Highlights
-
•
The QIAreach™ Anti-SARS-CoV-2 Total Test (Anti-CoV2) is a rapid, qualitative serological test.
-
•
Anti-CoV2 uses nanoparticle fluorescence technology to detect total antibody against SARS-CoV-2.
-
•
Anti-CoV2 had 100 % sensitivity (PPA; 95 % CI 88.4–100 %) and 100 % specificity (NPA; 95 % CI 95.2–100 %).
-
•
No cross-reactivity observed with seasonal coronaviruses or other respiratory pathogens tested.
-
•
Anti-CoV2 provides accurate qualitative detection of total antibodies against SARS-CoV-2.
Keywords: SARS-CoV-2, COVID-19, Antibody, Serological testing
Abstract
In 2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) caused a global pandemic. Disease diagnosis, appropriate clinical management and infection control are all important factors in controlling the spread of SARS-CoV-2. The QIAreach™ Anti-SARS-CoV-2 Total Test (Anti-CoV2) is a rapid, qualitative serological test, using proprietary nanoparticle fluorescence technology to detect total antibody (IgA, IgM, and IgG) against SARS-CoV-2. Here we report the results of the US Food and Drug Administration (FDA) clinical agreement study. Thirty positive plasma or serum samples were taken from consenting individuals with polymerase chain reaction (PCR)-confirmed SARS-CoV-2 infection ≥14 days from symptom onset. Seventy-five samples from before the believed circulation of SARS-CoV-2 (November 1, 2019) were used to assess specificity. Positive percent agreement (PPA) and negative percent agreement (NPA) were calculated along with the corresponding exact two-sided 95 % confidence intervals (CI) using an FDA Emergency Use Authorized PCR test as the reference method. Anti-CoV2 was shown to have 100 % sensitivity (PPA; 95 % CI 88.4–100 %) and 100 % specificity (NPA; 95 % CI 95.2–100 %). Against 157 pre-pandemic samples, no cross-reactivity was observed with seasonal coronaviruses or other respiratory pathogens tested. Additionally, no interference was observed when samples were spiked with: conjugated bilirubin 0.4 mg/ml; unconjugated bilirubin 0.4 mg/ml; hemoglobin 5 mg/ml; prednisolone 0.12 mg/ml; triglycerides 15 mg/ml. In conclusion, Anti-CoV2 provides accurate qualitative detection of total antibodies against SARS-CoV-2.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a novel beta-coronavirus that has caused a global outbreak of respiratory disease, Coronavirus Disease 2019 (COVID-19), with significant morbidity, mortality, and excess healthcare costs [1,2]. An important aspect of controlling and slowing the spread of this pandemic is the availability of reliable and accurate methods for testing both symptomatic and asymptomatic individuals [[3], [4], [5]]. Rapid detection of cases and contacts, along with appropriate clinical management and infection control efforts, are critical to public health and disease control [4,5]. Despite researchers working around the clock, much is yet to be discovered regarding SARS-CoV-2 transmission dynamics, ability to confer antibody production and immunity, and even prevalence of the disease within our communities.
In patients infected with SARS-CoV-2, rapid, successive seroconversion of specific immunoglobulin A (IgA), immunoglobulin M (IgM) and immunoglobulin (IgG) typically occur within 14 days post onset of symptoms (DPO), with IgA responses appearing earlier, larger and more sustained than IgM [[6], [7], [8], [9], [10], [11]]. Strength of antibody responses likely correlates with disease severity [[12], [13], [14], [15]]. In a study of 259 symptomatic North American patients infected with SARS-CoV-2, ELISA-based detection of IgG, IgA, or IgM antibody responses to the receptor binding domain of the SARS-CoV-2 spike protein were all accurate in identifying infected individuals 14–28 DPO, with 100 % specificity and a sensitivity of 97 %, 91 %, and 81 %, respectively [8]. In the same study, IgG responses persisted through 75 DPO [8].
Currently, antibody testing is not recommended as the sole basis for diagnosis of acute SARS-CoV-2 infection, and as such no antibody tests are authorized by the US Food and Drug Administration (FDA) for this purpose [16]. However, antibody tests may be used in conjunction with molecular tests as a diagnostic aid, particularly in patients with delayed presentation and where viral genomic load is below the limit of detection for PCR assays, and to facilitate contact tracing, surveillance and sero-epidemiologic studies [[16], [17], [18]]. They are important for detecting past infection, including those without symptoms, as well as identifying convalescent plasma donors, and for verifying successful vaccinations once one is developed [[16], [17], [18], [19]].
Lateral flow assays (LFAs) are portable, easy to use and provide a quick readout, making them ideal point-of-care (POC) serological tests [20]. However, current LFAs are run individually, sensitivity and specificity varies between assays, and there is subjectivity by individual readers when calling faint bands [20]. Worryingly, clinical performance and sensitivity issues for some COVID-19 LFAs have been noted, and a FDA “removed” test list has been created [21]. In a meta-analysis of 40 studies, pooled sensitivity for LFAs was substantially lower than ELISA platforms measuring IgG or IgM; 66.0 % (95 % confidence interval [CI] 49.3–79.3 %) and 84.3 % (95 % CI 75.6–90.9 %), respectively [22]. Additionally, detection of IgM is less specific than IgG for many LFAs, in line with previous evidence highlighting disproportionate false-positive results with IgM tests [[23], [24], [25]]. Despite potential performance concerns, the need for rapid COVID-19 testing has led the FDA to grant Emergency Use Authorization (EUA) to a number of COVID-19 serological tests [25].
