Abstract
Immune responses differ between women and men, and type I interferon (IFN) responses following Toll-like receptor 7 (TLR7) stimulation are higher in women. The precise mechanisms driving these sex differences in immunity are unknown. To investigate possible genetic factors, we quantify escape from X chromosome inactivation (XCI) for TLR7 and four other genes (RPS6KA3, CYBB, BTK, and IL13RA1) at the single plasmacytoid dendritic cell (pDC) level. We observe escape from XCI for all investigated genes, leading to biallelic expression patterns. pDCs with biallelic gene expression have significantly higher mRNA levels of the respective genes. Unstimulated pDCs with biallelic TLR7 expression exhibit significantly higher IFNα/β mRNA levels, and IFNα exposure results in significantly increased IFNα/β protein production by pDCs. These results identify unanticipated heterogeneity in escape from XCI of several genes in pDCs and highlight the important contribution of X chromosome factors to sex differences in type I IFN responses, which might explain observed sex differences in human diseases.
Keywords: escape from X chromosome inactivation, IFNα, pDCs, sex differences, TLR7, type I IFN, XCI
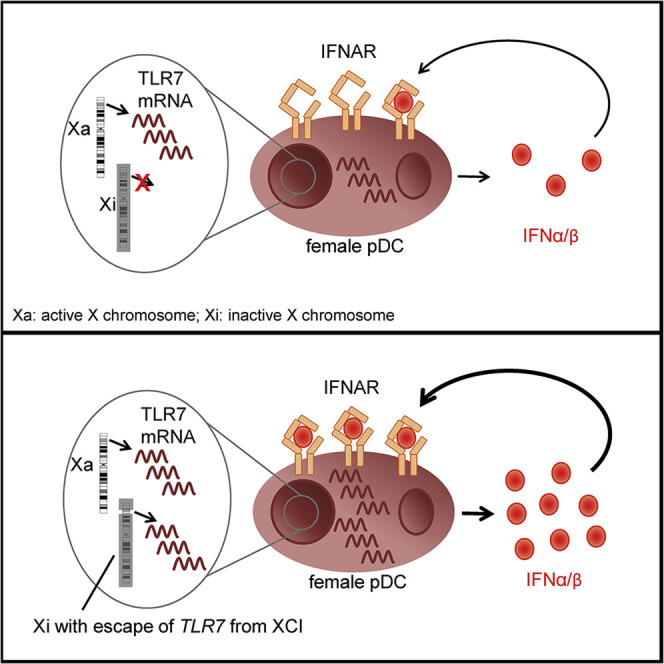
Graphical Abstract
Highlights
-
•
TLR7, CYBB, RPS6KA3, BTK, and IL13RA1 can escape XCI in human pDCs
-
•
Female pDCs with escape from XCI have higher mRNA levels of the respective genes
-
•
pDCs with escape from XCI of TLR7 have higher IFNα/β mRNA levels
In human female pDCs with two X chromosomes, Hagen et al. show that TLR7 escape from X chromosome inactivation (XCI) promotes higher TLR7 mRNA and higher IFNα/β mRNA at the single-cell level. This finding highlights the contribution of X chromosomal factors to sex differences in type I IFN responses.
Introduction
Immune responses differ between women and men, resulting in sex-specific differences in the manifestations of infectious and autoimmune diseases. A bias between females and males in the incidence and outcome of infectious diseases has been described for viral (HIV, hepatitis C virus [HCV], and influenza), bacterial (tuberculosis), and parasitic (amebiasis and leishmaniasis) infections (Sterling et al., 2001; Meditz et al., 2011; Grebely et al., 2014; Hertz and Schneider, 2019; Bernin and Lotter, 2014; Hoffmann et al., 2015). The current severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic also exhibits a sex difference with an estimated case mortality rate of 1.7 (male): 1 (female) in Europe (Bunders and Altfeld, 2020; Scully et al., 2020). Furthermore, immune responses toward vaccines differ between women and men, with most vaccines eliciting stronger immune responses in adult females (Flanagan et al., 2017). Females are also more likely to suffer from vaccine-induced adverse effects (Flanagan et al., 2017). The prevalence of autoimmune diseases evidences a clear sex bias, with the majority of autoimmune diseases being more prevalent in females than in males (Amur et al., 2012; Markle and Fish 2014). These data show that the ramifications of a sex bias in immune responses are manifested in the incidence and severity of infectious and autoimmune diseases, as well as in differences in vaccine-induced immune responses.
One signaling pathway that has been demonstrated to exhibit a strong sex bias in humans and mice is interferon α (IFNα) production of plasmacytoid dendritic cells (pDCs) following Toll-like receptor 7 (TLR7) stimulation (Meier et al., 2009; Ziegler et al., 2017; Seillet et al., 2012). This immunological sex difference has been suggested to contribute to the observed sex bias in the progression of HIV-1 disease (Addo and Altfeld, 2014) and the incidence and pathogenesis of systemic lupus erythematosus (SLE) (Souyris et al., 2019). pDCs circulate through lymphoid organs where they represent roughly 0.1%–0.5% of nucleated cells (Reizis, 2019), and they are the most potent producers of IFNα in the hematopoietic compartment (Siegal et al., 1999). In humans, type I IFNs consist of thirteen IFNα subtypes, IFNβ, IFNε, IFNκ, and IFNω that are located on chromosome 9 (Lazear et al., 2019). All type I IFNs bind to the same IFNα/β receptor (IFNAR) that consists of two subunits (IFNAR1 and IFNAR2), resulting in transcription of IFN-stimulated genes (ISGs) that restrict viral spreading (Ivashkiv and Donlin, 2014). Recently, it has been suggested that only a few pDCs initiate type I IFN secretion and that the other pDCs require IFNAR signaling for induction of type I IFN production (Wimmers et al., 2018).
The mechanisms contributing to higher IFNα production by female pDCs remain incompletely understood. Studies showed a positive regulation of IFNα production by estradiol (Seillet et al., 2012; Griesbeck et al., 2015), and further experiments suggested genetic (X chromosome linked) factors (Souyris et al., 2018) or a combination of both (Laffont et al., 2014). The location of the TLR7 gene on the X chromosome is a potential reason for genetically driven differences due to differential allosome allocation between XX females and XY males. In order to achieve dosage compensation between the sexes, one of the two X chromosomes in females is transcriptionally silenced, a process that is referred to as X chromosome inactivation (XCI) (Avner and Heard, 2001). Studies have indicated that some genes on the X chromosome can escape XCI in females, resulting in transcription of these genes from the active as well as the inactive X chromosome (Xi) (Carrel et al., 1999; Carrel and Willard, 2005; Souyris et al., 2018). The non-coding RNA XIST is necessary for establishing XCI (Loda and Heard, 2019), and it is thought that the Xi is in most settings stably preserved over time and cell divisions (Payer and Lee, 2008; Robert Finestra and Gribnau, 2017). Yet, relatively little is known about the occurrence and maintenance of escape from XCI at the single immune cell level.
Here, we demonstrate extensive heterogeneity in escape from XCI in female pDCs. Biallelic expression of the investigated X chromosome genes resulted in higher transcript levels of the respective mRNAs. IFNα and IFNβ mRNAs were more highly transcribed in biallelic than monoallelic TLR7-expressing pDCs, and exposure to the IFNα proteins resulted in higher levels of IFNα/β protein production. Taken together, these data demonstrate the important contribution of X chromosome factors to sex differences in type I IFN responses.
