Abstract
Basic helix‐loop‐helix (bHLH) transcription factors (TFs) are key regulators of plant specialized metabolites, including terpenoid indole alkaloids (TIAs) in Catharanthus roseus. Two previously characterized subgroup‐IVa bHLH TFs, BIS1 (bHLH Iridoid Synthesis 1) and BIS2 regulate iridoid biosynthesis in the TIA pathway. We reanalyzed the recently updated C. roseus genome sequence and discovered that BIS1 and BIS2 are clustered on the same genomic scaffold with a previously uncharacterized bHLH gene, designated as BIS3. Only a few bHLH gene clusters have been studied to date. Comparative analysis of 49 genome sequences from different plant lineages revealed the presence of analogous bHLH clusters in core angiosperms, including the medicinal plants Calotropis gigantea (giant milkweed) and Gelsemium sempervirens (yellow jessamine), but not in the analyzed basal angiosperm and lower plants. Similar to the iridoid pathway genes, BIS3 is highly expressed in roots and induced by methyl jasmonate. BIS3 activates the promoters of iridoid branch genes, geraniol synthase (GES), geraniol 10‐hydroxylase (G10H), 8‐hydroxygeraniol oxidoreductase (8HGO), iridoid synthase (IS), 7‐deoxyloganetic acid glucosyl transferase (7‐DLGT), and 7‐deoxyloganic acid hydroxylase (7DLH), but not iridoid oxidase (IO). Transactivation of the promoters was abolished when BIS3 is converted to a dominant repressor by fusing with the ERF‐associated amphiphilic repression (EAR) sequence. In addition, BIS3 acts synergistically with BIS1 and BIS2 to activate the G10H promoter in tobacco cells. Mutation of the known bHLH TF binding motif, G‐box (CACGTG) in the G10H promoter significantly reduced but did not abolish the transactivation by BIS3. Promoter deletion analysis of G10H suggests that the sequences adjacent to the G‐box are also involved in the regulation by BIS3. Overexpression of BIS3 in C. roseus flower petals significantly upregulated the expression of iridoid biosynthetic genes and increased loganic acid accumulation. BIS2 expression was significantly induced by BIS3 although BIS3 did not directly activate the BIS2 promoter. Our results advance our understanding of the regulation of plant specialized metabolites by bHLH TF clusters.
Keywords: bHLH gene cluster, Catharanthus roseus (Madagascar periwinkle), terpenoid indole alkaloids, transcriptional regulation
1. INTRODUCTION
Basic helix–loop–helix (bHLH) transcription factor (TF) genes are ancient regulatory genes present in all eukaryotic kingdoms and one of the largest TF families in plants (Feller et al., 2011). Plant bHLH TFs have been classified into 12 groups (I–XII), and each group is further subdivided into smaller orthologous subgroups (Heim et al., 2003). bHLH TFs have emerged as crucial components in the gene regulatory networks controlling many biological processes in plants, including light and phytohormone signaling (Kazan & Manners, 2013; Lee et al., 2006; Leivar et al., 2008; Yin et al., 2005), stress responses (Abe et al., 1997), shoot branching (Yang et al., 2012), and tissue and organ development (Kanaoka et al., 2008; Morohashi et al., 2007; Rajani & Sundaresan, 2001; Sorensen et al., 2003; Szécsi et al., 2006). In addition, bHLH TFs regulate the biosynthesis of plant specialized metabolites, such as nicotine in tobacco (Shoji & Hashimoto, 2011; Zhang et al., 2012), glucosinolates in Arabidopsis (Schweizer et al., 2013), cucurbitacin in cucumber (Shang et al., 2014), phytoalexins in rice (Yamamura et al., 2015), artemisinin in Artemisia annua (Shen et al., 2016), paclitaxel in Taxus cuspidata (Lenka et al., 2015), saponins in Medicago truncatula (Mertens, Pollier, et al., 2016; Ribeiro et al., 2020), amygdalin in almond (Sanchez‐Perez et al., 2019), and anthocyanins in many plant species (Patra et al., 2013; Xu et al., 2015). The bHLH TFs belonging to subgroup IIId, IIIe, and IIIf are well characterized for their roles in plant specialized metabolite biosynthesis. For instance, the subgroup‐IIIe (e.g., MYC2‐type) and ‐IIIf (e.g., GL3, EGL3, and TT8) bHLH TFs are positive regulators (Patra et al., 2013), whereas those of subgroup‐IIId (bHLH03, bHLH13, bHLH14, and bHLH17) are negative regulators, of flavonoid biosynthesis in Arabidopsis (Sasaki‐Sekimoto et al., 2013; Song et al., 2013). Two subgroup‐Ib bHLH, Bl (bitter leaf) and Bt (bitter fruit) regulate biosynthesis of cucurbitacin in cucumber (Shang et al., 2014). The group‐IV bHLH TFs are divided into four relatively less studied subgroups (IVa‐d). In Arabidopsis, members of subgroup‐IVa (bHLH18, bHLH19, bHLH20, and bHLH25) regulate iron homeostasis (Cui et al., 2018). A few subgroup‐IVa bHLH TFs are key regulators of terpene biosynthesis in plants. Triterpene Saponin Biosynthesis Activating Regulator 1 (TSAR1), TSAR2, and TSAR3 regulate the biosynthesis of the triterpene saponins in M. truncatula (Mertens, Pollier, et al., 2016; Ribeiro et al., 2020). In Chenopodium quinoa (quinoa), two TSAR‐like TFs, TSARL1 and TSARL2, regulate saponin biosynthesis (Jarvis et al., 2017). Information is limited on the subgroup‐IV bHLH TFs for the regulation of biosynthesis of specialized metabolites.
Cathuranthus roseus is the exclusive source of almost 200 terpenoid indole alkaloids (TIAs), including the therapeutic compounds vincristine and vinblastine (De Luca et al., 2014). TIA biosynthesis involves more than 30 different enzymes and takes place in at least four different cell types and subcellular compartments (Courdavault et al., 2014; Patra et al., 2013). TIAs are derived from two distinct pathways: the shikimate pathway generates the indole moiety, tryptamine, and the methylerythritol phosphate (MEP)‐derived iridoid pathway provides the terpenoid moiety secologanin. Condensation of tryptamine and secologanin yields strictosidine that serves as a precursor for numerous more complex TIAs (Thamm et al., 2016). Studies by independent research laboratories over the years have identified a number of TFs that regulate TIA biosynthesis in response to phytohormones and other abiotic factors. These TFs include transcriptional activators from the TF families of AP2/ERFs (ORCA2/3/4/5/6) (van der Fits & Memelink, 2000; Li et al., 2013; Menke et al., 1999; Paul et al., 2017, 2020; Singh et al., 2020), bHLH (CrMYC2, BIS1 and 2) (Schweizer et al., 2018; Van Moerkercke et al., 2015, 2016; Zhang et al., 2011), WRKY (CrWRKY1) (Suttipanta et al., 2011), and GATA (CrGATA1) (Liu et al., 2019), as well as transcriptional repressors from the families of basic leucine zipper (bZIP) GBF1/2 (Sui et al., 2018), bHLH (RMT1 and PIF1) (Liu et al., 2019; Patra et al., 2018) and zinc finger factors (ZCT1/2/3) (Pauw et al., 2004). However, compared to the discovery of almost the complete set of genes encoding TIA pathway enzymes, fewer regulatory proteins have been identified and characterized.
Genomic sequence analysis has revealed that a subset of TFs involved in specialized metabolite biosynthesis is present as clusters in plant genomes. In C. roseus, five AP2/ERFs, ORCA2, ORCA3, ORCA4, ORCA5, and ORCA6, reside on the same genomic scaffold to form a cluster (Kellner et al., 2015; Paul et al., 2017; Singh et al., 2020). Analogous AP2/ERF gene clusters have also been identified and characterized in tobacco for nicotine biosynthesis (Kajikawa et al., 2017; Shoji et al., 2010) and in tomato (Cardenas et al., 2016; Nakayasu et al., 2018; Thagun et al., 2016) and potato (Cardenas et al., 2016) for the biosynthesis of steroidal glycoalkaloids (SGAs). In petunia, an R2R3 MYB TF cluster controls anthocyanin biosynthesis (Zhang et al., 2019). A member of an almond bHLH TF cluster (bHLH1 to bHLH5) is involved in the biosynthesis of amygdalin (Sanchez‐Perez et al., 2019). Similarly, two bHLH factors, Bl and Bt, involved in cucurbitacin biosynthesis, are part of a small bHLH TF cluster (Shang et al., 2014). TF clusters comprise homologous TF genes in tandem orders with overlapping or unique functions. As demonstrated in ERF clusters in C. roseus and tobacco, a conserved mechanism allows the individual TFs within a cluster to regulate each other (Paul et al., 2020), with some individuals playing more dominant roles in regulating the pathway than others in the same cluster (Shoji & Yuan, 2021; Yuan, 2020). Our current understanding of the origin, numbers, and evolution of TF clusters remains scant. We, therefore, endeavored to explore the recently updated C. roseus genome sequence to identify additional candidate TF clusters involved in TIA biosynthesis.