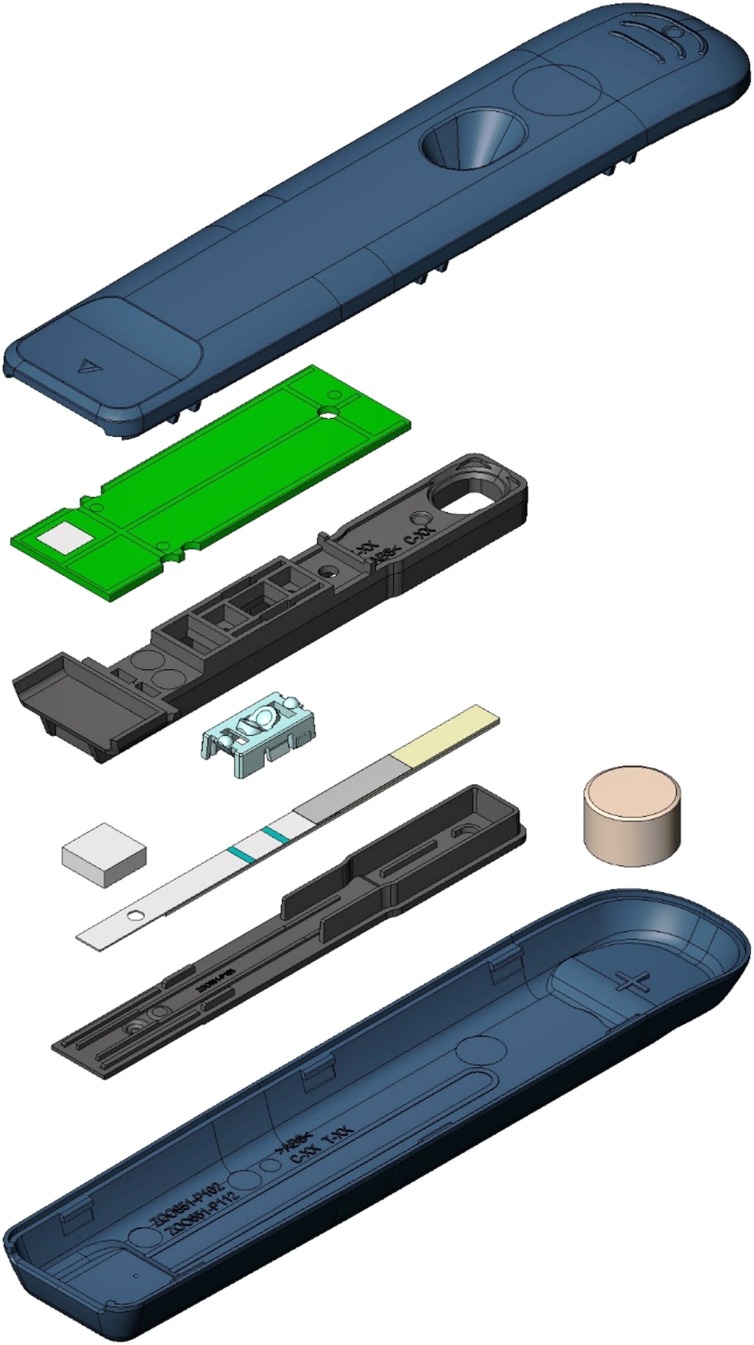
The QIAreach™ Anti-SARS-CoV-2 Total Test (Anti-CoV2) is a digital lateral flow serological test using patented nanoparticle fluorescence technology that qualitatively detects total antibodies to SARS-CoV-2 in human serum and plasma. Anti-CoV2 combines the benefits of traditional LFAs with added objective digital, rather than subjective, visual readouts. Unlike current LFAs, its unique digital detection cartridge (eStick, Fig. 1 ) and 8-port eHub design allows for low to higher volume testing of up to 32 tests per hub per hour to support real-time bio-surveillance and contact tracing in both epidemic and endemic settings.
Fig. 1.
Exploded Anti-CoV2 digital detection cartridge (eStick).
Casing (blue) contains LFA strip with optoelectronic technology and a microprocessor that converts a fluorescent signal into a qualitative readout for the detection of SARS-CoV-2 specific antibodies in patient test samples
The aim of this study was to validate Anti-CoV2 to ensure it meets the clinical performance, user needs, and supports the FDA EUA and other regulatory body submissions.
2. Materials and methods
2.1. Study samples
For the sensitivity analysis, plasma or serum samples from consenting subjects with PCR-confirmed SARS-CoV-2 infection were retrospectively obtained. Samples were collected ≥14 days from symptom onset. For the specificity analysis, plasma or serum samples collected prior to the outbreak of the COVID-19 pandemic (prior to November 1, 2019) were used. Samples were excluded if sample volume was <150 μL, samples or containers were physically damaged, or if samples were improperly collected or stored per instructions for use. Sample sizes were based on FDA EUA recommendations [24]; a total of 30 PCR-confirmed positive samples and 75 negative samples were tested in this study. The samples were blinded and randomized to the Anti-CoV2 operators prior to testing.