Results
Assessment of Escape from XCI in Different X Chromosome Genes
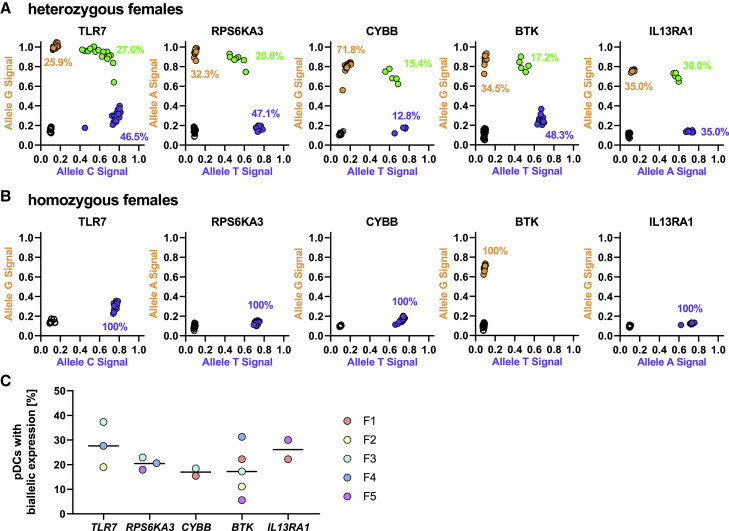
Previous studies have described escape from XCI in mice and humans, but the extent of escape from XCI for different X chromosome genes in pDCs and the resulting consequences for immune responses are not well understood. To investigate escape from XCI at the single pDC level, we took advantage of single nucleotide polymorphisms (SNPs) within the mature mRNA of genes encoded by the X chromosome, enabling differentiation of mRNA encoded by the two X chromosomes in heterozygous females (Berghöfer et al., 2006; Souyris et al., 2018). We selected TLR7 and four other genes (RPS6KA3, CYBB, BTK, and IL13RA1), based on their expression levels in pDCs, location on the X chromosome, and presence of a SNP in the mature mRNA with a minor allele frequency (MAF) of over 20% (Table S1; Figure 1 A). After identifying healthy female individuals with heterozygosity of the respective SNPs, freshly isolated peripheral blood mononuclear cells (PBMCs) were stimulated for 2 h with the TLR7/8 agonist CL097. pDCs were subsequently sorted (Figure S1A), and cDNA of single pDCs was obtained using the Fluidigm C1 technology (Figure 1B). pDCs expressing the respective mRNA derived from one (monoallelic) or two (biallelic) X chromosomes were identified using the Fluidigm Biomark HD platform based on detection of mRNA containing the major or minor allele nucleotide, or both, as shown for representative female individuals heterozygous for the respective SNPs in the X chromosome genes TLR7, RPS6KA3, CYBB, BTK, and IL13RA1 in Figure 2 A. Female individuals homozygous for either major allele (TLR7, RPS6KA3, CYBB, and IL13RA1) or the minor allele (BTK), thus only displaying one population in the escape plot, were used as controls (Figure 2B). Any cell without detectable mRNA for the respective gene was excluded from further analysis. The percentage of pDCs with biallelic expression for the investigated genes ranged from 5.6% to 37.3%, with the broadest range in pDCs with escape from XCI observed for BTK and TLR7 (BTK: 5.6%–31.3%; TLR7: 19.0–37.3%; Figure 2C). Overall these data show that TLR7, RPS6KA3, CYBB, BTK, and IL13RA1 can escape XCI in pDCs and that escape from XCI can be assessed at the single immune cell level.
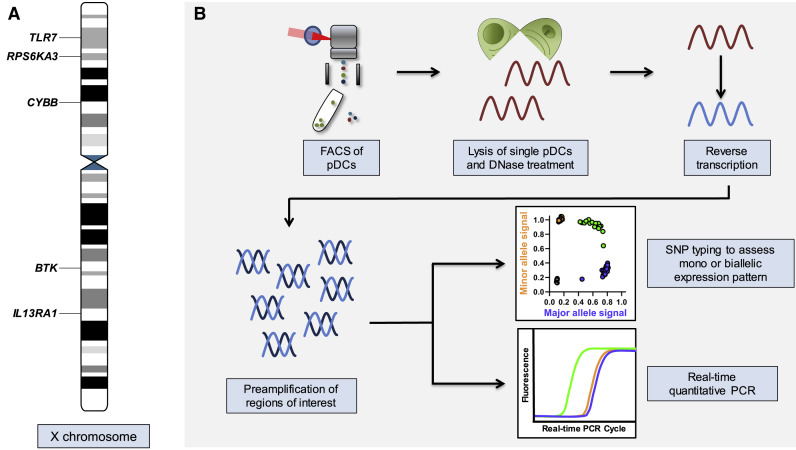
Figure 1.
Schematic Representation of Investigated Genes on the X Chromosome and Experimental Workflow
(A) Schematic representation of the X chromosome with the location of the five genes that were assessed in this study. The information was obtained from the Ensembl genome database project (https://www.ensembl.org/index.html) (Yates et al., 2020).
(B) Experimental workflow to assess escape from X chromosome inactivation (XCI) and gene expression at the single pDC level. Single pDCs were sorted and loaded into a C1 integrated fluidic circuit (IFC) from Fluidigm. Inside the C1 IFC, single pDCs were lysed. A DNase treatment was performed, and subsequently, mRNA was reverse transcribed to cDNA, including a preamplification of regions of interest. Harvested cDNA was then used in the Fluidigm Biomark platform for SNP typing of the cDNA to determine the mono- or biallelic expression pattern of the individual cells, and in separate experiments, the identical cDNA was used for quantitative real-time PCR to quantify mRNA expression of selected genes.
Figure 2.
Escape from XCI in TLR7, RPS6KA3, CYBB, BTK, and IL13RA1 in Human pDCs
(A) Representative escape plots of female individuals heterozygous for the respective SNPs with each dot representing one pDC. All cells in one plot were derived from one female individual at the same time point. Blue dots are cells with monoallelic expression of the major allele, orange dots are cells with monoallelic expression of the minor allele, and green dots represent cells with biallelic expression (major and minor allele) in the same cell, reflecting escape from XCI. Black circles represent cells without any detectable mRNA. Percentages were calculated using all cells with detectable mRNA. The expression pattern was determined using the following SNPs: TLR7, rs3853839; RPS6KA3, rs7051161; CYBB, rs5964151; BTK, rs700; and IL13RA1, rs2495636 (n = 1 per gene).
(B) Representative escape plots of female individuals homozygous for the respective SNPs to ensure specificity of the cDNA SNP typing. Black circles represent cells without any detectable mRNA. Percentages were calculated using all cells with detectable mRNA. The expression patterns were determined using the following SNPs: TLR7, rs3853839; RPS6KA3, rs7051161; CYBB, rs5964151; BTK, rs700; and IL13RA1, rs2495636 (n = 1 per gene).
(C) Distribution of pDCs with biallelic expression for the five investigated genes across five females. The black bar represents the median percentage of pDCs with biallelic expression. Percentages were calculated using all cells with detectable mRNA. Dots with the same color represent the same female individual. F, female. For each gene and female, the percentage of biallelic expressing pDCs are as follows: TLR7: F2: 19%, F3: 37.3%, F4: 27.6%; RPS6KA3: F3: 22.9%, F4: 20.6%, F5: 17.9%; CYBB: F1: 15.4%, F3: 18.5%; BTK: F1: 22.2%, F2: 11.1%, F3: 17.2%, F4: 31.3%, F5: 5.6%; and IL13RA1: F1: 22.2%, F5: 30%.
Female pDCs Present with a Heterogeneous XCI Profile
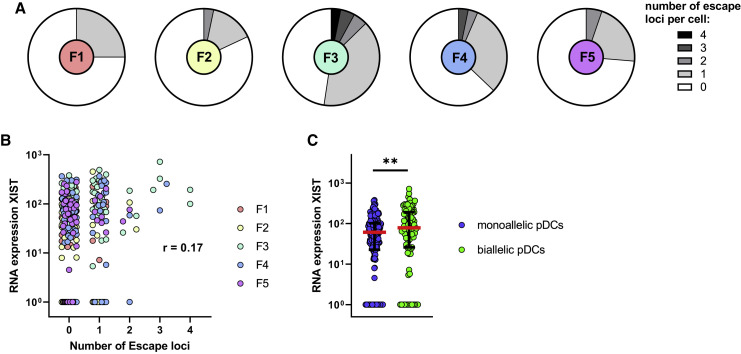
To investigate the diversity of escape from XCI within each female, we assessed the number of escape loci within single pDCs and observed that all except female 1 (F1) had at least two or more cells with escape from XCI in multiple loci (Figure 3 A). We furthermore detected pDCs in F2, F3, and F4 that had escape from XCI for every loci assessed. To investigate the influence of the non-coding RNA XIST within single pDCs, cDNA of the identical single pDCs in which escape from XCI was determined was used for XIST RNA quantification. The expression of XIST RNA did not correlate with the number of escape loci per cell (r = 0.17) (Figure 3B); however, there was a significant difference in XIST RNA expression levels comparing pDCs with at least one locus of biallelic expression compared to pDCs without biallelic expression (Figure 3C). In summary, different pDCs from the same individual had a heterogeneous profile of escape from XCI. We did not observe a correlation between the number of escape loci and XIST expression, but XIST RNA had higher expression in pDCs with biallelic expression.
Figure 3.
pDCs Have a Heterogeneous Profile of Escape from XCI
(A) Distribution of the number of XCI escape loci per female. The following number of loci was assessed per female: F1 = 3, F2 = 2, F3 = 4, F4 = 3, and F5 = 3. Single pDCs without any detectable mRNA in any of the loci were excluded. pDCs were sorted from 2-h CL097-stimulated PBMCs.
(B) Single-cell correlation of the number of escape loci per cell with the expression of XIST RNA. pDCs derived from n = 5 females were used. pDCs were sorted from 2-h CL097-stimulated PBMCs. A repeated-measures correlation (rmcorr) was performed, and the obtained rmcorr value is displayed (Bakdash and Marusich, 2017).