Two bHLH TFs, BIS1, and BIS2, regulate the iridoid branch of the TIA pathway (Van Moerkercke et al., 2015, 2016). We discovered that BIS1 and BIS2 are present in the same genomic scaffold in the C. roseus genome along with a previously uncharacterized bHLH TF, designated as BIS3. Genomic sequence analyses revealed the presence of analogous bHLH clusters in a wide range of plant species, including Arabidopsis and several medicinal plants. Transcriptomic analysis revealed that spatial expression of clustered BIS genes and iridoid pathway genes is highly correlated and root‐specific. Similar to BIS1 and BIS2, BIS3 expression is induced by methyl jasmonate (MeJA). Transient overexpression of BIS3 in C. roseus flower petals significantly upregulated the iridoid pathway genes. In addition, BIS3 activates the key iridoid pathway gene promoters in plant cells. Our findings provide new insights into bHLH TF clusters involved in the biosynthesis of plant specialized metabolites.
2. MATERIALS AND METHODS
2.1. Plant materials
Catharanthus roseus (L.) G. Don var. “Little Bright Eye” (NE Seed) was used for gene expression, cloning, and transient overexpression in flower petals. Nicotiana tabacum var. Xanthi cell line was used for protoplast‐based transient assays. Tobacco cell suspension cultures were maintained at 26°C in dark by gentle shaking (120 rpm). C. roseus plants were maintained in the growth room under 16/8 hr photoperiod at 24°C.
2.2. Construction of plant expression vector and Agrobacterium infiltration of C. roseus petals
For transient overexpression in flower petals, BIS3 was amplified using PCR from C. roseus seedling cDNA and cloned into the pCAMBIA2301 vector containing the CaMV35S promoter and the rbcS terminator (Pattanaik, Kong, et al., 2010). Construction pCAMBIA2301‐ORCA5 was described previously (Paul et al., 2020). The pCAMBIA2301 vector alone was used as an empty vector (EV) control. The plasmids were transformed into Agrobacterium tumefaciens GV3101 by the freeze‐thaw method. All open flowers in the plants were removed, and flower buds opened the next day were used for transformation. Agrobacterium infiltration of C. roseus flower petals was performed as previously described (Schweizer et al., 2018). Flower petals uniformly infiltrated with Agrobacterium were collected after 48h for the measurement of gene expression and metabolites. Data presented here are from three independent biological replicates.
2.3. RNA isolation and cDNA synthesis
C. roseus seeds were surface‐sterilized as described previously (Patra et al., 2018) and then germinated on half‐strength solid Murashige and Skoog (MS) medium (Caisson Labs). Two‐week‐old axenic seedlings were immersed in half‐strength MS medium with 100 µM methyl jasmonate (MeJA) for 2 hr. Mock‐treated seedlings were used as control. Total RNA was isolated from the seedlings, digested with DNase, and used for first‐strand cDNA synthesis as described previously (Paul et al., 2017).
2.4. Tobacco protoplast isolation and electroporation
The reporter plasmids for transient protoplast assays were generated by cloning the GES, G10H, 8HGO, IS, IO, 7‐DLGT, and 7DLH promoters upstream of a firefly luciferase (LUC) reporter and rbcS terminator. The G‐box motif (CACGTG) in the G10H promoter was mutated using site‐directed mutagenesis as previously described (Pattanaik et al., 2010). The generation of deletion fragments of G10H promoter using PCR was described previously (Suttipanta et al., 2007). The GUS reporter in G10H promoter fragments was replaced by LUC. The effector plasmids were made by cloning BIS1, BIS2, and BIS3 into a modified pBS vector under the control of the CaMV35S promoter and rbcS terminator. The 12 amino acid (LDLDLELRLGFA) EAR (ERF‐associated amphiphilic repression) motif (also know as SRDX) (Hiratsu et al., 2003) was fused to the 3'‐end of BIS3 to generate BIS3‐SRDX. The ß‐glucuronidase (GUS) reporter driven by the CaMV35S promoter and rbcS terminator was used as an internal control. Protoplast isolation from tobacco cell suspension cultures and electroporation with plasmid DNA were performed as previously described. The reporter (promoter‐LUC) plasmid alone or in combination with the effectors were electroporated into tobacco protoplasts as previously described (Pattanaik, Werkman, et al., 2010). An aliquote of 750 µl containing approximately 2 × 106 protoplasts were used for each electroporation. Luciferase and GUS activities in transfected protoplasts were measured as described previously (Pattanaik, Werkman, et al., 2010). Each experiment was repeated three times.
2.5. Real‐time quantitative PCR
Real‐time quantitative PCR (RT‐qPCR) was performed as described previously (Suttipanta et al., 2011). All PCRs were performed in triplicate and repeated at least three times. Total RNA isolated from C. roseus seedlings, or flower petals infiltrated with the empty‐vector, BIS3, or ORCA5 was digested with DNase and used for cDNA synthesis and RT‐qPCR as previously described (Patra et al., 2018; Paul et al., 2017). The comparative cycle threshold (Ct) method was used to measure transcript levels. In addition to the C. roseus Elongation Factor 1∞ (EF1∞), 40S Ribosomal Protein S9 (RPS9) gene, was used as a second internal control (Liscombe et al., 2010). The primers are listed in Table S1.
2.6. Bioinformatic analysis
Transcriptomic data from five different tissues (flower, immature leaf, mature leaf, stem, and root) were obtained from the NCBI sequence read archive database (accession number SRA030483). Raw reads were processed and reads per kilobase of transcript per million mapped reads (RPKM) value was calculated as described previously (Singh et al., 2015). The gene expression correlation matrices for the BISs were calculated and visualized with the corrplot R package (Wei & Simko, 2017). Heatmap analyses of BISs expression in different tissues were carried out using the pheatmap package with euclidean distance and complete linkage as distance measure and clustering methods (Kolde & Kolde, 2015).
The Neighbor‐Joining (NJ) tree was constructed based on a MAFFT v7 alignment and visualized by Evolview v3 (Subramanian et al., 2019). Gene location was visualized using the Gview (Petkau et al., 2010). PLAZA database, a versatile resource to study comparative genomics and to analyze the evolution of gene families in the green plant lineages (Van Bel et al., 2018), was used to get the information on the genomic organization of BIS homologs in other plant species.
2.7. Alkaloid analysis
Alkaloid extracted from flower petals infiltrated with empty vector or BIS3 were analyzed as previously described (Patra et al., 2018; Singh et al., 2020).
2.8. Statistical analysis
The data presented here were statistically analyzed by Student's t test or one‐way analysis of variance (ANOVA), and Tukey's Honestly Significant Difference (HSD) for multiple comparisons.
3. RESULTS
3.1. Genome sequence analysis revealed the BIS TF cluster in C. roseus genome
Previously, two subgroup‐IVa bHLH TFs, BIS1, and BIS2, have been reported as the regulators of iridoid branch of the TIA pathway in C. roseus (Van Moerkercke et al., 2015, 2016). The two known BIS genes were used as queries in our analysis to reveal the putative location of a bHLH gene cluster on scaffold 135. Further analysis showed that the >1,300‐kb long scaffold 135 contains three bHLH genes in a <100‐kb region of the scaffold. In addition to BIS1 and BIS2, an uncharacterized bHLH gene, designated here as BIS3 (GenBank accession no. MN646782), was found in close proximity to the same scaffold (Figure 1a).
FIGURE 1.
