M.-W., Kim N., Shin S., Cho S.I., Park H., Kim T.S., Park S.S., Choi E.H. Viral RNA load in mildly symptomatic and asymptomatic children with COVID-19, Seoul, South Korea. Emerg. Infect. Dis. 2020;26:2497–2499. doi: 10.3201/eid2610.202449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020;737:140405. doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X., Lau Y.C., Wong J.Y., Guan Y., Tan X., Mo X., Chen Y., Liao B., Chen W., Hu F., Zhang Q., Zhong M., Wu Y., Zhao L., Zhang F., Cowling B.J., Li F., Leung G.M. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- Huang J., Mao T., Li S., Wu L., Xu X., Li H., Xu C., Su F., Dai J., Shi J., Cai J., Huang C., Lin Xuan, Chen D., Lin Xiaoling, Sun B., Tang S. Long period dynamics of viral load and antibodies for SARS-CoV-2 infection: an observational cohort study. medRxiv. 2020 doi: 10.1101/2020.04.22.20071258. [DOI] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736:139652. doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen D.A., Wigginton K.R. Tracking COVID-19 with wastewater. Nat. Biotechnol. 2020;38:1151–1153. doi: 10.1038/s41587-020-0690-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavezzo E., Franchin E., Ciavarella C., Cuomo-Dannenburg G., Barzon L., Del Vecchio C., Rossi L., Manganelli R., Loregian A., Navarin N., Abate D., Sciro M., Merigliano S., De Canale E., Vanuzzo M.C., Besutti V., Saluzzo F., Onelia F., Pacenti M., Parisi S.G., Carretta G., Donato D., Flor L., Cocchio S., Masi G., Sperduti A., Cattarino L., Salvador R., Nicoletti M., Caldart F., Castelli G., Nieddu E., Labella B., Fava L., Drigo M., Gaythorpe K.A.M., Imperial College COVID-19 Response Team, Brazzale A.R., Toppo S., Trevisan M., Baldo V., Donnelly C.A., Ferguson N.M., Dorigatti I., Crisanti A. Suppression of a SARS-CoV-2 outbreak in the Italian municipality of Vo'. Nature. 2020;584:425–429. doi: 10.1038/s41586-020-2488-1. [DOI] [PubMed] [Google Scholar]
- Lodder W., de Roda Husman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020;5:533–534. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Mizumoto K., Kagaya K., Zarebski A., Chowell G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Eurosurveillance. 2020;25:2000180. doi: 10.2807/1560-7917.ES.2020.25.10.2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omori R., Matsuyama R., Nakata Y. The age distribution of mortality from novel coronavirus disease (COVID-19) suggests no large difference of susceptibility by age. Sci. Rep. 2020;10:16642. doi: 10.1038/s41598-020-73777-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2020. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D., Maresca M., Longobardi C., Mancon A., Romeri F., Pagani C., Cappelli F., Roscioli C., Moja L., Gismondo M.R., Salerno F. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 2020;744:140911. doi: 10.1016/j.scitotenv.2020.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stan Development Team . 2020. RStan: The R Interface to Stan. R Package Version 2.21.2. [Google Scholar]
- Tan W., Lu Y., Zhang J., Wang J., Dan Y., Tan Z., He X., Qian C., Sun Q., Hu Q., Liu H., Ye S., Xiang X., Zhou Y., Zhang W., Guo Y., Wang X.-H., He W., Wan X., Sun F., Wei Q., Chen C., Pan G., Xia J., Mao Q., Chen Y., Deng G. Viral kinetics and antibody responses in patients with COVID-19. medRxiv. 2020 doi: 10.1101/2020.03.24.20042382. [DOI] [Google Scholar]
- Teunis P.F.M., Sukhrie F.H.A., Vennema H., Bogerman J., Beersma M.F.C., Koopmans M.P.G. Shedding of norovirus in symptomatic and asymptomatic infections. Epidemiol. Infect. 2015;143:1710–1717. doi: 10.1017/S095026881400274X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Dutch National Institute for Public Health and the Environment (RIVM) Coronavirus monitoring in sewage research. 2020. https://www.rivm.nl/en/covid-19/sewage URL. (WWW Document)
- van Kampen J.J.A., van de Vijver D.A.M.C., Fraaij P.L.A., Haagmans B.L., Lamers M.M., Okba N., van den Akker J.P.C., Endeman H., Gommers D.A.M.P.J., Cornelissen J.J., Hoek R.A.S., van der Eerden M.M., Hesselink D.A., Metselaar H.J., Verbon A., de Steenwinkel J.E.M., Aron G.I., van Gorp E.C.M., van Boheemen S., Voermans J.C., Boucher C.A.B., Molenkamp R., Koopmans M.P.G., Geurtsvankessel C., van der Eijk A.A. Shedding of infectious virus in hospitalized patients with coronavirus disease-2019 (COVID-19): duration and key determinants. medRxiv. 2020 doi: 10.1101/2020.06.08.20125310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zheng J., Guo L., Yao H., Wang L., Xia X., Zhang W. Fecal viral shedding in COVID-19 patients: clinical significance, viral load dynamics and survival analysis. Virus Res. 2020;289:198147. doi: 10.1016/j.virusres.2020.198147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., Yin H., Xiao Q., Tang Y., Qu X., Kuang L., Fang X., Mishra N., Lu J., Shan H., Jiang G., Huang X. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020;5:434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Li X., Zhu B., Liang H., Fang C., Gong Y., Guo Q., Sun X., Zhao D., Shen J., Zhang H., Liu H., Xia H., Tang J., Zhang K., Gong S. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2020;26:502–505. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.-W., Wu X.-X., Jiang X.-G., Xu K.-J., Ying L.-J., Ma C.-L., Li S.-B., Wang H.-Y., Zhang S., Gao H.-N., Sheng J.-F., Cai H.-L., Qiu Y.-Q., Li L.-J. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: retrospective case series. BMJ. 2020;368:m606. doi: 10.1136/bmj.m606. [DOI] [PMC free article] [PubMed] [Google Scholar]