(C) Comparison of expression of XIST RNA for monoallelic pDCs (pDCs without any detected locus of escape) (blue) to biallelic pDCs (green, pDCs with at least one escape loci/loci with biallelic expression). pDCs derived from n = 5 females were used. pDCs were sorted from 2-h CL097-stimulated PBMCs. Median (red bar) with interquartile range (black bars) is shown. A mixed effects linear regression model with a random intercept was used to take into account intra-sample correlations. ∗∗p < 0.01.
pDCs with Escape from XCI Have Significantly Higher mRNA Levels of the Respective Escaped Genes Than pDCs with Monoallelic Expression
To determine whether biallelic expression of X chromosome genes resulted in higher mRNA expression levels of the respective genes within the individual cell, we analyzed cDNA that was used to determine mono- versus biallelic expression patterns also for quantification of mRNA levels by using the Fluidigm Biomark HD platform (Figure 1B). In all three female individuals heterozygous for the TLR7 SNP rs3853839, significantly higher TLR7 mRNA transcript levels were observed in pDCs with biallelic than those with monoallelic TLR7 expression (Figure 4 ; Figure S2A). Of note, monoallelic- and biallelic-TLR7-expressing pDCs from female individuals did not show any significant difference in mRNA expression of the reference gene GAPDH or the pDC genes HLA-DRA and IL3RA (CD123) (Figure S2B).
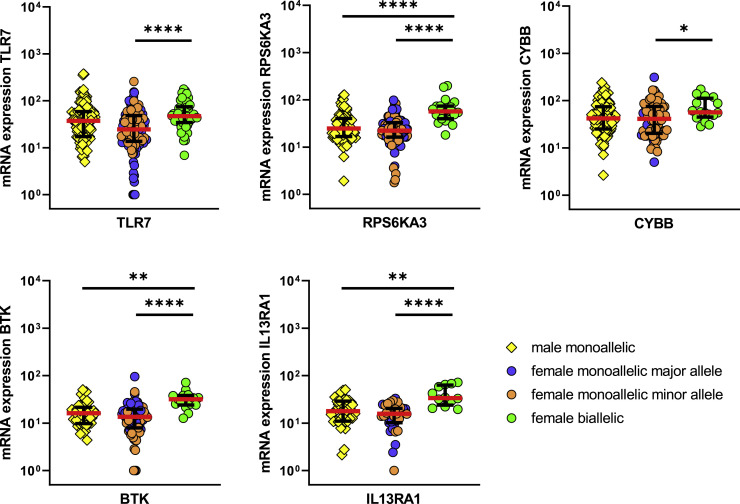
Figure 4.
pDCs with Biallelic Expression Have Higher mRNA Levels of the Respective Genes
Comparison of mRNA expression levels for the five investigated genes TLR7, RPS6KA3, CYBB, BTK, and IL13RA1 between male monoallelic pDCs (yellow squares), female monoallelic pDCs (blue circles, monoallelic expression of the major allele; orange circles, monoallelic expression of the minor allele), and female biallelic pDCs (green circles), thus pDCs with escape from XCI in the respective locus. pDCs were sorted from 2-h CL097-stimulated PBMCs. For each gene, the cells from multiple individuals were combined. The male pDCs were derived from n = 3 males. Because heterozygosity of the respective SNPs was necessary for assessment of expression patterns, genes were assessed within different numbers of females: TLR7, n = 3 females; RPS6KA3, n = 3 females; CYBB, n = 2 females; BTK, n = 5 females; and IL13RA1, n = 2 females. Expression patterns were determined using the following SNPs: TLR7, rs3853839; RPS6KA3, rs7051161; CYBB, rs5964151; BTK, rs700; and IL13RA1, rs2495636. Median (red bars) with interquartile range (black bars) is shown. A mixed effects linear regression model with a random intercept was used to take into account intra-sample correlations. Only significant differences are displayed. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗∗p < 0.0001.
Expanding the analysis to a comparison of mRNA levels of RPS6KA3, CYBB, BTK, and IL13RA1 between monoallelic- and biallelic-expressing pDCs, higher mRNA expression levels in pDCs with escape from XCI in the respective genes were consistently observed (Figure 4). For the genes RPS6KA3, BTK, and IL13RA1, female pDCs with monoallelic expression did not have higher mRNA levels than pDCs from males, whereas significant differences in mRNA levels between pDCs from males and biallelic-expressing pDCs from females were observed. We furthermore measured the TLR7 protein levels and observed significantly higher TLR7 protein levels in female pDCs than those in male pDCs (Figure S2C), as previously described for human PBMCs (Souyris et al., 2018). Altogether, these data demonstrate that escape from XCI in TLR7, RPS6KA3, CYBB, BTK, and IL13RA1 led to higher mRNA expression levels of the respective genes in pDCs with a biallelic expression pattern.
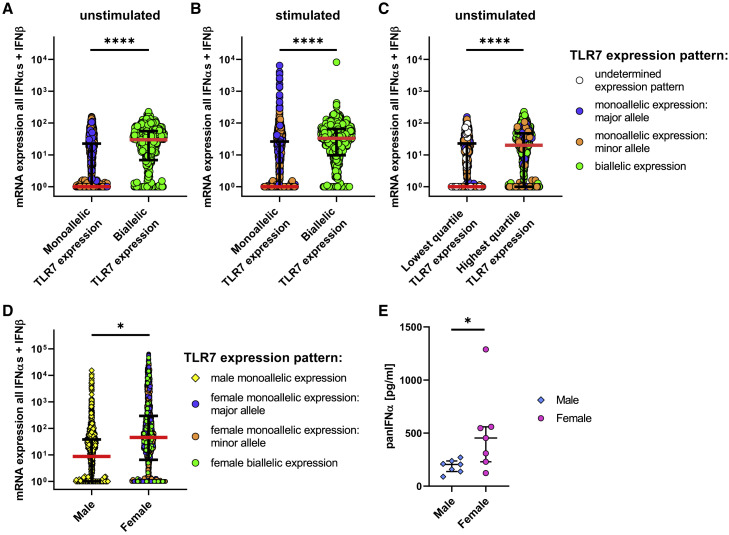
pDCs with Biallelic mRNA Expression of TLR7 Have Significantly Higher IFNα and IFNβ mRNA Expression
To further examine the functional consequences of escape of TLR7 from XCI for pDC function, we assessed whether escape of TLR7 from XCI and the resulting higher TLR7 mRNA levels were associated with higher mRNA transcript levels of IFNα subtypes and IFNβ in unstimulated pDCs. We observed a significant difference in IFNα and IFNβ mRNA expression levels between female pDCs with monoallelic compared to pDCs with biallelic TLR7 expression (Figure 5 A). The difference between monoallelic- and biallelic-TLR7-expressing pDCs was furthermore statistically significant for all IFNα subtypes and IFNβ mRNA expression individually (Figure S3). Following 2-h stimulation of pDCs, significant differences between monoallelic- and biallelic-TLR7-expressing pDCs for all IFNα subtypes and IFNβ mRNA persisted; however, stimulation led to an upregulation of IFNα and IFNβ mRNA levels, in particular in monoallelic-TLR7-expressing pDCs (Figure 5B). When we compared the lowest with the highest quartile of TLR7-mRNA-expressing pDCs, unstimulated pDCs with the highest TLR7 mRNA expression encoded significantly higher levels of IFNα and IFNβ mRNAs than pDCs with the lowest TLR7 mRNA expression levels (Figure 5C), and this difference was lost after 2 h of stimulation (data not shown). This result indicates that during initial IFNα and IFNβ production, pDCs with biallelic TLR7 expression and high TLR7 mRNA levels have higher type I IFN production; however, the TLR7-independent auto- and paracrine IFNAR feedback loops diminished these differences between low- and high-TLR7-expressing pDCs following stimulation.
Figure 5.
pDCs with Escape from XCI in TLR7 Transcribe Significantly Higher Levels of IFNα and IFNβ mRNA
(A and B) Comparison of mRNA expression levels of isolated, unstimulated pDCs for all IFNα subtypes and IFNβ combined (A) and mRNA expression levels of isolated, 2-h CL097-stimulated pDCs for all IFNα subtypes and IFNβ combined (B). Female monoallelic TLR7-expressing pDCs are displayed with blue circles (monoallelic expression of the major allele) or orange circles (monoallelic expression of the minor allele), and female biallelic TLR7-expressing pDCs are displayed with green circles (pDCs with escape from XCI in TLR7). Expression patterns were determined using the following TLR7 SNP: rs3853839. Individual pDCs from n = 3 females are displayed. Median (red bar) with interquartile range (black bars) is shown. A mixed effects linear regression model with a random intercept was used to take into account intra-sample correlations. ∗∗∗∗p < 0.0001.
(C) Comparison of mRNA expression of isolated, unstimulated pDCs of all IFNα subtypes and IFNβ combined between the lowest quartile of TLR7-expressing pDCs with the highest quartile of TLR7-expressing pDCs. Female monoallelic TLR7-expressing pDCs are displayed with blue circles (monoallelic expression of the major allele) or orange circles (monoallelic expression of the minor allele), and female biallelic TLR7-expressing pDCs are displayed with green circles (pDCs with escape from XCI in TLR7). White circles are pDCs without any detectable mRNA for TLR7. Expression patterns were determined using the following TLR7 SNP: rs3853839. Individual pDCs from n = 3 females are displayed. Median (red bar) with interquartile range (black bars) is shown. A mixed effects linear regression model with a random intercept was used to take into account intra-sample correlations. ∗∗∗∗p < 0.0001.