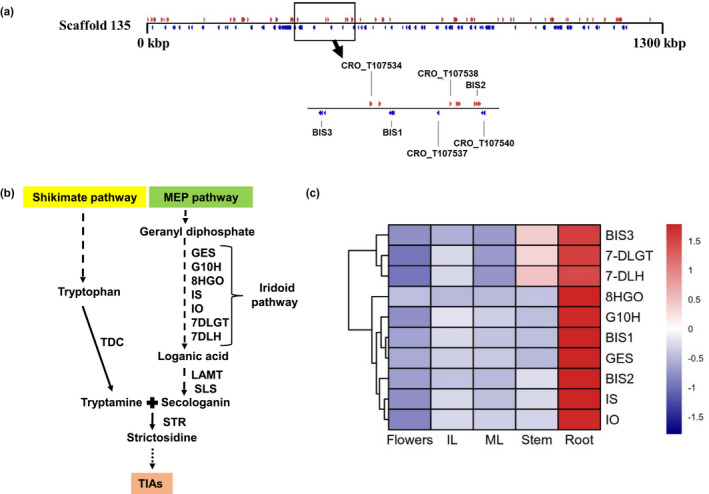
Characterization of the BISs gene cluster. (a) Organization of the BIS gene clusters in C. roseus genome. Genes and their orientations are depicted using RED and BLUE color arrows: RED, genes on the sense strand; BLUE, genes on the antisense strand. (b) A simplified diagram of TIA pathway in C. roseus: 7DLGT, 7‐deoxyloganetic acid glucosyl transferase; 7DLH, 7‐deoxyloganic acid hydroxylase; 8HGO, 8‐hydroxygeraniol oxidoreductase; G10H, geraniol 10‐hydroxylase; GES, geraniol synthase; LAMT, loganic acid O‐methyl transferase; IO, iridoid oxidase; IS, iridoid synthase; SLS, secologanin synthase; STR, strictosidine synthase; TDC, tryptophan decarboxylase (c) Expression of BISs and iridoid pathway genes in various tissues. Heatmap showing the normalized RNA‐seq (Accession no. SRA030483) expression values in reads per kilobase of transcript per million mapped reads (RPKM) transformed into Z‐score. Hierarchical clustering was conducted in R using the pheatmap package based on Euclidean distance with complete linkage rule. The color key at the side represents row‐wise Z‐score
3.2. Subgroup‐IVa bHLH TF cluster is common in core angiosperms
To determine the origin, distribution, and species‐specific expansions or reductions of the subgroup‐IVa bHLH TF cluster during the course of green plant evolution, we performed a comparative genomic analysis of 49 green plants from different lineages that include 42 eudicots, two monocots (rice and maize), the basal angiosperm (Amborella trichopoda), one lycophyte (Selaginella moellendorffii), liverwort (Marchantia polymorpha), moss (Physcomitrella patens), and one chlorophyte (Chlamydomonas reinhardtii) (Figure 2a). We did not find such bHLH gene cluster in lower plant lineages, such as C. reinhardtii, M. polymorpha, P. patens, S. moellendorffii, and A. trichopoda. Subgroup‐IVa bHLH TF clusters were found in most of the analyzed monocot and dicot genomes, with a few exceptions, such as Erythranthe guttata (Phrymaceae) and Populus trichocarpa (Salicaceae). The numbers of genes in the subgroup‐IVa bHLH clusters vary greatly from two (Chenopodium quinoa, Calotropis gigantea), three (Arabidopsis and C. roseus) to seven (Trifolium pratense, Eucalyptus grandis). The bHLH genes in a cluster are either arranged in tandem (as in Arabidopsis) or interrupted by one or more genes (as in C. roseus) (Figure 2b). These findings indicate that subgroup‐IVa bHLH clusters possibly have appeared after the separation of the mesangiosperms (core angiosperms) from the basal angiosperm, A. trichopoda, and other lower plants, and further expansion or reduction occurred at the genus level in the core angiosperm lineage.
FIGURE 2.
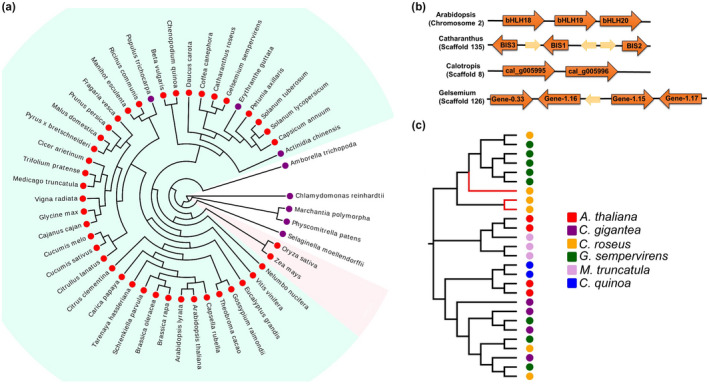
The clade IV bHLHs in plants (a) Species tree showing the distribution of bHLH subgroup IVa gene cluster in different plants. The presence and absence of gene clusters are indicated by red and purple circles, respectively. The clade with monocot plants is highlighted with a light pink background, while dicot‐specific clades are with aquamarine background. (b) A schematic representation of the subgroup IVa bHLH clusters in Catharanthus roseus and their orthologs in Calotropis gigantea, Gelsemium sempervirens, and Arabidopsis thaliana. (c) Phylogenetic analysis of bHLH subgroup IVa transcription factors from C. roseus, C. gigantea, G. sempervirens, A. thaliana, Medicago truncatula and Chenopodium quinoa. The BISs‐specific clade is highlighted in red color
3.3. Phylogenetic analysis of subgroup‐IVa bHLH cluster
In Arabidopsis, three (bHLH18, bHLH19, bHLH20) of the four subgroup‐IVa bHLH TFs form a cluster that regulates the iron deficiency responses and homeostasis by interacting with other bHLH TFs (Cui et al., 2018). In C. roseus, the subgroup‐IVa contains six bHLH TFs, and only BIS1, BIS2, and BIS3 are clustered in the same genomic scaffold, while the other three are present in different scaffolds. We also found subgroup‐IVa bHLH gene clusters in the genomes of two recently sequenced medicinal plants, C. gigantea (Apocynaceae) and Gelsemium sempervirens (Gelsemiaceae). C. gigantea is the source of the anticancer and antimalarial cardenolides (Hoopes et al., 2018) and found to contain a cluster of two bHLH genes. G. sempervirens is related to C. roseus and produces oxyindole alkaloids (Franke et al., 2019). G. sempervirens genome harbors a cluster of three bHLH genes that are homologous to BISs (Figure 2b). Unlike the C. roseus and G. sempervirens bHLH clusters, which are interrupted by other non‐homologous genes, the two bHLH genes in C. gigantea are arranged in a tandem array (Figure 2b). In C. roseus, one gene (CRO_T107534) is located in between BIS1 and BIS3 and two genes (CRO_T107537, CRO_T107538) are present in between BIS1 and BIS2. CRO_T107534, CRO_T107537, and CRO_T107538 are annotated as hypothetical proteins in C. roseus genome. Next, we performed phylogenetic analysis of the subgroup‐IVa bHLH TFs from Arabidopsis, C. roseus, M. truncatula, C. quinoa, G. sempervirens, and C. gigantean. The three members of the BIS cluster are grouped together in a sub‐clade, whereas three other non‐clustered C. roseus IVa bHLH TFs are present in different sub‐clades (Figure 2c).
3.4. BIS TFs share limited sequence identity but a similar expression profile
Amino acid sequence analysis showed that all three BIS contain a highly conserved bHLH DNA binding domain (Figure S1). BIS3 shares approximately 39% amino acid sequence identity with BIS1 and 85% identity with BIS2. Analysis of the genomic sequence revealed that both BIS1 and BIS2 harbor four exons and three introns whereas the BIS3 gene harbors five exons interrupted by four introns (Figure S1). Next, we analyzed the expression profiles of the BIS cluster genes and the iridoid pathway genes (Figure 1b). We used the publicly available transcriptomic data from different tissues (immature leaf, mature leaf, stem, root and flower; accession number SRA030483) to examine gene expression. Similar to BIS1 and BIS2, expression of BIS3 was significantly higher in the roots compared to other tissues (Figure 1c). Co‐expression analysis showed that, similar to all BISs, iridoid pathway genes are highly expressed in roots (Figure 1c).
3.5. BIS3 expression is induced by methyl jasmonate
MeJA is a major elicitor of a wide range of plant specialized metabolites, including anthocyanins, nicotine, TIA, glucosinolates (GS), benzophenanthridine alkaloids, flavonoids, and artemisinin (Wasternack & Strnad, 2019). Members of the subgroup‐IVa bHLH TF families are induced by MeJA in M. truncatula and C. roseus (Mertens et al., 2016). To determine the temporal expression patterns of the BIS3 in response to MeJA, we treated C. roseus seedlings with 100 µM MeJA for 2 hr and measured gene expression using RT‐qPCR. As shown in Figure 3, the expression of all three BISs was significantly induced (~3‐ to 4‐fold) by MeJA. The sequential conversion of geranyl diphosphate to loganic acid is catalyzed by seven enzymes. The transcript levels of genes encoding these seven enzymes were also induced by 2‐ to 4‐fold after 2 hr of MeJA treatment (Figure 3). Collectively, these findings suggest that MeJA positively affects BIS3 expression along with other iridoid pathway genes and regulators.
FIGURE 3.