Cross-reactivity was evaluated by testing SARS-CoV-2 seronegative specimens from subjects with antibodies to other coronaviruses, other viral and bacterial infections and autoantibodies that could potentially cause false positive results. For the cross-reactivity analysis, negative SARS-CoV-2 plasma or serum samples collected prior to the outbreak of the COVID-19 pandemic (prior to November 1, 2019) were used.
2.2. Sample collection and storage
All specimens were collected and processed based on standard procedures used by manufacturers and vendors; plasma specimens for this study were collected using either heparin or EDTA tubes, while serum specimens for this study were collected using serum tubes. Specimens were stored frozen (at or below −20 °C) after collection, and shipped on dry ice then stored at ≤−20 °C upon arrival.
2.3. Interference study
The effect of potentially interfering substances on Anti-CoV2 performance was evaluated by spiking endogenous and exogenous interfering substances into SARS-CoV-2 negative plasma and low titer SARS-CoV-2 antibody positive plasma at the highest Clinical and Laboratory Standards Institute recommended concentrations. Notably, hemoglobin levels above 5 mg/ml (significantly reddish brown colored samples) can potentially interfere with the Anti-CoV2 optical measurement system. Anti-CoV2 eStick firmware features built-in controls to determine unacceptably high levels of hemolysate and will return an invalid result in the form of an error code if interference is present.
2.4. QIAreach™ Anti-SARS-CoV-2 Total Test
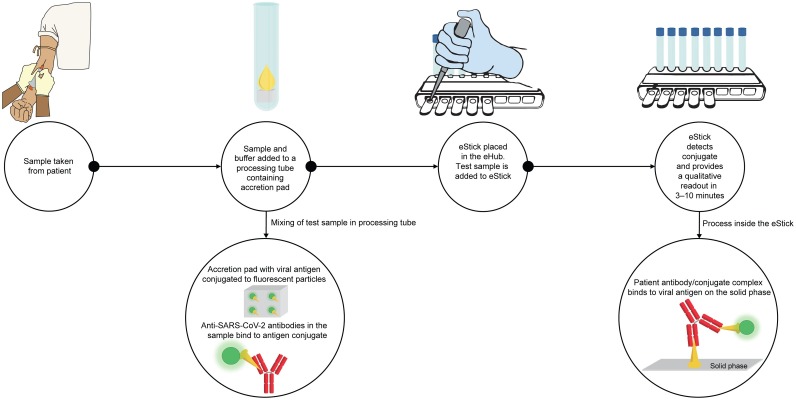
Anti-CoV2 was performed according to the manufacturer’s instructions. Anti-CoV2 Diluent Buffer is first added to the Anti-CoV2 Processing Tube and reconstitutes a SARS-CoV-2 viral S1 protein-nanoparticle conjugate. Patient serum or plasma is then added to the Processing Tube and mixed with the resuspended conjugate. The sample is then transferred from the Processing Tube to a single-use eStick sample port. Antibody responses are measured via nanoparticle fluorescence. The eStick contains optoelectronic technology and a microprocessor that converts a fluorescent signal into a qualitative readout for the detection of SARS-CoV-2 specific antibodies in patient test samples (Fig. 2 ). Upon test completion, positive or negative test results from the eStick firmware are reported on the OLED display from each connection port of the eHub. When using the manufacturer’s software to backup or transfer test results, test data is automatically transferred to an attached computer using the eHub connection port.
Fig. 2.
Assay workflow for Anti-CoV2. Specific anti-SARS-CoV-2 antibodies (red) in patient sample bind to SARS-CoV-2 antigens (orange) conjugated to fluorescent particles (green). After transfer of the sample mixture to the eStick, the antibody-conjugate complex binds to the SARS-CoV-2 antigen on the solid phase. The eStick detects the presence of SARS-CoV-2 antibodies and provides a qualitative readout to the eHub in 3–10 min.
The comparator methods for sensitivity analysis were FDA EUA-authorized PCR tests for the diagnosis of SARS-CoV-2 infection. Serology results from each individual were compared with results from one of three tests: cobas® SARS-CoV-2 Test (Roche, Switzerland), Abbott RealTime SARS-CoV-2 on the m2000 RealTime system (Abbott, United States), or Xpert® Xpress SARS-CoV-2 (Cepheid, United States).
2.5. Statistical analysis
The positive percent agreement (PPA; sensitivity) and negative percent agreement (NPA; specificity) were calculated along with the corresponding exact two-sided 95 % CI.
Calculation of positive predictive value (PPV) was based on the assumption that the prevalence of SARS-CoV-2 infection was 5 % at the time of sample collection. Calculation of negative predictive value (NPV) was based on the assumption that the prevalence of SARS-CoV-2 infection is 5 %. Interference criteria was based on 90 % overall qualitative agreement of negative and positive samples to an expected result.
3. Results
3.1. Samples
All 30 specimens tested for sensitivity were from a cohort of SARS−CoV-2 RT-PCR positive patients who experienced symptoms consistent with COVID-19, with days between symptom onset and sample collection ranging from 14 to 58 days. All samples met the eligibility criteria. The breakdown of the sample collection is given in Table 1 .
Table 1.