(D) Comparing mRNA expression of pDCs of all IFNα subtypes and IFNβ combined between female pDCs and male pDCs. pDCs were sorted from 2-h CL097-stimulated PBMCs. Expression patterns were determined using the following TLR7 SNP: rs3853839. Male pDCs (yellow squares), female monoallelic-TLR7-expressing pDCs (blue circles, monoallelic expression of the major allele; orange circles, monoallelic expression of the minor allele) and female biallelic-TLR7-expressing pDCs (green circles, pDCs with escape from XCI in TLR7). Expression patterns were determined using the following SNP: TLR7: rs3853839. Individual pDCs from n = 3 females and n = 3 males are displayed. Median (red bar) with interquartile range (black bars) is shown. A mixed effects linear regression model with a random intercept was used to take into account intra-sample correlations. ∗p < 0.05.
(E) PBMCs of males (n = 7) and females (n = 7) were stimulated for 2 h with CL097. The collected supernatant was analyzed for panIFNα levels with an ELISA. Female results are shown independent of TLR7 expression pattern. Median with interquartile range is shown. Mann-Whitney test was used for statistical analysis. ∗p < 0.05.
Following 2-h TLR7/8 stimulation of PBMCs, the mRNA expression level for all IFNα subtypes and IFNβ was significantly higher in pDCs from females than that from males, independent of whether there was monoallelic or biallelic expression of TLR7 (Figure 5D), highlighting the important role of the initial TLR7-dependent induction of IFNα/β production for these sex differences in type I IFN responses. To investigate whether the difference in mRNA levels following 2 h of stimulation was also present at the protein level, we measured the amount of panIFNα protein that was secreted in the supernatant following 2-h TLR7/8 stimulation of human PBMCs. PBMCs of females produced significantly more IFNα protein than males (female median = 453 pg/ml; male median = 205 pg/ml) (Figure 5E), which is in line with previous studies (Berghöfer et al., 2006; Meier et al., 2009; Ziegler et al., 2017). To demonstrate that the amount of IFNα that was secreted within 2 h of stimulation can indeed initiate an IFNAR feedback loop, we pre-treated PBMCs with IFNα2 (1 U/ml; 5 pg/ml) and IFNα4 (1 U/ml; 1.9 pg/ml) and observed that the IFNα pre-treatment led to a significant increase in IFNα and IFNβ protein production by human pDCs (Figure S4). Overall, these data demonstrate that IFNα and IFNβ mRNAs have significantly higher expression in pDCs with biallelic expression of TLR7 and that higher initial IFNα secretion by female pDCs can increase the IFNα and IFNβ protein production probably due to an auto- and paracrine IFNAR feedback loop.
Discussion
Sex differences in immune responses have been described in infectious and autoimmune diseases (Markle and Fish, 2014), as well as in response to vaccinations (Flanagan et al., 2017). Next to modulation of immune function by sex hormones, differences in the number of X chromosomes between women and men can contribute to this immunological sex bias (Libert et al., 2010). Previous studies have shown that 10%–25% of genes on the X chromosome can escape from XCI (Carrel et al., 1999; Carrel and Willard, 2005), with variability between different tissues and cells (Tukiainen et al., 2017). With a size of 156.04 mega base pairs (Mbps), the X chromosome is the 8th biggest chromosome (Yates et al., 2020) and contains a number of genes important for immune function and regulation (Fish, 2008), suggesting that escape from XCI in females might impact immune function. Investigating escape from XCI at the single pDC level, we show that biallelic expression of X chromosome genes occurred frequently, contributed to transcriptional heterogeneity of pDCs, and resulted in higher mRNA levels of the respective genes. IFNα and IFNβ mRNAs were more highly transcribed in female pDCs with biallelic expression of TLR7, and exposure of pDCs to IFNα protein significantly enhanced IFNα and IFNβ protein production. Taken together, these data demonstrate the important role of X chromosome factors in the regulation of the type I IFN responses and suggest that female pDCs with biallelic expression of TLR7 and stronger induction of type I IFNs are responsible for initiating sex-specific differences in IFNα and IFNβ protein production.
A number of recent studies have demonstrated escape from XCI (Wang et al., 2016) with consequences for sex-specific immune function (Souyris et al., 2018; Oghumu et al., 2019), but the mechanisms underlying escape from XCI of specific genes and their functional consequences at the single-cell level are not well understood. We investigated the expression of five X-chromosome-encoded genes in pDCs at the single-cell level, including TLR7 for which escape from XCI has been previously reported (in human B cells, pDCs, and monocytes; Souyris et al., 2018), and in four additional genes (RPS6KA3, CYBB, BTK, and IL13RA1) selected based on their expression levels in pDCs, location on the X chromosome, and presence of a SNP with a high MAF in the mature mRNA. Our data show that the five investigated genes escaped XCI in pDCs of every female that was investigated and that escape from XCI occurred in 5%–37% of pDCs. The degree of escape from XCI of these different genes varied between different pDCs, resulting in heterogeneous X chromosome gene expression within female pDC populations. In line with a previous study reporting enhanced TLR7 transcripts in biallelic-TLR7-expressing B cells and higher TLR7 protein expression in human PBMCs (Souyris et al., 2018), we observed that pDCs with escape from XCI in TLR7 had significantly higher TLR7 mRNA transcript levels than female monoallelic-TLR7-expressing pDCs, and we confirmed significantly higher TLR7 protein levels in pDCs from females than in those from males.
Investigation of the other four studied X chromosome genes consistently demonstrated higher mRNA expression in female pDCs with biallelic expression of the respective genes than in female pDCs with monoallelic expression. Available data on the regulation of escape from XCI in pDCs is very limited. One study in mice described that XIST expression varies between different immune cells and that XIST is nearly absent in murine pDCs (Syrett et al., 2019). In contrast, we detected XIST RNA in the majority of human pDCs. These dissimilarities to the study by Syrett et al. (2019) might be due to differences in the factors regulating XCI in humans and mice (Migeon, 2017). Interestingly, we observed a significant difference in XIST RNA expression between pDCs with or without biallelic expression. However, it has been suggested that the inactivation activity of XIST is limited to a short time frame during early development (Wutz and Jaenisch, 2000), and multiple studies have proposed that XIST is not required for the maintenance of the inactivation of the X chromosome (Csankovszki et al., 1999; Rack et al., 1994), indicating that additional studies are required to investigate the role of XIST in regulating escape from XCI in humans. In summary, our results show transcription from multiple genes located on the second X chromosome in women, with significant consequences for the mRNA levels within the respective cell, contributing to a transcriptional heterogeneity of individual pDCs in female individuals.
Previous studies have shown that pDCs of females produce significantly more IFNα than pDCs of males following TLR7 stimulation (Berghöfer et al., 2006; Meier et al., 2009; Seillet et al., 2012; Griesbeck et al., 2015; Ziegler et al., 2017) and implicated these sex differences in type I IFN production in the pathogenesis of infectious and autoimmune diseases that exhibit clear differences between women and men (Farzadegan et al., 1998; Meditz et al., 2011; Sterling et al., 2001; Amur et al., 2012). Remarkably, the incidence of SLE, an autoimmune disease primarily observed in women, is also elevated in males with Klinefelter syndrome (47, XXY) (Scofield et al., 2008). Women with Turner syndrome (45, X0) appear to have a lower risk for SLE (Cooney et al., 2009), whereas the incidence for females with triple X syndrome (47, XXX) is 2.5 times higher than for women without the syndrome (46, XX) (Liu et al., 2016), suggesting an X chromosome dosage effect in the pathogenesis of this autoimmune disease (Scofield et al., 2008). SLE patients have elevated IFNα levels in their blood (Friedman et al., 1982; Kim et al., 1987) and significantly higher expression of ISGs (Crow et al., 2003), and there are case reports of human patients developing SLE following IFNα therapy, highlighting the connection between type I IFNs and SLE (Rönnblom et al., 1990; Fukuyama et al., 2000). A mouse strain with an autoimmune phenotype resembling human SLE has furthermore been shown to have an additional cluster of X-linked genes containing murine Tlr7 (Izui et al., 1994; Subramanian et al., 2006), and depletion or impairment of pDCs in vivo improved clinical symptoms of SLE (Rowland et al., 2014; Sisirak et al., 2014). These data highlight an important role of X-linked factors in autoimmune diseases that might be explained by escape of TLR7 from XCI and result in higher induction of type I IFN mRNA in these pDCs. The current SARS-CoV-2 pandemic also displays a sex bias with 1.7 times higher case mortality rates in males than in females reported in Europe (Gebhard et al., 2020). A case study with four young men (under 35) without major preexisting diseases reported severe to fatal COVID-19 progression linked to loss-of-function mutations of TLR7 (van der Made et al., 2020), highlighting the importance of functional TLR7 signaling in the control of SARS-CoV-2 infection. It was furthermore suggested that early treatment with IFNα2 during initial SARS-CoV-2 infection was associated with reduced mortality (Wang et al., 2020). Our data showing higher IFNα and IFNβ mRNA levels in female pDCs with biallelic TLR7 expression suggest that higher type I IFN responses in females might potentially contribute to a better control of SARS-CoV-2 infection in women (Bunders and Altfeld, 2020).