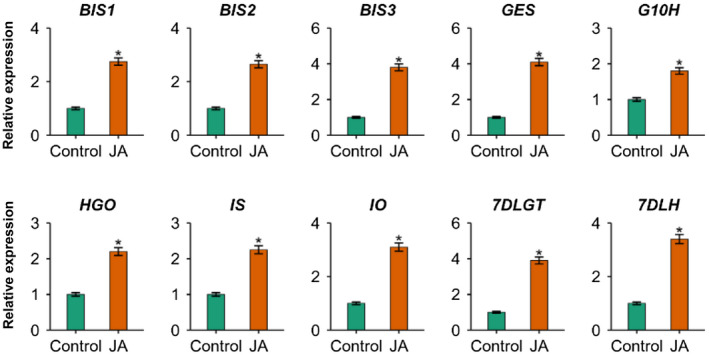
Expression of BISs and iridoid pathway genes in response to MeJA. Ten‐day‐old C. roseus seedlings were treated with 100 μM MeJA (JA) for 2 hr and gene expression in the whole seedling was measured by RT‐qPCR. Mock‐treated seedlings were used as controls. Data represent means ± SDs of three biological samples. Statistical significance was calculated using the Student's t test: *p < .05
3.6. BIS3 transactivates the iridoid pathway gene promoters in tobacco cells
To determine whether BIS3 can activate the iridoid biosynthetic genes, we performed promoter transactivation assays using BIS3 in tobacco protoplasts (Figure 4). The promoters of GES, G10H, HGO, IS, IO, 7DLGT or 7DLH fused to firefly luciferase (LUC) reporter gene was electroporated into tobacco protoplasts alone or in combination with BIS3. Luciferase activities were significantly higher (4.8‐ to 15‐fold) when BIS3 was coexpressed with GES, G10H, HGO, IS 7DLGT or 7DLH promoter‐reporter plasmid (Figure 4). We did not detect a significant change in the transactivation of the IO promoter by BIS3 (Figure 4). To further elucidate the function of BIS3 in the activation of iridoid pathway genes, we converted BIS3 to a dominant repressor (BIS3‐SRDX) by fusing it with the 12 amino acid (LDLDLELRLGFA) EAR (ERF‐associated amphiphilic repression) motif (also know as SRDX) (Hiratsu et al., 2003). As shown in Figure 4, BIS3‐SRDX was unable to transactivate the GES, G10H, 8HGO, IS, 7DLGT or 7DLH promoters in the protoplast assay. The basal activity of the promoters is presumably due to basal transcription factors present in tobacco cells. To determine whether BIS3 acts synergistically with BIS1 or BIS2 to regulate the iridoid pathway genes, we compared the transactivation activity of the G10H promoter by BIS1, BIS2 or BIS3 alone or in combination in tobacco cells. As expected, BIS1 and BIS2 significantly activated the G10H promoter in tobacco cells; however, the transactivation activity was increased significantly when BIS3 was co‐electroporated with BIS1 or BIS2 (Figure S2), suggesting that BIS cluster TFs act together to regulate the G10H promoter.
FIGURE 4.
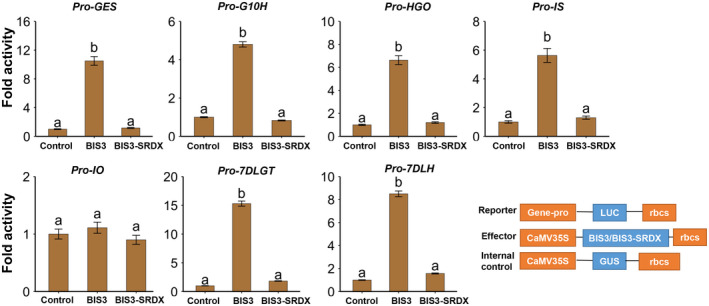
Transactivation of iridoid pathway promoters by BIS3 in tobacco cells. Schematic diagrams of plasmids used in transactivation assay. The GES, G10H, 8HGO, IS, IO, 7DLGT, and 7DLH promoters fused to luciferase (LUC) reporter were electroporated into tobacco protoplasts either alone or with an effector plasmid (BIS3). The CaMV35S‐GUS reporter was used as an internal control. Luciferase activity was normalized against GUS activity. Control represents the reporter alone without effectors. Data represent means ± SDs of three biological samples. Different letters denote statistical differences as assessed by one‐way ANOVA and Tukey HSD test, p < .05. Schematic diagrams of the plasmid constructs used in transactivation assay (bottom panel)
3.7. The G‐box motif and adjacent sequences in the G10H promoter are critical for transactivation by BIS3
The conversion geraniol to 10‐hydroxy geraniol, catalyzed by G10H, is the first committed step in iridoid biosynthesis in C. roseus (Thamm et al., 2016). We have previously isolated and characterized the G10H promoter in C. roseus hairy roots and transgenic tobacco plants (Suttipanta et al., 2007). The 533 bp G10H promoter (−497 to +40 relative to the transcription start site; TSS) used in our study contains only one canonical G‐box motif (CACGTG) at position −185 to −180 relative to the transcription start site (TSS). bHLH TFs are known to bind E‐box (CANNTG), G‐box or variants in the target promoters (Patra et al., 2018). Recent studies have shown that the subgroup‐IV bHLH TFs also bind a variant of G‐box motif, N‐box (CACGAG), in the promoters (Mertens, Pollier, et al., 2016; Yamamura et al., 2015). Although the G10H promoter does not contain an N‐box motif (Suttipanta et al., 2007), it is significantly activated by BIS1 (Figure S2) (Van Moerkercke et al., 2015), BIS2 (Van Moerkercke et al., 2016) and BIS3 (this study; Figure 4 and Figure S2). We, therefore, mutated the G‐box (CACGTG) to CAAAAA and evaluated the transactivation of the mutant promoter by BIS3 in tobacco cells. Mutation of G‐box motif reduced but did not abolish the transactivation of the G10H promoter by BIS3. To further determine the sequence motifs involved in BIS3 regulation, we generated two deletion fragments of the G10H promoter (−177 to +40 and −103 to +40 relative to TSS) that do not contain known bHLH binding motifs and measured their transactivation activities by BIS3 in tobacco cells. Similar to mutation, deletion of the G‐box motif in D1‐G10H (−177 to +40) did not abolished transactivation by BIS3 (Figure 5). However, further deletion D2‐G10H (−103 to +40) of the G10H promoter almost abolished the transactivation, suggesting that additional sequences (~70 bps adjacent to the G‐box) in the promoter potentially contribute to the transactivation of G10H by BIS3.
FIGURE 5.
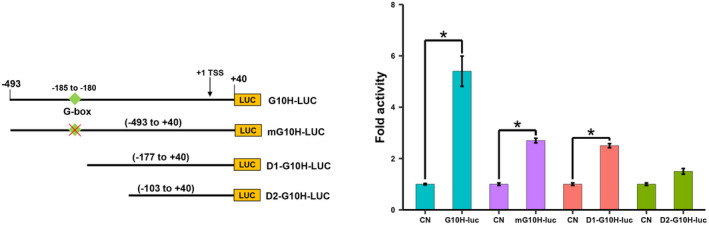
Transactivation of the G10H promoter mutant and deletion fragment by BIS3 in tobacco cells. Schematic diagrams of the G10H promoter fragments used in transactivation assay (left panel). The mutant or deleted fragments of G10H promoters fused to luciferase (LUC) reporter were electroporated into tobacco protoplasts either alone or with an effector plasmid (BIS3). CaMV35S‐GUS reporter served as an internal control. Luciferase activity was normalized against GUS activity. Control represents the reporter alone without effectors. Data represent means ± SDs of three biological samples. Statistical significance was calculated using the Student's t test: *p < .05; CN, control (reporter only)
3.8. BIS3 overexpression in C. roseus flower petal upregulates TIA pathway gene expression
To further substantiate the role of BIS3 in the regulation of the TIA pathway, we transiently overexpressed BIS3 in C. roseus flower petals as previously described (Schweizer et al., 2018; Singh et al., 2020) and measured the expression of TIA pathway genes by RT‐qPCR. As shown in Figure 6a, expression of all iridoid pathway genes were significantly upregulated (25‐ to 4,500‐fold) compared to the empty vector (EV) control. Induced expression of IS was the highest, while that of 7DLGT was the lowest. Although BIS3 does not activate the IO promoter in the tobacco protoplast assay (Figure 4), IO expression was increased by 600‐fold compared to EV in flower petals overexpressing BIS3 (Figure 6a). The conversion of loganic acid to secologanin is catalyzed by loganic acid O‐methyltransferase (LAMT) and secologanin synthase (SLS), which are regulated the ORCAs (Paul et al., 2020; Van Moerkercke et al., 2015). BIS3 overexpression moderately upregulated LAMT (5‐fold) and SLS (1.5‐fold) expression. In addition, expression of TDC (tryptophan decarboxylase) and STR (strictosidine synthase), which are direct targets of ORCAs, was increased in BIS3‐overexpressing petals compared to EV (Figure 6b). To determine whether BIS3 regulates or is regulated by ORCAs, we measured the expression of three BIS in ORCA5‐overexpressing flower petals. Expression of BIS1 and BIS2 was induced by 4‐ and 15‐fold, respectively, whereas BIS3 expression remained unchanged (Figure S3a). In addition, we measured the expression of all five ORCAs in BIS3‐overexpressing flower petals. Expression of ORCA2 showed modest induction (~ 6‐fold), whereas that of ORCA3 and ORCA5 reduced significantly; ORCA4 and ORCA6 expression was not significantly altered by BIS3 (Figure S3b).