Sample collection details for PCR-confirmed SARS-CoV-2 positive samples.
| Days between blood draw and first date of symptom onset | ≤7 | 8–14 | 15–21 | 22–28 | 29–35 | 36–42 | 43–49 | 50–56 | 57–60 |
|---|---|---|---|---|---|---|---|---|---|
| Number of samples | 0 | 1 | 7 | 5 | 5 | 7 | 3 | 0 | 2 |
3.2. Clinical agreement
In 30 PCR-confirmed SARS-CoV-2 samples taken ≥14 DPO, antibodies to SARS-CoV-2 were detected in all samples using Anti-CoV2 resulting in 100 % sensitivity (PPA; 95 % CI 88.4–100 %) (Table 2 ). In 75 SARS-CoV-2 negative samples taken prior to the COVID-19 pandemic, no antibodies to SARS-CoV-2 were detected in all samples using Anti-CoV2 resulting in 100 % specificity (NPA; 95 % CI 95.2–100 %) (Table 2). The presumed PPV and NPV with Anti-CoV2 are 100 % (95 % CI 49.2–100 %) and 100 % (95 % CI 99.4–100 %), respectively.
Table 2.
Positive and negative percent agreement for Anti-CoV2.
CI, confidence interval; NPA, negative percent agreement; PPA, positive percent agreement.
PPA = true positive/(true positive + false negative).
NPA = true negative/(true negative + false positive).
3.3. Cross-Reactivity
Anti-CoV2 was evaluated for potential cross-reactivity with antibodies to common pathogens including seasonal coronaviruses and other respiratory viruses and bacteria, and anti-nuclear antibodies (ANA). A total of 157 individual specimens collected from SARS-CoV-2 negative individuals prior to the COVID-19 pandemic were tested. No cross-reactivity was observed (Table 3 ).
Table 3.
Cross-reactivity summary for Anti-CoV2*.
| Pathogen | N | Number cross-reactive | Number non-reactive |
|---|---|---|---|
| Adenovirus | 27 | 0 | 27 |
| Anti-nuclear antibody (ANA) | 5 | 0 | 5 |
| Bordetella pertussis | 27 | 0 | 27 |
| Chlamydia pneumoniae | 41 | 0 | 41 |
| Enterovirus | 20 | 0 | 20 |
| Haemophilus influenzae | 5 | 0 | 5 |
| HCoV 229E | 17 | 0 | 17 |
| HCoV HKU1 | 5 | 0 | 5 |
| HCoV NL63 | 5 | 0 | 5 |
| HCoV OC43 | 14 | 0 | 14 |
| Hepatitis B- HBc | 5 | 0 | 5 |
| Hepatitis B- HBs | 5 | 0 | 5 |
| Hepatitis C | 5 | 0 | 5 |
| Influenza A | 68 | 0 | 68 |
| Influenza B | 72 | 0 | 72 |
| Legionella pneumophila | 12 | 0 | 12 |
| Mycoplasma pneumoniae | 77 | 0 | 77 |
| Parainfluenza virus | 99 | 0 | 99 |
| Respiratory syncytial virus | 81 | 0 | 81 |
ANA, anti-nuclear antibody; HBc, hepatitis B core antibody; HBs, hepatitis B surface antibody; HCoV, human coronavirus.
Cross reactivity is evaluated separately for each pathogen category. Several specimens contained antibodies to multiple pathogens, resulting in some specimens listed more than once, but in separate pathogen categories.
3.4. Interference
Ten replicates of each sample were tested for interference. Interference was not observed when samples were spiked with endogenous and exogenous interfering substances at the following levels: conjugated bilirubin 0.4 mg/ml; unconjugated bilirubin 0.4 mg/ml; hemoglobin 5 mg/ml; prednisolone 0.12 mg/ml; triglycerides 15 mg/ml.
4. Discussion
In this study, we assessed the diagnostic performance of Anti-CoV2 for the detection of antibodies against SARS-CoV-2, based on the FDA EUA serological testing guidelines. FDA acceptance criteria for EUA submission are: minimum clinical sensitivity of ≥90 % for SARS-CoV-2 positive specimens and a minimum clinical specificity of ≥95 % for SARS-CoV-2 negative specimens [26]. Using 30 PCR-confirmed SARS-CoV-2 samples and 75 negative SARS-CoV-2 samples, Anti-CoV2 was shown to have 100 % sensitivity (PPA) and 100 % specificity (NPA). There was no evidence of interference from potentially interfering substances at the concentrations listed above, and no cross-reactivity was observed against samples containing antibodies to common pathogens including seasonal coronaviruses and other respiratory viruses such as influenza A and B.
The high specificity of Anti-CoV2 arises due to the multiple binding steps that have to occur for the antibodies to be detected. First, the antibody must bind to the conjugated antigen in the Processing Tube, and then the antibody-conjugated antigen complex must bind to the antigen on the solid phase within the eStick. Both the capture and detection antigen is the Spike protein S1 subunit; however, no comprehensive assessment was conducted to determine specific viral epitopes within S1. The chance of detecting non-specific binding of antibody is low due to this antigen-antibody-antigen complex requirement (Fig. 2).
No antibody tests are authorized by the FDA to be the sole diagnostic test for COVID-19 [16]. However, serological testing does have applications in sero-epidemiological studies, real-time bio-surveillance, contact tracing, detecting prior infection, identifying convalescent plasma donors, and for verifying successful vaccinations once one is developed [[16], [17], [18]]. Since the start of the COVID-19 outbreak, many manufacturers have developed serological tests with variable results, and initially with limited oversight from regulators [27]. Indeed, some tests are reported to have inadequate sensitivity and specificity [21,22]. This variability has led to uncertainty in the reliability of serological testing within the medical community, which emphasizes the need for quality tests that are properly validated [28].