Quantifying IFNα/β mRNA expression in pDCs, we observed that unstimulated pDCs with biallelic expression of TLR7 exhibit higher mRNA levels of IFNα/β. Furthermore, the highest quartile of TLR7-expressing pDCs also expressed higher levels of IFNα/β mRNA, indicating that the initial IFNα/β response importantly depends on TLR7 mRNA expression levels. These differences in IFNα/β mRNA levels between the lowest and the highest quartile of TLR7-expressing pDCs diminished following stimulation, likely due to the auto- and paracrine IFNAR signaling that is independent of TLR7. Unfortunately, current methods do not allow determination of whether an individual pDC with biallelic expression of TLR7 and higher TLR7 mRNA expression also produces more IFNα/β protein. However, these data suggest that female pDCs with biallelic and thus higher expression of TLR7 mRNA might belong to the group of pDCs described to independently initiate IFNα secretion (Wimmers et al., 2018). The observation that unstimulated pDCs harbor mRNA for type I IFNs is in line with published data describing that pDCs constitutively produce small amounts of type I IFNs as a self-priming mechanism for TLR stimulation (Kim et al., 2014). Taken together, these data suggest that pDCs as professional type I IFN producers already have a basal level of type I IFN mRNA expression, with pDCs with biallelic TLR7 expression exhibiting higher mRNA levels of IFNα/β.
In summary, the results from these studies support a model in which a subset of female pDCs with escape from XCI in TLR7 and resulting higher TLR7 mRNA levels can respond with stronger initial IFNα/β production, initiating an auto- and paracrine IFNAR feedback loop that drives higher IFNα/β production in females. Our results furthermore show the significant impact of escape from XCI at the single-cell level on transcriptional and functional heterogeneity of immune cells, which can contribute to the described differences in immune-mediated diseases between women and men.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD11c-PE/Cy7 (Bu15) | Biolegend | Cat#: 337216 |
| RRID: AB_2129790 | ||
| CD123-BV711 (9F5) | BD | Cat#: 563161 |
| RRID: AB_2738038 | ||
| CD123-FITC (6H6) | Biolegend | Cat#: 306014 |
| RRID: AB_2124259 | ||
| CD14-APC/Cy7 (HCD14) | Biolegend | Cat#: 325620 |
| RRID: AB_830693 | ||
| CD19-BUV395 (SJ25C1) | BD | Cat#: 563549 |
| RRID: AB_2738272 | ||
| CD19-BUV737 (SJ25C1) | BD | Cat#: 564303 |
| RRID: AB_2716867 | ||
| CD3-BUV737 (UCHT1) | BD | Cat#: 564307 |
| RRID: AB_2744390 | ||
| CD56-BUV395 (NCAM16.2) | BD | Cat#: 563554 |
| RRID: AB_2687886 | ||
| IFNα2b-V450 (7N4-1) | BD | Cat#: 561382 |
| RRID: AB_10716058 | ||
| IFNα-PE (LT27:295) | Miltenyi Biotec | Cat#: 130-099-098 |
| RRID: AB_871560 | ||
| IFNβ-FITC (MMHB-3) | PBL Assay Science | Cat#: PBL-21400-3 |
| RRID: AB_387831 | ||
| HLA-DR-BV605 (L243) | Biolegend | Cat#: 307640 |
| RRID: AB_2561913 | ||
| TLR7-APC (4G6) |
Novus Biologicals |
Cat#: NBP2-25274APC |
| RRID: N/A | ||
| Biological Samples | ||
| Human blood | Healthy individuals from the University Medical Center Hamburg-Eppendorf | Cat#: N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 1x DNA Suspension Buffer | Teknova | Cat#: T0221 |
| 2x Sso Fast Eva Green Supermix with Low ROX | Bio-Rad | Cat#: 1725210 |
| 2x Fast Probe Master Mix | Biotium | Cat#: 31005 |
| ACK Lysing Buffer | Lonza | Cat#: 10-548E |
| Biocoll-Trennlösung | Biochrom | Cat#: L6115 |
| Brefeldin A | Sigma-Aldrich | Cat#: B7651-5MG |
| CL097 | Invivogen | Cat#: tlrl-c97 |
| Dulbecco’s phosphate buffered saline (PBS) | Sigma-Aldrich | Cat#: D8537 |
| Dymethyl sulfoxide (DMSO) | Sigma-Aldrich | Cat#: D5879-100ML |
| Ethylenediaminetetraacetic acid (EDTA) solution | Sigma-Aldrich | Cat#: 03690-100ML |
| Fetal bovine serum (FBS) superior | Biochrom | Cat#: S0615 |
| Fixation Medium (Medium A) | ThermoFisher Scientific | Cat#: GAS001S100 |
| Human Recombinant IFNα2a | Stemcell | Cat#: 78076.1 |
| Permeabilization Medium (Medium B) | ThermoFisher Scientific | Cat#: GAS002S100 |
| Recombinant Human IFNα4b | PBL Assay Science | Cat#: 11180-1 |
| ROX Reference Dye | ThermoFisher Scientific | Cat#: 12223012 |
| GIBCO RPMI 1640 Medium | Life Technologies | Cat#: 21875091 |
| Critical Commercial Assays | ||
| C1 Single-Cell Reagent Kit for Preamp | Fluidigm | Cat#: 100-5319 |
| DNeasy Blood & Tissue Kit (250) | QIAGEN | Cat#: 69506 |
| GE 96.96 Dynamic Array DNA Binding Dye Sample & Assay Loading Reagent Kit with Control Line Fluid | Fluidigm | Cat#: 100-3415 |
| Human pan IFN-alpha ELISA Kit | Stemcell | Cat#: 02000 |
| QIAGEN Multiplex PCR Kit | QIAGEN | Cat#: 206143 |
| Plasmacytoid Dendritic Cell Isolation Kit II, human | Miltenyi Biotec | Cat#: 130-097-415 |
| Single Cell-to-CT qRT-PCR Kit | Invitrogen / ThermoFisher Scientific | Cat#: 4458237 |
| SNP Type 192.24 Genotyping Reagent Kit with Control Line Fluid | Fluidigm | Cat#: 100-4136 |
| Zombie Aqua Fixable Viability Kit | Biolegend | Cat#: 423102 |
| Oligonucleotides | ||
| See Tables S2–S4 for the list of Oligonucleotides. | N/A | |
| Software and Algorithms | ||
| FlowJo10 | FlowJo LLC | https://www.flowjo.com |
| Fluidigm Real-Time PCR Analysis, Version 4.3.1 | Fluidigm | https://www.fluidigm.com/ |
| Fluidigm SNP Genotyping Analysis; Version 4.3.2 | Fluidigm | https://www.fluidigm.com/ |
| GraphPad Prism 8 | GraphPad Software, Inc | https://www.graphpad.com:443/ |
| Other | ||
| 192.24 Dynamic Array IFC for SNP Genotyping | Fluidigm | BMK-M-192.24GT |
| 48.48 Dynamic Array IFC for Genotyping | Fluidigm | BMK-M-48.48GT |
| 96.96 Dynamic Array IFC for Gene Expression | Fluidigm | BMK-M-96.96 |
| C1 Single-Cell Preamp IFC, 5–10 μm | Fluidigm | 100-5757 |
| S-Monovette 9 ml K3 EDTA | Sarstedt | 02.1066.001 |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Marcus Altfeld (marcus.altfeld@leibniz-hpi.de)
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
This study did not generate/analyze datasets or code.
Experimental Model and Subject Details
Healthy individuals were recruited at the University Medical Center Hamburg-Eppendorf. The ethical commission “Ärztekammer” Hamburg approved the study (PV4780). The age of the donors ranged from 20 – 36. A similar number of females and males were included for the experiments to study sex-specific differences and the sex of each individual donor is indicated in the figure legend. Experiments investigating the effect of escape from XCI were only performed in female individuals, due to the requirement of two X chromosomes. Informed consent was provided by each individual prior to enrollment in the study.
Method Details
Isolation and freezing of PBMCs
Blood from healthy individuals was drawn in EDTA tubes (Sarstedt) and the processing of the blood was started within 90 minutes after venipuncture (Meier et al., 2008). PBMCs were isolated using Ficoll-Paque (Biochrom) density centrifugation. Remaining erythrocytes were lysed in 3 mL of ACK Lying Buffer (Lonza) for 3 min. Freshly isolated PBMCs were immediately used for experiments or frozen in heat inactivated FBS (Biochrom) with 10% DMSO (Sigma-Aldrich) at −80°C and then transferred the next day into liquid nitrogen storage.
Purification of pDCs
pDCs were purified from frozen PBMCs via negative selection using the Plasmacytoid Dendritic Cell Isolation Kit II, human (Miltenyi Biotec) according to manufactures instructions. For up to 25 Mio PBMCs 25% of the recommended amount of were used (e.g., 25 μl of Non-pDC Biotin-Antibody Cocktail II for up to 25 Mio PBMCs). As buffer PBS (Sigma-Aldrich) + 0.5% FBS (Biochrom) + 2mM EDTA (Sigma-Aldrich) was used.