FIGURE 6.
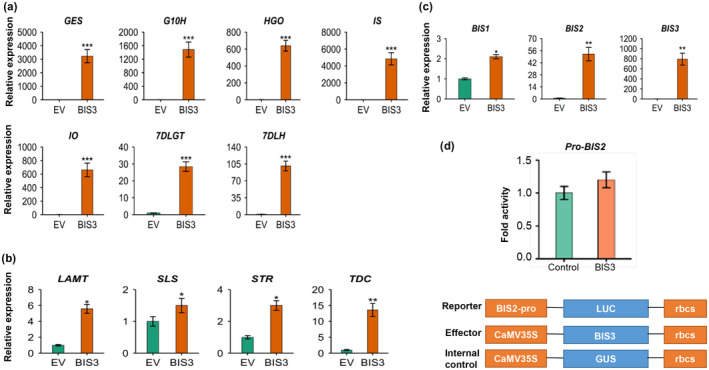
Expression of TIA pathway genes in BIS3‐overexpressing flower petals. Expression of iridoid (a), other TIA pathway (b) and BIS1, BIS2, and BIS3 (c) in BIS3‐overexpressing flower petals as measured by RT‐qPCR. (d) Transactivation of BIS2 promoter by BIS3 in tobacco cells. BIS2 promoter fused to luciferase (LUC) reporter were electroporated into tobacco protoplasts either alone or with BIS3. CaMV35S‐GUS reporter served as an internal control. Luciferase activity was normalized against GUS activity. Control represents the reporter alone without effectors.The error bars represent the means ± SD from three biological replicates. Statistical significance was calculated using the Student's t test: *p < .05; **p < .01, ***p < .001
Previous studies have shown that BIS2 expression was induced in BIS1‐overexpressing hairy roots and flower petals (Schweizer et al., 2018; Van Moerkercke et al., 2016). To determine whether BIS3 overexpression activates similar amplification loops, we measured the expression of BIS1 and BIS2 in BIS3‐overexpressing flower petals. BIS2 expression was increased by more than 50‐fold, whereas that of BIS1 was induced by 2‐fold in BIS3‐overexpressing flower petals (Figure 6c). To further determine whether BIS3 can directly activate BIS2 expression, we cloned the BIS2 promoter and fused it to the LUC reporter. Transient expression assay showed that BIS3 was unable to transactivate the BIS2 promoter in tobacco cells (Figure 6d). In addition, we measured the TIA pathway metabolites, such as loganic acid, secologanin, tabersonine, catharanthine and ajmalicine, in control and BIS3‐overexpressing flower petals. Accumulation of loganic acid increased significantly in BIS3‐overexpressing flowers, while the slight increase of secologanin, catharanthine, and tabersonine contents was not statistically significant compared to the control. Ajmalicine content was reduced in BIS3‐overexpressing flowers (Figure S4).
4. DISCUSSION
The advances in genome sequencing have significantly accelerated gene discovery and knowledge on genome organization, especially in non‐model plants. Unlike those of prokaryotes, genes in eukaryotic genomes are randomly distributed among the chromosomes. However, an increasing number of comparative genomic studies reveal that metabolic gene clusters are common in plants (Nutzmann et al., 2018). With a few exceptions (e.g., cytochrome P450 genes), a majority of the metabolic gene clusters comprise non‐homologous genes encoding enzymes for species‐specific metabolites. Genome sequence analysis has also identified TF gene clusters comprising of two or more homologous genes. Compared to the AP2/ERF clusters, clusters formed by other TFs, e.g., bHLH or MYB, are less characterized. Here, we discovered that the previously characterized BIS1 and BIS2 are joined by BIS3 to form a subgroup‐IVa bHLH cluster (Figure 1). Comparative genomic analysis of primitive and higher plants revealed that subgroup‐IVa bHLH clusters appear to be present in the core angiosperms but not in lower plants (Figure 2). Genome‐wide analysis and evolutionary studies on bHLH gene families of different plant lineages suggest that expansion and diversity of bHLH genes have likely occurred after the split between green algae and land plant species, followed by a second expansion after the split between moss and vascular plants (Carretero‐Paulet et al., 2010; Feller et al., 2011). Therefore, bHLH clusters possibly have originated later in the evolution of higher plants through repeated gene duplication.
Transcriptomic resources have accelerated the identification of genes encoding enzymes or regulators in TIA and other metabolic pathways in plants. Candidate gene identification can often be achieved by coexpression analysis, based on the hypothesis that functionally related genes such as genes in a metabolic pathway, either in a cluster or randomly distributed in the genome, exhibit similar spatiotemporal expression profiles (Mutwil, 2020). In addition, previous studies suggest that transcription factors (TFs) regulating metabolic pathways often coexpress with genes encoding enzymes in the pathway (De Geyter et al., 2012). Supporting this notion, our coexpression analysis revealed that the enzyme‐encoding and regulatory genes in the iridoid branch of the TIA pathway are highly and preferentially expressed in roots (Figure 1). Biosynthesis of plant specialized metabolites, often acting as defense molecules under adverse conditions, is induced in response to many biotic and abiotic factors. The phytohormone JA and its methyl esters MeJA are key elicitors of diverse specialized metabolites, including SGAs, TIAs, terpenes, and nicotine (Cardenas et al., 2016; Lenka et al., 2015; Mertens, Pollier, et al., 2016; Patra et al., 2018; Paul et al., 2017; Shoji et al., 2010; Thagun et al., 2016). Here, we showed that similar to BIS1 and BIS2, expression of BIS3 was induced by MeJA (Figure 3). The JA‐response and similar spatiotemporal expression patterns as the iridoid pathway genes suggest that the BIS cluster coordinates the regulation of the TIA pathway.
The involvement of BIS3 in the regulation of the TIA pathway was further substantiated by the promoter activation assay in plant cells and transient overexpression in C. roseus flower petals. BIS3 significantly activates the promoters of GES, G10H, 8HGO, IS, 7DLGT and 7DLH, key genes in the iridoid pathway. Transactivation activity of the G10H promoter by BIS1 (Figure S2) appears to be higher than that by BIS3 (Figure 4). This is possibly due to the sequence differences in the activation and DNA‐binding domains of the two TFs. Similar observations of differential activation of target gene promoters have been made for the members of the ORCA TF cluster in C. roseus (Paul et al., 2017) and NIC2 ERFs in tobacco (Shoji et al., 2010), suggesting that individual TFs within a cluster may play different regulatory roles (Shoji & Yuan, 2021; Yuan, 2020). Similar to BIS1(Van Moerkercke et al., 2015), BIS3 does not activate the IO promoter in tobacco cells; however, overexpression of BIS1 (Van Moerkercke et al., 2015), BIS2 (Van Moerkercke et al., 2016) or BIS3 (this study; Figure 6a) significantly upregulated IO expression in C. roseus hairy roots or flower petals. This is possibly due to 1) the presence of regulatory or enhancer elements further upstream in the IO promoter sequence and 2) the existence of a regulatory loop involving another factor that is activated by BIS TFs. BIS1 acts synergistically with BIS2 to activate some but not all iridoid pathway gene promoters in tobacco cells (Van Moerkercke et al., 2016). Here we demonstrated that BIS3 overexpression induced the expression of BIS1 and BIS2. Additionally, BIS3 acts synergistically with BIS1 or BIS2 to activate the G10H promoter in tobacco cells (Figure S2).