In an assessment of ten LFAs (not including Anti-CoV-2), sensitivity for the detection of IgG or IgM at >20 DPO ranged from 81.8 %–100 %, with specificity 84.3 %–100 % [23]. In a separate study of six POC LFAs, sensitivity at >21 DPO ranged from 0 % to 100 %, with specificity 96 %–100 % [29], and another assessment of five POC LFAs in a low prevalence setting reported sensitivities ranging from 51.8 %–67.9 %, with specificity 95.6 %–100.0 % [30]. Furthermore, in a meta-analysis of 40 studies, pooled sensitivity for LFAs was substantially lower than ELISA platforms measuring IgG or IgM; 66.0 % (95 % CI 49.3–79.3 %) and 84.3 % (95 % CI 75.6–90.9 %), respectively [20]. In a study evaluating LFAs using finger-prick self-testing, sensitivity ranged from 21 % to 92 % versus PCR-confirmed cases, and sensitivity was observed to be significantly inferior to ELISA in 8 out of 11 LFAs assessed [31].
The high sensitivity and specificity of Anti-CoV2 compares favorably with other LFAs. The high sensitivity of Anti-CoV2 is due in part to its detection of total antibody, which will have a higher concentration in sera than each class, or class combinations, meaning Anti-CoV2 is potentially less prone to false negative results compared with other LFAs that only detect one or two classes of antibody. Multiple studies have shown evidence of seroconversion to only one class of antibody, meaning tests designed to detect the presence of one class of Ig may result in false-negatives [6,8,9,23]. Seroconversion to total Ig has also been shown to occur more quickly than seroconversion to IgG or IgM [6,9], and in a separate study, IgA was persistently higher throughout the observation period compared with IgM [11]. Another study demonstrated that IgA testing provided better diagnostic accuracy early in disease [10]. LFAs that detect IgM and/or IgG are therefore, inherently less sensitive than LFAs that detect total antibody. In this regard, The Infectious Diseases Society of America only recommends the use of IgG or total Ig for clinical and epidemiological purposes due to the lack of evidence in support of IgM only, IgA only or IgG and IgM combination tests [32].
Anti-CoV2 is simple to perform, providing rapid, precise results with minimal training and equipment. Additionally, the system overcomes the limitations in common LFA visual tests that are key to scaling testing to meet the demands inherent in a global pandemic. Simultaneous testing of multiple samples allows for increased testing volume, electronic test interpretation eliminates the subjectivity inherent in common LFA visual tests and the restrictive in-person time window required to read results, thereby reducing the number of skilled workers needed to scale up testing. The portability and ability to use the system at POC, or in near patient settings, are also of potential interest for many low- and middle-income countries, where access to molecular testing for diagnosing SARS-CoV-2 infection is insufficient. In these areas, rapid POC testing without the need for laboratories could provide an alternative triage option [3]. Anti-CoV2 could provide a reliable indication of a previous infection with SARS-CoV-2 for use in bio-surveillance, contact tracing, identification of convalescent plasma donors, sero-epidemiologic studies, and verification of vaccine efficacy.
There are some limitations to this study. Although the sample size in this study was in line with the FDA EUA serological testing guidelines, further evaluation on a larger sample set, including samples from individuals categorized by symptom severity and/or hospitalization would be of interest. In a comparative analysis of five widely used serological assays (not including Anti-CoV2), sensitivity was 100 % for all assays in hospitalized patients, but much lower for non-hospitalized patients; 69 %–91.6 % [33]. Additionally, in our study, only samples taken between 14 and 60 DPO were used in the sensitivity analysis; therefore, it is unclear what the clinical performance of this assay will be on samples taken either side of this time frame. Additionally, Anti-CoV2 needs to be evaluated in sero-epidemiological studies, as well as against other tests including LFAs and ELISA-based platforms.
In conclusion, QIAreach™ Anti-SARS-CoV-2 Total Test provides highly accurate detection of total antibodies against SARS-CoV-2.
Author contributions
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published. All authors contributed to data analysis, data interpretation and writing of this report.
Funding
This study has been funded by QIAGEN QSI.
Declaration of Competing Interest
Francis Stieber is an employee of QIAGEN LLC.
Jenny Howard is an employee of QIAGEN LLC.
Sonia N. Rao is an employee of QIAGEN LLC.
L. Masae Kawamura is an employee of QIAGEN LLC.
Davide Manissero is an employee of QIAGEN Manchester Ltd.
Joanna Love is an employee of QIAGEN Manchester Ltd.
Mei Yang is an employee of QIAGEN LLC.
Robin Uchiyama is an employee of QIAGEN LLC.
Sean Parsons is an employee of Ellume Limited.
Chris Miller is an employee of Ellume Limited.
Harmony Douwes is an employee of Ellume Limited.
Luke Fairburn is an employee of Ellume Limited.
Aaron McDonald is an employee of Ellume Limited.
Jeffrey Boyle is an employee of QIAGEN LLC.
Disclosure information
Francis Stieber is an employee of QIAGEN LLC. Jenny Howard is an employee of QIAGEN LLC. Sonia N. Rao is an employee of QIAGEN LLC. L. Masae Kawamura is an employee of QIAGEN LLC. Davide Manissero is an employee of QIAGEN Manchester Ltd. Joanna Love is an employee of QIAGEN Manchester Ltd. Mei Yang is an employee of QIAGEN LLC. Robin Uchiyama is an employee of QIAGEN LLC. Sean Parsons is an employee of Ellume Limited. Chris Miller is an employee of Ellume Limited. Harmony Douwes is an employee of Ellume Limited. Luke Fairburn is an employee of Ellume Limited. Aaron McDonald is an employee of Ellume Limited. Jeffrey Boyle is an employee of QIAGEN LLC.