TLR stimulation and IFNα pre-treatment
2 million freshly isolated PBMCs per ml were stimulated in GIBCO RPMI 1640 Medium (Life technologies) supplemented with 10% heat inactivated FBS (Biochrom) (R10) with 1 μg/ml of TLR7/8 agonist CL097 (Invivogen). For analysis of the supernatant the stimulation was performed for 2 h. For flow cytometric analysis the stimulation was done for 6 h in the presence of 5 μg/ml brefeldin A (BFA) (Sigma-Aldrich), as previously described (Meier et al., 2009; Ziegler et al., 2017). For IFNα pre-treatment experiments PBMCs were cultured for 2 h in R10 with or without IFNα2a (Stemcell) or IFNα4b (PBL Assay Science) at the indicated concentrations prior to stimulation with CL097 (in the presence of BFA). An unstimulated, but BFA-treated control sample was included for every experiment. For subsequent mRNA analysis, the stimulation of PBMCs was performed for 2 h. For mRNA analysis pDCs were purified from frozen PBMCs and stimulated at 50 000 pDCs per ml with 1 μg/ml of CL097 for 2 h or pDCs were purified from frozen PBMCs and left unstimulated.
Cell staining, flow Cytometry and cell sorting
Prior to staining, PBMCs / isolated pDCs were washed with PBS (Sigma-Aldrich). Staining was performed for 20 minutes (RT / dark) with the indicated surface antibodies and Zombie Aqua (Biolegend). The antibodies used for staining are shown in the Key Resources Table. For mRNA analysis PBMCs / isolated pDCs were left unfixed and stored on ice until pDCs were sorted as single live CD3-CD19-CD56-CD11c-CD14-CD123+HLA-DR+ cells using a FACSAria Fusion. Gating strategies are shown in Figure S1A.
For intracellular cytokine staining and intracellular staining of TLR7, PBMCs were fixed with Medium A (Life Technologies). Intracellular staining was done using Medium B (Life Technologies) with the indicated antibodies for 20 minutes. IFNα2 was quantified with clone 7N4-1 (BD) which primarily detects IFNα2. PolyIFNα was quantified with clone LT27:295 (Miltenyi Biotec), which detects IFNα2 and several other IFNα subtypes (Svensson et al., 1996). Gating strategies are shown in Figure S1A (TLR7) and Figure S1B (cytokines). Frozen PBMCs were used for the staining of TLR7. For staining of TLR7 an FMO control was included for every sample and the median fluorescent intensity (MdFI) of TLR7 was normalized to the FMO control of the same donor (MdFI TLR7 / MdFI FMO). Cells were analyzed using a LSRFortessa Flow Cytometer (BD) in the core facility Fluorescence Cytometry at the HPI, and FCS data were analyzed using FlowJo version 10.
panIFNα ELISA
panIFNα concentrations were measured in duplicates in cell culture supernatant following 2 h stimulation of freshly isolated PBMCs with CL097 (1 μg/ml) (Invivogen) using Human pan IFN-alpha ELISA Kit (Stemcell) according to manufactures’ instructions. The harvested supernatant was stored at –80°C until the measurement.
Genotyping of healthy donors
Genomic DNA (gDNA) of donors was obtained using DNeasy Blood & Tissue Kit (QIAGEN) according to protocol. The genotyping was performed with the Biomark HD (Fluidigm) using the 48.48 Dynamic Array IFC for Genotyping (Fluidigm). A specific target amplification (STA) of gDNA was performed prior to SNP genotyping. STA and SNP genotyping was done according to company protocol. The primers used for preamplification and SNP genotyping are shown in Table S2 and Table S4, respectively.
Single cell gene expression analysis
A platform that consisted of the following Fluidigm devices was used for single cell analysis: C1, Juno and Biomark HD (Fluidigm).
C1
Single-Cell Preamp IFCs, 5–10 μm (Fluidigm) were used for the generation of cDNA. After cell capture, all 96 capture sites were visualized under a microscope and only capture sites containing one single cell were included into further analysis. Single Cell Lysis Solution and DNase I solution (both included in the Single Cell-to-CT qRT-PCR Kit, Invitrogen / Thermo Fisher Scientific) were mixed in a ratio of 10:1 before being added to the Lysis final mix. Generation of cDNA was performed following company protocol. Pre-amplification of mRNA surrounding the respective SNPs was done with the outer primers of the SNP / DG primers to create an amplicon that could be analyzed with both the SNP and DG primers (Tables S2 and S3). A negative control was included with every C1 run.
Biomark HD
Gene expression analysis was performed with Biomark HD using 96.96 Dynamic Array IFC for Gene Expression. SNP typing of mRNAs was performed using 192.24 Dynamic Array IFCs for SNP genotyping following company protocol but using the cDNA generated in the C1 instead of gDNA. A water control was included in every Biomark run. Primers used for real-time quantitative PCR and SNP genotyping are shown in Table S3 and Table S4 respectively.
Data processing
The data processing was performed as described previously (Hess et al., 2020). In short, cells lacking detectable mRNA in two out of the three reference genes (B2M, RPL13A, GAPDH) were excluded from further analysis. Since it has been previously reasoned against normalization of expression data from single cells (McDavid et al., 2013; Arsenio et al., 2014), the gene expression results were not normalized to any reference gene (McDavid et al., 2013; Arsenio et al., 2014). A limit of detection (LOD) was defined as a ct value of 24 (Livak et al., 2013; Hipp et al., 2017), subsequently all ct values higher than the LOD were set to 24. mRNA expression levels were defined as 2(LOD-ct) (Livak et al., 2013; Hipp et al., 2017). Thus the value of 1 indicates a cell without detectable mRNA for the respective gene. In the Real-Time PCR analysis software (Fluidigm) the definition of the peak detection ranges of melting curves were set as the median of the temperature peak ± 1.2°C. The median of the temperature peak of all non-failed results in the factory setting for the gene of interest for all analyzed cells was used. The remaining settings for melting curve analysis were left unchanged in the software (Peak Sensitivity: 7; Peak Ratio Threshold: 0.8). The ct value was set to the LOD for all reactions that were marked as failed under these settings. The softwares Fluidigm SNP Genotyping Analysis (Version 4.3.2) and Fluidigm Real-Time PCR Analysis (Version 4.3.1) were used for analysis.
Quantification and Statistical Analysis
Graphs and plots were generated with GraphPad Prism 8 (GraphPad Software). Statistical analyses were performed using GraphPad Prism 8 and R (version 3.5). Mann-Whitney test was employed for comparison between two unpaired groups. Wilcoxon matched-pairs signed rank test was used for comparison between two paired groups. For comparisons including single cells from different individuals, a mixed effects linear regression model with a random intercept was used to take into account the intra-sample correlations. Correlations for data including single cells from different donors were performed by employing the method of calculating the correlation of repeated-measurements (package rmcorr) taking into account the intra-individual associations as described previously (Bakdash and Marusich, 2017). Detailed information can be found in the respective figure legends.
Acknowledgments
We thank the blood cohort organizers and all the volunteers for the blood donations. We would also like to thank Arne Düsedau at the HPI Technology Platform Flow Cytometry/FACS for his help with cell sorting. The study was supported by the Landesforschungsförderung Hamburg: LFF-FV 45 Geschlechtsdimorphismus im Immunsystem and by the German Center for Infection Research (DZIF): TTU 01.933. S.H.H. was supported by the Leibniz Center Infection. The publication of this article was funded by the Open Access Fund of the Leibniz Association.
Author Contributions
S.H.H. and M.A. designed the study. S.H.H. and F.H. performed experiments and analyzed the data. J.H. gave critical experimental support. S.H.H. performed basic statistical tests. S.D. and H.S. performed mixed effects linear regression models and correlations with repeated measurements. L.R. provided important statistical guidance. S.M.Z. gave important intellectual input. S.H.H. and M.A. wrote the manuscript. All authors reviewed the manuscript and gave approval for publication.
Declaration of Interests
The authors declare that they have no conflict of interest.
Published: December 8, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108485.