Mutation or deletion of the G‐box motif in the G10H promoter significantly reduced but not abolished its transactivation by BIS3 (Figure 5). Contraditive results have been reported for promoter activation by clade‐IV bHLH TFs in different plants, suggesting that the activation is most likely promoter‐context dependent (Mertens, Van Moerkercke, et al., 2016; Yamamura et al., 2015). For instance, mutation of the N‐box motif in the C. roseus IS promoter reduced but did not abolish its transactivation by BIS1 (Mertens, Van Moerkercke, et al., 2016). In rice, CPS2 (copalyl diphosphate synthase 2) and CYP99A2 (cytochrome P450 monooxygenase 99A2), involved in phytoalexin biosynthesis, are regulated by the clade‐IV bHLH TF DPF. Mutation of the N‐box motif in the CPS2 promoter, but not the CYPA99A2 promoter, completely eliminated the transactivation by DPF (Yamamura et al., 2015). In the M. truncatula saponin pathway, mutation of the N‐box motif in the CYP93E2 promoter completely abolished its activation by the bHLH factor TSAR1 (Mertens, Pollier, et al., 2016). These findings suggest that G‐ or N‐box motifs are necessary but not exclusive for the activation of the C. roseus G10H and IS promoters by BISs and of the rice CYP99A2 promoter by DPF. Other sequences in the promoters are likely involved in the regulation, as we demonstrated in the promoter deletion assay (Figure 5). The involvement of BIS3 in the regulation of the iridoid pathway was further demonstrated by overexpressing BIS3 in C. roseus flower petals (Figure 6). BIS3 overexpression significantly activated the expression of all seven iridoid pathway genes. The accumulation of loganic acid increased significantly in BIS3‐overexpressing flowers. Collectively, our observations authenticate the role of BIS3 in the regulation of the iridoid branch of the TIA pathway in C. roseus. Other TIA pathway metabolites, such as secologanin, tabersonine and catharanthine, did not show significant increase in BIS3‐overexpressing flowers (Figure S4). This is possibly due to the modest increase of expression of the indole pathway genes (TDC, LAMT, and STR) in BIS3‐overexpressing flowers compared to that of iridoid pathway genes (Figure 6). In addition, expression of ORCAs was either reduced or not significantly altered in BIS3‐overexpressing flowers (Figure S3). Similar to BIS3, BIS1 overexpression in C. roseus flowers did not result in increased accumulation of tabersonine and catharanthine in flowers (Schweizer et al., 2018).
In conclusion, our work establishes the three‐member BIS TF cluster in C. roseus. Positive amplification and negative regulatory loops are evident in many metabolic and phytohormone signaling pathways. A regulatory loop between BIS3 and BIS1 or BIS2 seems to exist although likely to involve additional regulatory factors (Figure 6c,d). Although BIS2 expression was significantly induced in BIS3‐overexpressing petals, BIS3 was unable to directly activate BIS2 promoter in tobacco cells. This is similar to the previous study where BIS1 is unable to activate the BIS2 promoter, although BIS2 expression was significantly induced by BIS1, possibly due to 1) the presence of regulatory or enhancer elements further upstream in the promoter sequence and 2) the existence of a regulatory loop involving another factor that is activated by BIS TFs. The relationship among BIS1, BIS2, and BIS3 with regard to mutual regulation remains to be further explored. To date, only a few bHLH TF clusters have been functionally characterized. The characterization of the BIS cluster extended our understanding of bHLH TF clusters. Furthermore, the regulation of saponin biosynthesis in M. truncatula (Mertens, Pollier, et al., 2016; Ribeiro et al., 2020) and C. quinoa (Jarvis et al., 2017) as well as TIA biosynthesis in C. roseus (Van Moerkercke et al., 2015, 2016) (this study) by subgroup‐IVa bHLH TF clusters, suggests that an evolutionarily conserved regulatory mechanism modulates biosynthesis of specialized metabolites in plants.
CONFLICT OF INTEREST
The authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
L.Y., S.P., and S.K.S. designed the research; S.K.S., B.P., P.P., Y.L., and S.P. performed experiments; S.K.S., B.P., and S.P. analyzed data; and L.Y., S.K.S., and S.P. wrote the paper.
Supporting information
Fig S1‐S4‐Table S1
Singh SK, Patra B, Paul P, Liu Y, Pattanaik S, Yuan L. BHLH IRIDOID SYNTHESIS 3 is a member of a bHLH gene cluster regulating terpenoid indole alkaloid biosynthesis in Catharanthus roseus . Plant Direct. 2021;5:e00305 10.1002/pld3.305
Funding information
This work is supported partially by the Harold R. Burton Endowed Professorship to L.Y. and by the National Science Foundation under Cooperative Agreement no. 1355438 to L.Y.
Contributor Information
Sitakanta Pattanaik, Email: spatt2@uky.edu.
Ling Yuan, Email: lyuan3@uky.edu.
REFERENCES
- Abe, H. , Yamaguchi‐Shinozaki, K. , Urao, T. , Iwasaki, T. , Hosokawa, D. , & Shinozaki, K. (1997). Role of arabidopsis MYC and MYB homologs in drought‐ and abscisic acid‐regulated gene expression. The Plant Cell, 9, 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas, P. D. , Sonawane, P. D. , Pollier, J. , Vanden Bossche, R. , Dewangan, V. , Weithorn, E. , Tal, L. , Meir, S. , Rogachev, I. , Malitsky, S. , Giri, A. P. , Goossens, A. , Burdman, S. , & Aharoni, A. (2016). GAME9 regulates the biosynthesis of steroidal alkaloids and upstream isoprenoids in the plant mevalonate pathway. Nature Communications, 7, 10654 10.1038/ncomms10654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carretero‐Paulet, L. , Galstyan, A. , Roig‐Villanova, I. , Martinez‐Garcia, J. F. , Bilbao‐Castro, J. R. , & Robertson, D. L. (2010). Genome‐wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiology, 153, 1398–1412. 10.1104/pp.110.153593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courdavault, V. , Papon, N. , Clastre, M. , Giglioli‐Guivarc'h, N. , St‐Pierre, B. , & Burlat, V. (2014). A look inside an alkaloid multisite plant: The Catharanthus logistics. Current Opinion in Plant Biology, 19, 43–50. [DOI] [PubMed] [Google Scholar]
- Cui, Y. , Chen, C. L. , Cui, M. , Zhou, W. J. , Wu, H. L. , & Ling, H. Q. (2018). Four IVa bHLH transcription factors are novel interactors of FIT and mediate JA inhibition of iron uptake in Arabidopsis. Molecular Plant, 11, 1166–1183. [DOI] [PubMed] [Google Scholar]
- De Geyter, N. , Gholami, A. , Goormachtig, S. , & Goossens, A. (2012). Transcriptional machineries in jasmonate‐elicited plant secondary metabolism. Trends in Plant Science, 17, 349–359. 10.1016/j.tplants.2012.03.001 [DOI] [PubMed] [Google Scholar]
- De Luca, V. , Salim, V. , Thamm, A. , Masada, S. A. , & Yu, F. (2014). Making iridoids/secoiridoids and monoterpenoid indole alkaloids: Progress on pathway elucidation. Current Opinion in Plant Biology, 19, 35–42. [DOI] [PubMed] [Google Scholar]
- Feller, A. , Machemer, K. , Braun, E. L. , & Grotewold, E. (2011). Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. The Plant Journal, 66, 94–116. 10.1111/j.1365-313X.2010.04459.x [DOI] [PubMed] [Google Scholar]
- Franke, J. , Kim, J. , Hamilton, J. P. , Zhao, D. , Pham, G. M. , Wiegert‐Rininger, K. , Crisovan, E. , Newton, L. , Vaillancourt, B. , Tatsis, E. , Buell, C. R. , & O'Connor, S. E. (2019). Gene discovery in gelsemium highlights conserved gene clusters in monoterpene indole alkaloid biosynthesis. ChemBioChem, 20, 83–87. [DOI] [PubMed] [Google Scholar]
- Heim, M. A. , Jakoby, M. , Werber, M. , Martin, C. , Weisshaar, B. , & Bailey, P. C. (2003). The basic helix‐loop‐helix transcription factor family in plants: A genome‐wide study of protein structure and functional diversity. Molecular Biology and Evolution, 20, 735–747. 10.1093/molbev/msg088 [DOI] [PubMed] [Google Scholar]
- Hiratsu, K. , Matsui, K. , Koyama, T. , & Ohme‐Takagi, M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. The Plant Journal, 34, 733–739. [DOI] [PubMed] [Google Scholar]
- Hoopes, G. M. , Hamilton, J. P. , Kim, J. , Zhao, D. , Wiegert‐Rininger, K. , Crisovan, E. , & Buell, C. R. (2018). Genome assembly and annotation of the medicinal plant calotropis gigantea, a producer of anticancer and antimalarial cardenolides, G3: Genes, Genomes, Genetics, 8, 385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis, D. E. , Ho, Y. S. , Lightfoot, D. J. , Schmockel, S. M. , Li, B. , Borm, T. J. , Ohyanagi, H. , Mineta, K. , Michell, C. T. , Saber, N. , Kharbatia, N. M. , Rupper, R. R. , Sharp, A. R. , Dally, N. , Boughton, B. A. , Woo, Y. H. , Gao, G. , Schijlen, E. G. , Guo, X. , … Tester, M. (2017). The genome of Chenopodium quinoa. Nature, 542, 307–312. [DOI] [PubMed] [Google Scholar]
- Kajikawa, M. , Sierro, N. , Kawaguchi, H. , Bakaher, N. , Ivanov, N. V. , Hashimoto, T. , & Shoji, T. (2017). Genomic insights into the evolution of the nicotine biosynthesis pathway in tobacco. Plant Physiology, 174, 999–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaoka, M. M. , Pillitteri, L. J. , Fujii, H. , Yoshida, Y. , Bogenschutz, N. L. , Takabayashi, J. , Zhu, J.‐K. , & Torii, K. U. (2008). SCREAM/ICE1 and SCREAM2 specify three cell‐state transitional steps leading to Arabidopsis stomatal differentiation. The Plant Cell, 20, 1775–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan, K. , & Manners, J. M. (2013). MYC2: The master in action. Molecular Plant, 6, 686–703. [DOI] [PubMed] [Google Scholar]
- Kellner, F. , Kim, J. , Clavijo, B. J. , Hamilton, J. P. , Childs, K. L. , Vaillancourt, B. , Cepela, J. , Habermann, M. , Steuernagel, B. , Clissold, L. , McLay, K. , Buell, C. R. , & O'Connor, S. E. (2015). Genome‐guided investigation of plant natural product biosynthesis. The Plant Journal, 82, 680–692. [DOI] [PubMed] [Google Scholar]
- Kolde, R. , & Kolde, M. R. (2015). Package ‘pheatmap’. R Package, 1.