CRediT authorship contribution statement
Francis Stieber: Conceptualization, Methodology, Validation, Formal analysis, Writing - original draft, Writing - review & editing. Jenny Howard: Conceptualization, Validation, Formal analysis, Writing - original draft, Writing - review & editing, Supervision. Sonia N. Rao: Conceptualization, Writing - original draft, Writing - review & editing. L. Masae Kawamura: Conceptualization, Writing - original draft, Writing - review & editing. Davide Manissero: Conceptualization, Methodology, Writing - original draft, Writing - review & editing. Joanna Love: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing - review & editing, Supervision. Mei Yang: Methodology, Validation, Formal analysis, Writing - review & editing. Robin Uchiyama: Conceptualization, Methodology, Validation, Formal analysis, Writing - review & editing. Sean Parsons: Conceptualization, Methodology, Validation, Formal analysis, Resources, Writing - original draft, Supervision, Funding acquisition. Chris Miller: Conceptualization, Methodology, Software, Resources, Validation, Writing - review & editing. Harmony Douwes: Methodology, Validation, Investigation, Data curation, Formal analysis, Writing - review & editing. Aaron McDonald: Conceptualization, Methodology, Data curation, Validation, Formal analysis, Writing - review & editing. Luke Fairburn: Software, Project administration, Methodology, Validation, Formal analysis, Writing - review & editing. Jeffrey Boyle: Conceptualization, Methodology, Validation, Formal analysis, Writing - review & editing, Resources, Supervision.
Acknowledgments
The authors would like to acknowledge Sarah Johnston, MBiolSci, of Ashfield Healthcare Communications, part of UDG Healthcare plc, for medical writing support that was funded by QIAGEN Manchester Ltd.
References
- 1.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) Outbreak in China: summary of a report of 72 314 cases from the Chinese center for disease control and prevention. JAMA. 2020;323(13):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 2.Bchetnia M., Girard C., Duchaine C., Laprise C. The outbreak of the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): a review of the current global status. J Infect Public Health. 2020 doi: 10.1016/j.jiph.2020.07.011. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peeling R.W., Wedderburn C.J., Garcia P.J., Boeras D., Fongwen N., Nkengasong J., Sall A., Tanuri A., Heymann D.L. Serology testing in the COVID-19 pandemic response. Lancet Infect. Dis. 2020;20(9):e245–e249. doi: 10.1016/S1473-3099(20)30517-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.European Commission . Vol. 18. European Union; 2020. https://ec.europa.eu/info/sites/info/files/covid19_-_eu_recommendations_on_testing_strategies_v2.pdf (COVID 19: EU Recommendations for Testing Strategies). March Available at: [Accessed August 2020] [Google Scholar]
- 5.European Centre for Disease Prevention and Control . Vol. 1. ECDC; Stockholm: 2020. https://www.ecdc.europa.eu/sites/default/files/documents/Overview-rapid-test-situation-for-COVID-19-diagnosis-EU-EEA.pdf (An Overview of the Rapid Test Situation for COVID-19 Diagnosis in the EU/EEA). Available at: April [Accessed August 2020] [Google Scholar]
- 6.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., Wang X., Yuan J., Li T., Li J., Qian S., Hong C., Wang F., Liu Y., Wang Z., He Q., Li Z., He B., Zhang T., Fu Y., Ge S., Lie L., Zhang J., Xia N., Zhang Z. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clinical Infect Dis. 2020 doi: 10.1093/cid/ciaa344. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu H.Q., Sun B.Q., Fang Z.F., Zhao J.C., Liu X.Y., Li Y.M., Sun X.Z., Liang H.F., Zhong B., Huang Z.F., Zheng P.Y., Tian L.F., Qu H.Q., Liu D.C., Wang E.Y., Xiao X.J., Li S.Y., Ye F., Li Guan, Hu D.S., Hakonarson H., Liu Z.G., Zhong N.S. Distinct features of SARS-CoV-2-specific IgA response in COVID-19 patients. Eur. Respir. J. 2020;56(2) doi: 10.1183/13993003.01526-2020. DOI: 0.1183/13993003.01526-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iyer A.S., Jones F.K., Nodoushani A., Kelly M., Becker M., Slater D., Mills R., Teng Em Kamruzzaman M., Garcia-Beltran W.F., Astudillo M., Yang D., Miller T.E., Oliver E., Fischinger S., Atyeo C., Iafrate A.J., Calderwood S.B., Lauer S.A., Yu J., Li Z., Feldman J., Hauser B.M., Caradonna T.M., Branda J.A., Turbett S.E., LaRocque R.C., Mellon G., Barouch D.H., Schmidt A.G., Azman A.S., Alter G., Ryan E.T., Harris J.B., Charles R.C. Dynamics and significance of the antibody response to SARS-CoV-2 infection. medRxiv. 2020 DOI: 2020.07.18.20155374. [Google Scholar]