Supplemental Information
References
- Addo M.M., Altfeld M. Sex-Based Differences in HIV Type 1 Pathogenesis. J. Infec. Dis. 2014;209:S86–S92. doi: 10.1093/infdis/jiu175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amur S., Parekh A., Mummaneni P. Sex differences and genomics in autoimmune diseases. J. Autoimmun. 2012;38:J254–J265. doi: 10.1016/j.jaut.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Arsenio J., Kakaradov B., Metz P.J., Kim S.H., Yeo G.W., Chang J.T. Early specification of CD8+ T lymphocyte fates during adaptive immunity revealed by single-cell gene-expression analyses. Nat. Immunol. 2014;15:365–372. doi: 10.1038/ni.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avner P., Heard E. X-chromosome inactivation: counting, choice and initiation. Nat. Rev. Genet. 2001;2:59–67. doi: 10.1038/35047580. [DOI] [PubMed] [Google Scholar]
- Bakdash J.Z., Marusich L.R. Repeated Measures Correlation. Front. Psychol. 2017;8:456. doi: 10.3389/fpsyg.2017.00456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghöfer B., Frommer T., Haley G., Fink L., Bein G., Hackstein H. TLR7 Ligands Induce Higher IFN-Alpha Production in Females. J. Immunol. 2006;177:2088–2096. doi: 10.4049/jimmunol.177.4.2088. [DOI] [PubMed] [Google Scholar]
- Bernin H., Lotter H. Sex Bias in the Outcome of Human Tropical Infectious Diseases: Influence of Steroid Hormones. J. Infect. Dis. 2014;209:S107–S113. doi: 10.1093/infdis/jit610. [DOI] [PubMed] [Google Scholar]
- Bunders M.J., Altfeld M. Implications of Sex Differences in Immunity for SARS-CoV-2 Pathogenesis and Design of Therapeutic Interventions. Immunity. 2020;53:487–495. doi: 10.1016/j.immuni.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrel L., Willard H.F. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- Carrel L., Cottle A.A., Goglin K.C., Willard H.F. A first-generation X-inactivation profile of the human X chromosome. Proc. Natl. Acad. Sci. USA. 1999;96:14440–14444. doi: 10.1073/pnas.96.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney C.M., Bruner G.R., Aberle T., Namjou-Khales B., Myers L.K., Feo L., Li S., D’Souza A., Ramirez A., Harley J.B., Scofield R.H. 46,X,del(X)(q13) Turner’s syndrome women with systemic lupus erythematosus in a pedigree multiplex for SLE. Genes Immun. 2009;10:478–481. doi: 10.1038/gene.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow M.K., Kirou K.A., Wohlgemuth J. Microarray analysis of interferon-regulated genes in SLE. Autoimmunity. 2003;36:481–490. doi: 10.1080/08916930310001625952. [DOI] [PubMed] [Google Scholar]
- Csankovszki G., Panning B., Bates B., Pehrson J.R., Jaenisch R. Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat. Genet. 1999;22:323–324. doi: 10.1038/11887. [DOI] [PubMed] [Google Scholar]
- Farzadegan H., Hoover D.R., Astemborski J., Lyles C.M., Margolick J.B., Markham R.B., Quinn T.C., Vlahov D. Sex differences in HIV-1 viral load and progression to AIDS. Lancet. 1998;352:1510–1514. doi: 10.1016/S0140-6736(98)02372-1. [DOI] [PubMed] [Google Scholar]
- Fish E.N. The X-files in immunity: sex-based differences predispose immune responses. Nat. Rev. Immunol. 2008;8:737–744. doi: 10.1038/nri2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan K.L., Fink A.L., Plebanski M., Klein S.L. Sex and Gender Differences in the Outcomes of Vaccination over the Life Course. Annu. Rev. Cell Dev. Biol. 2017;33:577–599. doi: 10.1146/annurev-cellbio-100616-060718. [DOI] [PubMed] [Google Scholar]
- Friedman R.M., Preble O., Black R., Harrell S. Interferon production in patients with systemic lupus erythematosus. Arthritis Rheum. 1982;25:802–803. doi: 10.1002/art.1780250717. [DOI] [PubMed] [Google Scholar]
- Fukuyama S., Kajiwara E., Suzuki N., Miyazaki N., Sadoshima S., Onoyama K. Systemic lupus erythematosus after alpha-interferon therapy for chronic hepatitis C: a case report and review of the literature. Am. J. Gastroenterol. 2000;95:310–312. doi: 10.1111/j.1572-0241.2000.01715.x. [DOI] [PubMed] [Google Scholar]
- Gebhard C., Regitz-Zagrosek V., Neuhauser H.K., Morgan R., Klein S.L. Impact of sex and gender on COVID-19 outcomes in Europe. Biol. Sex Differ. 2020;11:29. doi: 10.1186/s13293-020-00304-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebely J., Page K., Sacks-Davis R., van der Loeff M.S., Rice T.M., Bruneau J., Morris M.D., Hajarizadeh B., Amin J., Cox A.L. The effects of female sex, viral genotype, and IL28B genotype on spontaneous clearance of acute hepatitis C virus infection. Hepatology. 2014;59:109–120. doi: 10.1002/hep.26639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesbeck M., Ziegler S., Laffont S., Smith N., Chauveau L., Tomezsko P., Sharei A., Kourjiian G., Porichis F., Hart M. Sex Differences in Plasmacytoid Dendritic Cell Levels of IRF5 Drive Higher IFN-α Production in Women. J. Immunol. 2015;195:5327–5336. doi: 10.4049/jimmunol.1501684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz D., Schneider B. Sex differences in tuberculosis. Semin. Immunopathol. 2019;41:225–237. doi: 10.1007/s00281-018-0725-6. [DOI] [PubMed] [Google Scholar]
- Hess L.U., Martrus G., Ziegler A.E., Langeneckert A.E., Salzberger W., Goebels H., Sagebiel A.F., Hagen S.H., Poch T., Ravichandran G. The Transcription Factor Promyelocytic Leukemia Zinc Finger Protein Is Associated With Expression of Liver-Homing Receptors on Human Blood CD56bright Natural Killer Cells. Hepatol. Commun. 2020;4:409–424. doi: 10.1002/hep4.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipp N., Symington H., Pastoret C., Caron G., Monvoisin C., Tarte K., Fest T., Delaloy C. IL-2 imprints human naive B cell fate towards plasma cell through ERK/ELK1-mediated BACH2 repression. Nat. Commun. 2017;8:1443. doi: 10.1038/s41467-017-01475-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann J., Otte A., Thiele S., Lotter H., Shu Y., Gabriel G. Sex differences in H7N9 influenza A virus pathogenesis. Vaccine. 2015;33:6949–6954. doi: 10.1016/j.vaccine.2015.08.044. [DOI] [PubMed] [Google Scholar]
- Ivashkiv L.B., Donlin L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izui S., Merino R., Fossati L., Iwamoto M. The role of the Yaa gene in lupus syndrome. Int. Rev. Immunol. 1994;11:211–230. doi: 10.3109/08830189409061728. [DOI] [PubMed] [Google Scholar]
- Kim T., Kanayama Y., Negoro N., Okamura M., Takeda T., Inoue T. Serum levels of interferons in patients with systemic lupus erythematosus. Clin. Exp. Immunol. 1987;70:562–569. [PMC free article] [PubMed] [Google Scholar]
- Kim S., Kaiser V., Beier E., Bechheim M., Guenthner-Biller M., Ablasser A., Berger M., Endres S., Hartmann G., Hornung V. Self-priming determines high type I IFN production by plasmacytoid dendritic cells. Eur. J. Immunol. 2014;44:807–818. doi: 10.1002/eji.201343806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laffont S., Rouquié N., Azar P., Seillet C., Plumas J., Aspord C., Guéry J.-C. X-Chromosome Complement and Estrogen Receptor Signaling Independently Contribute to the Enhanced TLR7-Mediated IFN-α Production of Plasmacytoid Dendritic Cells from Women. J. Immunol. 2014;193:5444–5452. doi: 10.4049/jimmunol.1303400. [DOI] [PubMed] [Google Scholar]
- Lazear H.M., Schoggins J.W., Diamond M.S. Shared and Distinct Functions of Type I and Type III Interferons. Immunity. 2019;50:907–923. doi: 10.1016/j.immuni.2019.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libert C., Dejager L., Pinheiro I. The X chromosome in immune functions: when a chromosome makes the difference. Nat. Rev. Immunol. 2010;10:594–604. doi: 10.1038/nri2815. [DOI] [PubMed] [Google Scholar]
- Liu K., Kurien B.T., Zimmerman S.L., Kaufman K.M., Taft D.H., Kottyan L.C., Lazaro S., Weaver C.A., Ice J.A., Adler A.J. X Chromosome Dose and Sex Bias in Autoimmune Diseases: Increased Prevalence of 47,XXX in Systemic Lupus Erythematosus and Sjögren’s Syndrome. Arthritis Rheumatol. 2016;68:1290–1300. doi: 10.1002/art.39560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Wills Q.F., Tipping A.J., Datta K., Mittal R., Goldson A.J., Sexton D.W., Holmes C.C. Methods for qPCR gene expression profiling applied to 1440 lymphoblastoid single cells. Methods. 2013;59:71–79. doi: 10.1016/j.ymeth.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loda A., Heard E. Xist RNA in action: Past, present, and future. PLoS Genet. 2019;15:e1008333. doi: 10.1371/journal.pgen.1008333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markle J.G., Fish E.N. SeXX matters in immunity. Trends Immunol. 2014;35:97–104. doi: 10.1016/j.it.2013.10.006. [DOI] [PubMed] [Google Scholar]