- Lee, S. , Lee, S. , Yang, K.‐Y. , Kim, Y.‐M. , Park, S.‐Y. , Kim, S. Y. , & Soh, M.‐S. (2006). Overexpression of PRE1 and its homologous genes activates gibberellin‐dependent responses in Arabidopsis thaliana. Plant and Cell Physiology, 47, 591–600. [DOI] [PubMed] [Google Scholar]
- Leivar, P. , Monte, E. , Al‐Sady, B. , Carle, C. , Storer, A. , Alonso, J. M. , Ecker, J. R. , & Quail, P. H. (2008). The Arabidopsis phytochrome‐interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. The Plant Cell, 20, 337–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenka, S. K. , Nims, N. E. , Vongpaseuth, K. , Boshar, R. A. , Roberts, S. C. , & Walker, E. L. (2015). Jasmonate‐responsive expression of paclitaxel biosynthesis genes in Taxus cuspidata cultured cells is negatively regulated by the bHLH transcription factors TcJAMYC1, TcJAMYC2, and TcJAMYC4. Frontiers in Plant Science, 6, 115 10.3389/fpls.2015.00115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. Y. , Leopold, A. L. , Sander, G. W. , Shanks, J. V. , Zhao, L. , & Gibson, S. I. (2013). The ORCA2 transcription factor plays a key role in regulation of the terpenoid indole alkaloid pathway. BMC Plant Biology, 13, 155 10.1186/1471-2229-13-155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscombe, D. K. , Usera, A. R. , & O'Connor, S. E. (2010). Homolog of tocopherol C methyltransferases catalyzes N methylation in anticancer alkaloid biosynthesis. Proceedings of the National Academy of Sciences of the United States of America, 107, 18793–18798. 10.1073/pnas.1009003107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Patra, B. , Pattanaik, S. , Wang, Y. , & Yuan, L. (2019). GATA and phytochrome interacting factor transcription factors regulate light‐induced vindoline biosynthesis in Catharanthus roseus . Plant Physiology, 180, 1336–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke, F. L. , Champion, A. , Kijne, J. W. , & Memelink, J. (1999). A novel jasmonate‐ and elicitor‐responsive element in the periwinkle secondary metabolite biosynthetic gene Str interacts with a jasmonate‐ and elicitor‐inducible AP2‐domain transcription factor, ORCA2. The EMBO Journal, 18, 4455–4463. 10.1093/emboj/18.16.4455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens, J. , Pollier, J. , Vanden Bossche, R. , Lopez‐Vidriero, I. , Franco‐Zorrilla, J. M. , & Goossens, A. (2016). The bHLH transcription factors TSAR1 and TSAR2 regulate triterpene saponin biosynthesis in Medicago truncatula. Plant Physiology, 170, 194–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens, J. , Van Moerkercke, A. , Vanden Bossche, R. , Pollier, J. , & Goossens, A. (2016). Clade IVa basic helix‐loop‐helix transcription factors form part of a conserved jasmonate signaling circuit for the regulation of bioactive plant terpenoid biosynthesis. Plant and Cell Physiology, 57, 2564–2575. 10.1093/pcp/pcw168 [DOI] [PubMed] [Google Scholar]
- Morohashi, K. , Zhao, M. , Yang, M. , Read, B. , Lloyd, A. , Lamb, R. , & Grotewold, E. (2007). Participation of the Arabidopsis bHLH factor GL3 in trichome initiation regulatory events. Plant Physiology, 145, 736–746. 10.1104/pp.107.104521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutwil, M. (2020). Computational approaches to unravel the pathways and evolution of specialized metabolism. Current Opinion in Plant Biology, 55, 38–46. [DOI] [PubMed] [Google Scholar]
- Nakayasu, M. , Shioya, N. , Shikata, M. , Thagun, C. , Abdelkareem, A. , Okabe, Y. , Ariizumi, T. , Arimura, G. I. , Mizutani, M. , Ezura, H. , Hashimoto, T. , & Shoji, T. (2018). JRE4 is a master transcriptional regulator of defense‐related steroidal glycoalkaloids in tomato. The Plant Journal, 94, 975–990. 10.1111/tpj.13911 [DOI] [PubMed] [Google Scholar]
- Nutzmann, H. W. , Scazzocchio, C. , & Osbourn, A. (2018). Metabolic gene clusters in eukaryotes. Annual Review of Genetics, 52, 159–183. 10.1146/annurev-genet-120417-031237 [DOI] [PubMed] [Google Scholar]
- Patra, B. , Pattanaik, S. , Schluttenhofer, C. , & Yuan, L. (2018). A network of jasmonate‐responsive bHLH factors modulate monoterpenoid indole alkaloid biosynthesis in Catharanthus roseus . New Phytologist, 217, 1566–1581. [DOI] [PubMed] [Google Scholar]
- Patra, B. , Schluttenhofer, C. , Wu, Y. , Pattanaik, S. , & Yuan, L. (2013). Transcriptional regulation of secondary metabolite biosynthesis in plants. Biochimica Et Biophysica Acta, 1829, 1236–1247. [DOI] [PubMed] [Google Scholar]
- Pattanaik, S. , Kong, Q. , Zaitlin, D. , Werkman, J. R. , Xie, C. H. , Patra, B. , & Yuan, L. (2010). Isolation and functional characterization of a floral tissue‐specific R2R3 MYB regulator from tobacco. Planta, 231, 1061–1076. 10.1007/s00425-010-1108-y [DOI] [PubMed] [Google Scholar]
- Pattanaik, S. , Werkman, J. R. , Kong, Q. , & Yuan, L. (2010). Site‐directed mutagenesis and saturation mutagenesis for the functional study of transcription factors involved in plant secondary metabolite biosynthesis. Methods in Molecular Biology, 643, 47–57. [DOI] [PubMed] [Google Scholar]
- Paul, P. , Singh, S. K. , Patra, B. , Liu, X. , Pattanaik, S. , & Yuan, L. (2020). Mutually regulated AP2/ERF gene clusters modulate biosynthesis of specialized metabolites in plants. Plant Physiology, 182, 840–856. 10.1104/pp.19.00772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, P. , Singh, S. K. , Patra, B. , Sui, X. , Pattanaik, S. , & Yuan, L. (2017). A differentially regulated AP2/ERF transcription factor gene cluster acts downstream of a MAP kinase cascade to modulate terpenoid indole alkaloid biosynthesis in Catharanthus roseus . New Phytologist, 213, 1107–1123. [DOI] [PubMed] [Google Scholar]
- Pauw, B. , Hilliou, F. A. , Martin, V. S. , Chatel, G. , de Wolf, C. J. , Champion, A. , Pre, M. , van Duijn, B. , Kijne, J. W. , van der Fits, L. , & Memelink, J. (2004). Zinc finger proteins act as transcriptional repressors of alkaloid biosynthesis genes in Catharanthus roseus. Journal of Biological Chemistry, 279, 52940–52948. [DOI] [PubMed] [Google Scholar]
- Petkau, A. , Stuart‐Edwards, M. , Stothard, P. , & Van Domselaar, G. (2010). Interactive microbial genome visualization with GView. Bioinformatics, 26, 3125–3126. 10.1093/bioinformatics/btq588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajani, S. , & Sundaresan, V. (2001). The Arabidopsis myc/bHLH gene ALCATRAZ enables cell separation in fruit dehiscence. Current Biology, 11, 1914–1922. [DOI] [PubMed] [Google Scholar]
- Ribeiro, B. , Lacchini, E. , Bicalho, K. , Mertens, J. , Arendt, P. , Vanden Bossche, R. , Calegario, G. , Gryffroy, L. , Ceulemans, E. , Buitink, J. , Goossens, A. , & Pollier, J. (2020). A seed‐specific regulator of triterpene saponin biosynthesis in Medicago truncatula. The Plant Cell, 32, 2020–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez‐Perez, R. , Pavan, S. , Mazzeo, R. , Moldovan, C. , Aiese Cigliano, R. , Del Cueto, J. , Ricciardi, F. , Lotti, C. , Ricciardi, L. , Dicenta, F. , Lopez‐Marques, R. L. , & Moller, B. L. (2019). Mutation of a bHLH transcription factor allowed almond domestication. Science, 364, 1095–1098. 10.1126/science.aav8197 [DOI] [PubMed] [Google Scholar]