- 9.Lou B., Li T.D., Zheng S.F., Su Y.Y., Li Z.Y., Liu W., Yu F., Ge S.X., Zou Q.D., Yuan Q., Lin S., Hong C.M., Yao X.Y., Zhang X.J., Wu D.H., Zhou G.L., Hou W.H., Li T.D., Zhang Y.L., Zhang S.Y., Fan J., Zhang X.J., Xia N.S., Chen Y. Serology characteristics of SARS-CoV-2 infection since exposure and post symptom onset. Eur. Respir. J. 2020;56(2) doi: 10.1183/13993003.00763-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma H., Zeng W., He H., Zhao D., Jiang D., Zhou P., Cheng L., Li Y., Ma X., Jin T. Serum IgA, IgM, and IgG responses in COVID-19. Cell. Mol. Immunol. 2020;17:773–775. doi: 10.1038/s41423-020-0474-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Padoan A., Sciacovelli L., Basso D., Negrini D., Zuin S., Cosma C., Faggian D., Matricardi P., Plebani M. IgA-Ab response to spike glycoprotein of SARS-CoV-2 in patients with COVID-19: a longitudinal study. Clinical Chimica Acta. 2020;507:164–166. doi: 10.1016/j.cca.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rijkers G., Murk J.L., Wintermans B., van Looy B., van den Berge M., Veenemans J., Stohr J., Reusken C., van der Pol P., Reimerink J. Differences in antibody kinetics and functionality between severe and mild SARS-CoV-2 infections. J Infect Dis. 2020 doi: 10.1101/2020.06.09.20122036. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long Q-x, Deng H-j, Chen J., Hu J., B-z Liu, Liao P., Lin Y., Yu L., Mo Z., Xu Y., Gong F., Wu G., Zhang X., Chen Y., Li Z., Wang K., Zhang X., Tian W., Niu C., Yang Q., Xiang J., Du H., Liu H., Lang C., Luo X., Wu S., Cui X., Zhou Z., Wang J., Xue C., Li X., Wang L., Tang X., Zhang Y., Qiu J., Liu X., Li J., Zhang D., Zhang F., Cai X., Wang D., Hu Y., Ren J., Tang N., Liu P., Li Q., Huang A. Antibody responses to SARS-CoV-2 in COVID-19 patients: the perspective application of serological tests in clinical practice. medRxiv. 2020 doi: 10.1101/2020.03.18.20038018. [DOI] [Google Scholar]
- 14.Wang Y., Zhang L., Sang L., Ye F., Ruan S., Zhong B., Song T., Alshukairi A.N., Chen R., Zhang Z., Gan M., Zhu A., Huang Y., Luo L., Mok C.K.P., Al Gethamy M.M., Tan H., Li Z., Huang X., Li F., Sun J., Zhang Y., Wen L., Li Y., Chen Z., Zhuang Z., Zhuo J., Chen C., Kuang L., Wang J., Lv H., Jiang Y., Li M., Lin Y., Deng Y., Tang L., Liang J., Huang J., Perlman S., Zhong N., Zhao J., Malik Peiris J.S., Li Y., Zhao J. Kinetics of viral load and antibody response in relation to COVID-19 severity. J Clin Invest. 2020 doi: 10.1172/JCI138759. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan W., Lu Y., Zhang J., Wang J., Dan Y., Tan Z., He X., Qian C., Sun Q., Hu Q., Liu H., Ye S., Xiang X., Zhou Y., Zhang W., Gui Y., Wang X.H., He W., Wan X., Sun F., Wei Q., Chen C., Pan G., Xia J., Mao Q., Chen Y., Deng G. Viral kinetics and antibody responses in patients with COVID-19. medRxiv. 2020 doi: 10.1101/2020.03.24.20042382. [DOI] [Google Scholar]
- 16.US Food and Drug Administration . FDA: Division of Industry and Consumer Education; 2020. Important Information on the Use of Serological (Antibody) Tests for COVID-19 - Letter to Health Care Providers.https://www.fda.gov/medical-devices/letters-health-care-providers/important-information-use-serological-antibody-tests-covid-19-letter-health-care-providers Available at: [Accessed August 2020] [Google Scholar]
- 17.Infectious Diseases Society of America . 2020. IDSA COVID-19 Antibody Testing Primer. IDSA.https://www.idsociety.org/globalassets/idsa/public-health/covid-19/idsa-covid-19-antibody-testing-primer.pdf May 4Available at: [Accessed August 2020] [Google Scholar]
- 18.Petherick A. Developing antibody tests for SARS-CoV-2. Lancet. 2020;395(10230):1101–1102. doi: 10.1016/S0140-6736(20)30788-1. DOI: 10.1042/EBC20150012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.CDC Overview of Testing for SARS-CoV-2 (COVID-19) https://www.cdc.gov/coronavirus/2019-ncov/hcp/testing-overview.html Available at: [Accessed August 2020]
- 20.Koczula K.M., Gallotta A. Lateral flow assays. Essays Biochem. 2016;60(1):111–120. doi: 10.1042/EBC20150012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.US Food and Drug Administration . FDA: Division of Industry and Consumer Education; 2020. Certain COVID-19 Serology/Antibody Tests Should Not Be Used - Letter to Clinical Laboratory Staff and Health Care Providers.https://www.fda.gov/medical-devices/letters-health-care-providers/certain-covid-19-serologyantibody-tests-should-not-be-used-letter-clinical-laboratory-staff-and Available at: [Accessed August 2020] [Google Scholar]