- McDavid A., Finak G., Chattopadyay P.K., Dominguez M., Lamoreaux L., Ma S.S., Roederer M., Gottardo R. Data exploration, quality control and testing in single-cell qPCR-based gene expression experiments. Bioinformatics. 2013;29:461–467. doi: 10.1093/bioinformatics/bts714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meditz A.L., MaWhinney S., Allshouse A., Feser W., Markowitz M., Little S., Hecht R., Daar E.S., Collier A.C., Margolick J. Sex, race, and geographic region influence clinical outcomes following primary HIV-1 infection. J. Infect. Dis. 2011;203:442–451. doi: 10.1093/infdis/jiq085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier A., Fisher A., Sidhu H.K., Chang J.J., Wen T.F., Streeck H., Alter G., Silvestri G., Altfeld M. Rapid loss of dendritic cell and monocyte responses to TLR ligands following venipuncture. J. Immunol. Methods. 2008;339:132–140. doi: 10.1016/j.jim.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier A., Chang J.J., Chan E.S., Pollard R.B., Sidhu H.K., Kulkarni S., Wen T.F., Lindsay R.J., Orellana L., Mildvan D. Sex differences in the Toll-like receptor-mediated response of plasmacytoid dendritic cells to HIV-1. Nat. Med. 2009;15:955–959. doi: 10.1038/nm.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migeon B.R. Choosing the Active X: The Human Version of X Inactivation. Trends Genet. 2017;33:899–909. doi: 10.1016/j.tig.2017.09.005. [DOI] [PubMed] [Google Scholar]
- Oghumu S., Varikuti S., Stock J.C., Volpedo G., Saljoughian N., Terrazas C.A., Satoskar A.R. Cutting Edge: CXCR3 Escapes X Chromosome Inactivation in T Cells during Infection: Potential Implications for Sex Differences in Immune Responses. J. Immunol. 2019;203:789–794. doi: 10.4049/jimmunol.1800931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer B., Lee J.T. X chromosome dosage compensation: how mammals keep the balance. Annu. Rev. Genet. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- Rack K.A., Chelly J., Gibbons R.J., Rider S., Benjamin D., Lafreniére R.G., Oscier D., Hendriks R.W., Craig I.W., Willard H.F. Absence of the XIST gene from late-replicating isodicentric X chromosomes in leukaemia. Hum. Mol. Genet. 1994;3:1053–1059. doi: 10.1093/hmg/3.7.1053. [DOI] [PubMed] [Google Scholar]
- Reizis B. Plasmacytoid Dendritic Cells: Development, Regulation, and Function. Immunity. 2019;50:37–50. doi: 10.1016/j.immuni.2018.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert Finestra T., Gribnau J. X chromosome inactivation: silencing, topology and reactivation. Curr. Opin. Cell Biol. 2017;46:54–61. doi: 10.1016/j.ceb.2017.01.007. [DOI] [PubMed] [Google Scholar]
- Rönnblom L.E., Alm G.V., Oberg K.E. Possible induction of systemic lupus erythematosus by interferon-alpha treatment in a patient with a malignant carcinoid tumour. J. Intern. Med. 1990;227:207–210. doi: 10.1111/j.1365-2796.1990.tb00144.x. [DOI] [PubMed] [Google Scholar]
- Rowland S.L., Riggs J.M., Gilfillan S., Bugatti M., Vermi W., Kolbeck R., Unanue E.R., Sanjuan M.A., Colonna M. Early, transient depletion of plasmacytoid dendritic cells ameliorates autoimmunity in a lupus model. J. Exp. Med. 2014;211:1977–1991. doi: 10.1084/jem.20132620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield R.H., Bruner G.R., Namjou B., Kimberly R.P., Ramsey-Goldman R., Petri M., Reveille J.D., Alarcón G.S., Vilá L.M., Reid J. Klinefelter’s syndrome (47,XXY) in male systemic lupus erythematosus patients: support for the notion of a gene-dose effect from the X chromosome. Arthritis Rheum. 2008;58:2511–2517. doi: 10.1002/art.23701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully E.P., Haverfield J., Ursin R.L., Tannenbaum C., Klein S.L. Considering how biological sex impacts immune responses and COVID-19 outcomes. Nat. Rev. Immunol. 2020;20:442–447. doi: 10.1038/s41577-020-0348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seillet C., Laffont S., Trémollières F., Rouquié N., Ribot C., Arnal J.F., Douin-Echinard V., Gourdy P., Guéry J.C. The TLR-mediated response of plasmacytoid dendritic cells is positively regulated by estradiol in vivo through cell-intrinsic estrogen receptor α signaling. Blood. 2012;119:454–464. doi: 10.1182/blood-2011-08-371831. [DOI] [PubMed] [Google Scholar]
- Siegal F.P., Kadowaki N., Shodell M., Fitzgerald-Bocarsly P.A., Shah K., Ho S., Antonenko S., Liu Y.J. The nature of the principal type 1 interferon-producing cells in human blood. Science. 1999;284:1835–1837. doi: 10.1126/science.284.5421.1835. [DOI] [PubMed] [Google Scholar]
- Sisirak V., Ganguly D., Lewis K.L., Couillault C., Tanaka L., Bolland S., D’Agati V., Elkon K.B., Reizis B. Genetic evidence for the role of plasmacytoid dendritic cells in systemic lupus erythematosus. J. Exp. Med. 2014;211:1969–1976. doi: 10.1084/jem.20132522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souyris M., Cenac C., Azar P., Daviaud D., Canivet A., Grunenwald S., Pienkowski C., Chaumeil J., Mejía J.E., Guéry J.C. TLR7 escapes X chromosome inactivation in immune cells. Sci. Immunol. 2018;3:1–11. doi: 10.1126/sciimmunol.aap8855. [DOI] [PubMed] [Google Scholar]
- Souyris M., Mejía J.E., Chaumeil J., Guéry J.-C. Female predisposition to TLR7-driven autoimmunity: gene dosage and the escape from X chromosome inactivation. Semin. Immunopathol. 2019;41:153–164. doi: 10.1007/s00281-018-0712-y. [DOI] [PubMed] [Google Scholar]
- Sterling T.R., Vlahov D., Astemborski J., Hoover D.R., Margolick J.B., Quinn T.C. Initial plasma HIV-1 RNA levels and progression to AIDS in women and men. N. Engl. J. Med. 2001;344:720–725. doi: 10.1056/NEJM200103083441003. [DOI] [PubMed] [Google Scholar]
- Subramanian S., Tus K., Li Q.-Z., Wang A., Tian X.-H., Zhou J., Liang C., Bartov G., McDaniel L.D., Zhou X.J. A Tlr7 translocation accelerates systemic autoimmunity in murine lupus. Proc. Natl. Acad. Sci. USA. 2006;103:9970–9975. doi: 10.1073/pnas.0603912103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson H., Cederblad B., Lindahl M., Alm G. Stimulation of natural interferon-alpha/beta-producing cells by Staphylococcus aureus. J. Interferon Cytokine Res. 1996;16:7–16. doi: 10.1089/jir.1996.16.7. [DOI] [PubMed] [Google Scholar]
- Syrett C.M., Sindhava V., Sierra I., Dubin A.H., Atchison M., Anguera M.C. Diversity of Epigenetic Features of the Inactive X-Chromosome in NK Cells, Dendritic Cells, and Macrophages. Front. Immunol. 2019;9:3087. doi: 10.3389/fimmu.2018.03087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukiainen T., Villani A.C., Yen A., Rivas M.A., Marshall J.L., Satija R., Aguirre M., Gauthier L., Fleharty M., Kirby A. Landscape of X chromosome inactivation across human tissues. Nature. 2017;550:244–248. doi: 10.1038/nature24265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Made C.I., Simons A., Schuurs-Hoeijmakers J., van den Heuvel G., Mantere T., Kersten S., van Deuren R.C., Steehouwer M., van Reijmersdal S.V., Jaeger M. Presence of Genetic Variants Among Young Men With Severe OVID-19. JAMA. 2020;324:1–11. doi: 10.1001/jama.2020.13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Syrett C.M., Kramer M.C., Basu A., Atchison M.L., Anguera M.C. Unusual maintenance of X chromosome inactivation predisposes female lymphocytes for increased expression from the inactive X. Proc. Natl. Acad. Sci. USA. 2016;113:E2029–E2038. doi: 10.1073/pnas.1520113113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Zhan Y., Zhu L., Hou Z., Liu F., Song P., Qiu F., Wang X., Zou X., Wan D. Retrospective Multicenter Cohort Study Shows Early Interferon Therapy Is Associated with Favorable Clinical Responses in COVID-19 Patients. Cell Host Microbe. 2020;28:455–464.e2. doi: 10.1016/j.chom.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmers F., Subedi N., van Buuringen N., Heister D., Vivié J., Beeren-Reinieren I., Woestenenk R., Dolstra H., Piruska A., Jacobs J.F.M. Single-cell analysis reveals that stochasticity and paracrine signaling control interferon-alpha production by plasmacytoid dendritic cells. Nat. Commun. 2018;9:3317. doi: 10.1038/s41467-018-05784-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A., Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol. Cell. 2000;5:695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- Yates A.D., Achuthan P., Akanni W., Allen J., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., Azov A.G., Bennett R. Ensembl 2020. Nucleic Acids Res. 2020;48:D682–D688. doi: 10.1093/nar/gkz966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler S.M., Beisel C., Sutter K., Griesbeck M., Hildebrandt H., Hagen S.H., Dittmer U., Altfeld M. Human pDCs display sex-specific differences in type I interferon subtypes and interferon α/β receptor expression. Eur. J. Immunol. 2017;47:251–256. doi: 10.1002/eji.201646725. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate/analyze datasets or code.