- 22.Bastos M.L., Tavaziva G., Abidi S.K., Campbell J.R., Haraoui L.P., Johnston J.C., Lan Z., Law S., MacLean E., Trajman A., Menzies D., Benedetti A., Khan F.A. Diagnostic accuracy of serological tests for covid-19: systematic review and meta-analysis. BMJ. 2020;370:m2516. doi: 10.1136/bmj.m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whitman J.D., Hiatt J., Mowery C.T., Shy B.R., Yu R., Yamamoto T.N., Rathore U., Goldgof G.M., Whitty C., Woo J.M., Gallman A.E., Miller T.E., Levine A.G., Nguyen D.N., Bapat S.P., Balcerek J., Bylsma S.A., Lyons A.M., Li S., Wong A.W., EM Gilli S.-Buck, Steinhart Z.B., Lee Y., Apathy R., Lipke M.J., Smith J.A., Zheng T., Boothby I.C., Isaza E., Chan J., Acenas D.D., 2nd, Lee J., Macrae T.A., Kyaw T.S., Wu D., Ng D.L., Gu W., York V.A., Eskandarian H.A., Callaway P.C., Warrier L., Moreno M.E., Levan J., Torres L., Farrington L.A., Loudermilk R., Koshal K., Zorn K.C., Garcia-Beltran W.F., Yang D.L., Astudillo M.G., Bernstein B.E., Gelfand J.A., Ryan E.T., Charles R.C., Iafrate A.J., Lennerz J.K., Miller S., Chiu C.Y., Stramer S.L., Wilson M.R., Mangdlik A., Ye C.J., Krogan N.J., Anderson M.S., Cyster J.G., Ernst J.D., Wu A.H.B., Lynch K.L., Bern C., Hsu P.D., Marson A. Test performance evaluation of SARS-CoV-2 serological assays. medRxiv. 2020 doi: 10.1101/2020.04.25.20074856. 2020.04.25.20074856. [DOI] [Google Scholar]
- 24.Steensels D., Oris E., Coninx L., Nuyens D., Delforge M.L., Vermeersch P., Heylen L. Hospital-wide SARS-CoV-2 antibody screening in 3056 staff in a tertiary center in Belgium. JAMA. 2020;324(2):195–197. doi: 10.1001/jama.2020.11160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landry M.L. Immunoglobulin M for acute infection: true or false? Clin. Vaccine Immunol. 2016;23(7):540–545. doi: 10.1128/CVI.00211-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.US Food and Drug Administration . FDA; 2020. EUA Authorized Serology Test Performance.https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/eua-authorized-serology-test-performance Available at: [Accessed August 2020] [Google Scholar]
- 27.US Food and Drug Administration . FDA; 2020. In Vitro Diagnostics EUAs.https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/vitro-diagnostics-euas Available at: [Accessed August 2020] [Google Scholar]
- 28.Torres R., Rinder H.M. Double-edged spike-are SARS-CoV-2 serologic tests safe right now? Lab. Med. 2020;51(3):236–238. doi: 10.1093/labmed/lmaa025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Charlton C.L., Kanji J.N., Johal K., Bailey A., Plitt S.S., MacDonald C., Kunst A., Buss E., Burnes L.E., Fonseca K., Berenger B.M., Schnabl K., Hu J., Stokes W., Zelyas N., Tipples G. Evaluation of six commercial mid to high volume antibody and six point of care lateral flow assays for detection of SARS-CoV-2 antibodies. J Clin Microbiol. 2020 doi: 10.1128/JCM.01361-20. in press JCM.01361-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bond K., Nicholson S., Lim S.M., Karapanagiotidis T., Williams E., Johnson D., Hoang T., Sia C., Purcell D., Mordant F., Lewin S.R., Catton M., Subbarao K., Howden B.P., Williamson D.A. Evaluation of serological tests for SARS-CoV-2: implications for serology testing in a low-prevalence setting. J Infect Dis. 2020 doi: 10.1093/infdis/jiaa467. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flower B., Brown J.C., Simmons B., Moshe M., Frise R., Penn R., Kugathasan R., Petersen C., Daunt A., Ashby D., Riley S., Atchinson C.J., Taylor G.P., Satkunarajah S., Naar L., Klaber R., Badhan A., Rosadas C., Khan M., Fernandez N., Sureda-Vives M., Cheeseman H.M., O’Hara J., Fontana G., Pallett S.J.C., Rayment M., Jones R., Moore L.S.P., McClure M.O., Cherepanov P., Tedder R., Ashrafian H., Shattock R., Ward H., Darzi A., Elliot P., Barclay W.S., Cooke G.S. Clinical and laboratory evaluation of SARS-CoV-2 lateral flow assays for use in a national COVID-19 seroprevalence survey. Thorax. 2020 doi: 10.1136/thoraxjnl-2020-215732. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanson K.E., Caliendo A.M., Arias C.A., Englund J.A., Hayden M.K., Lee M.J. Infectious Diseases Society of America Guidelines on the Diagnosis of COVID-19: Serologic Testing. https://www.idsociety.org/covid19guidelines/serology Available at: [Accessed August 2020] [DOI] [PMC free article] [PubMed]
- 33.Brochot E., Demey B., Handala L., François C., Duverlie G., Castelain S. Comparison of different serological assays for SARS-CoV-2 in real life. J. Clin. Virol. 2020;130 doi: 10.1016/j.jcv.2020.104569. [DOI] [PMC free article] [PubMed] [Google Scholar]