- Sasaki‐Sekimoto, Y. , Jikumaru, Y. , Obayashi, T. , Saito, H. , Masuda, S. , Kamiya, Y. , Ohta, H. , & Shirasu, K. (2013). Basic helix‐loop‐helix transcription factors JASMONATE‐ASSOCIATED MYC2‐LIKE1 (JAM1), JAM2, and JAM3 are negative regulators of jasmonate responses in Arabidopsis. Plant Physiology, 163, 291–304. 10.1104/pp.113.220129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer, F. , Colinas, M. , Pollier, J. , Van Moerkercke, A. , Vanden Bossche, R. , de Clercq, R. , & Goossens, A. (2018). An engineered combinatorial module of transcription factors boosts production of monoterpenoid indole alkaloids in Catharanthus roseus . Metabolic Engineering, 48, 150–162. [DOI] [PubMed] [Google Scholar]
- Schweizer, F. , Fernandez‐Calvo, P. , Zander, M. , Diez‐Diaz, M. , Fonseca, S. , Glauser, G. , Lewsey, M. G. , Ecker, J. R. , Solano, R. , & Reymond, P. (2013). Arabidopsis basic helix‐loop‐helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. The Plant Cell, 25, 3117–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang, Y. , Ma, Y. , Zhou, Y. , Zhang, H. , Duan, L. , Chen, H. , Zeng, J. , Zhou, Q. , Wang, S. , Gu, W. , Liu, M. , Ren, J. , Gu, X. , Zhang, S. , Wang, Y. , Yasukawa, K. , Bouwmeester, H. J. , Qi, X. , Zhang, Z. , … Huang, S. (2014). Biosynthesis, regulation, and domestication of bitterness in cucumber. Science, 346, 1084–1088. 10.1126/science.1259215 [DOI] [PubMed] [Google Scholar]
- Shen, Q. , Lu, X. , Yan, T. , Fu, X. , Lv, Z. , Zhang, F. , Pan, Q. , Wang, G. , Sun, X. , & Tang, K. (2016). The jasmonate‐responsive AaMYC2 transcription factor positively regulates artemisinin biosynthesis in Artemisia annua. New Phytologist, 210, 1269–1281. [DOI] [PubMed] [Google Scholar]
- Shoji, T. , & Hashimoto, T. (2011). Tobacco MYC2 regulates jasmonate‐inducible nicotine biosynthesis genes directly and by way of the NIC2‐locus ERF genes. Plant and Cell Physiology, 52, 1117–1130. 10.1093/pcp/pcr063 [DOI] [PubMed] [Google Scholar]
- Shoji, T. , Kajikawa, M. , & Hashimoto, T. (2010). Clustered transcription factor genes regulate nicotine biosynthesis in tobacco. The Plant Cell, 22, 3390–3409. 10.1105/tpc.110.078543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji, T. , & Yuan, L. (2021). ERF gene clusters: Working together to regulate metabolism. Trends in Plant Science, 26, 23–32. 10.1016/j.tplants.2020.07.015 [DOI] [PubMed] [Google Scholar]
- Singh, S. K. , Patra, B. , Paul, P. , Liu, Y. , Pattanaik, S. , & Yuan, L. (2020). Revisiting the ORCA gene cluster that regulates terpenoid indole alkaloid biosynthesis in Catharanthus roseus . Plant Science, 293, 110408. [DOI] [PubMed] [Google Scholar]
- Singh, S. K. , Wu, Y. , Ghosh, J. S. , Pattanaik, S. , Fisher, C. , Wang, Y. , Lawson, D. , & Yuan, L. (2015). RNA‐sequencing reveals global transcriptomic changes in Nicotiana tabacum responding to topping and treatment of axillary‐shoot control chemicals. Scientific Reports, 5, 18148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, S. , Qi, T. , Fan, M. , Zhang, X. , Gao, H. , Huang, H. , Wu, D. , Guo, H. , & Xie, D. (2013). The bHLH subgroup IIId factors negatively regulate jasmonate‐mediated plant defense and development. PLoS Genetics, 9, e1003653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen, A. M. , Krober, S. , Unte, U. S. , Huijser, P. , Dekker, K. , & Saedler, H. (2003). The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. The Plant Journal, 33, 413–423. [DOI] [PubMed] [Google Scholar]
- Subramanian, B. , Gao, S. , Lercher, M. J. , Hu, S. , & Chen, W.‐H. (2019). Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Research, 47, W270–W275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui, X. , Singh, S. K. , Patra, B. , Schluttenhofer, C. , Guo, W. , Pattanaik, S. , & Yuan, L. (2018). Cross‐family transcription factor interaction between MYC2 and GBFs modulates terpenoid indole alkaloid biosynthesis. Journal of Experimental Botany, 69, 4267–4281. [DOI] [PubMed] [Google Scholar]
- Suttipanta, N. , Pattanaik, S. , Gunjan, S. , Xie, C. H. , Littleton, J. , & Yuan, L. (2007). Promoter analysis of the Catharanthus roseus geraniol 10‐hydroxylase gene involved in terpenoid indole alkaloid biosynthesis. Biochimica Et Biophysica Acta, 1769, 139–148. [DOI] [PubMed] [Google Scholar]
- Suttipanta, N. , Pattanaik, S. , Kulshrestha, M. , Patra, B. , Singh, S. K. , & Yuan, L. (2011). The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiology, 157, 2081–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szécsi, J. , Joly, C. , Bordji, K. , Varaud, E. , Cock, J. M. , Dumas, C. , & Bendahmane, M. (2006). BIGPETALp, a bHLH transcription factor is involved in the control of Arabidopsis petal size. EMBO Journal, 25, 3912–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thagun, C. , Imanishi, S. , Kudo, T. , Nakabayashi, R. , Ohyama, K. , Mori, T. , Kawamoto, K. , Nakamura, Y. , Katayama, M. , Nonaka, S. , Matsukura, C. , Yano, K. , Ezura, H. , Saito, K. , Hashimoto, T. , & Shoji, T. (2016). Jasmonate‐responsive ERF transcription factors regulate steroidal glycoalkaloid biosynthesis in tomato. Plant and Cell Physiology, 57, 961–975. [DOI] [PubMed] [Google Scholar]
- Thamm, A. M. K. , Qu, Y. , & De Luca, V. (2016). Discovery and metabolic engineering of iridoid/secoiridoid and monoterpenoid indole alkaloid biosynthesis. Phytochemistry Reviews, 15, 339–361. [Google Scholar]
- Van Bel, M. , Diels, T. , Vancaester, E. , Kreft, L. , Botzki, A. , Van de Peer, Y. , Coppens, F. , & Vandepoele, K. (2018). PLAZA 4.0: An integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Research, 46, D1190–D1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Fits, L. , & Memelink, J. (2000). ORCA3, a jasmonate‐responsive transcriptional regulator of plant primary and secondary metabolism. Science, 289, 295–297. [DOI] [PubMed] [Google Scholar]
- Van Moerkercke, A. , Steensma, P. , Gariboldi, I. , Espoz, J. , Purnama, P. C. , Schweizer, F. , Miettinen, K. , Vanden Bossche, R. , De Clercq, R. , Memelink, J. , & Goossens, A. (2016). The basic helix‐loop‐helix transcription factor BIS2 is essential for monoterpenoid indole alkaloid production in the medicinal plant Catharanthus roseus . The Plant Journal, 88, 3–12. [DOI] [PubMed] [Google Scholar]
- Van Moerkercke, A. , Steensma, P. , Schweizer, F. , Pollier, J. , Gariboldi, I. , Payne, R. , Vanden Bossche, R. , Miettinen, K. , Espoz, J. , Purnama, P. C. , Kellner, F. , Seppanen‐Laakso, T. , O'Connor, S. E. , Rischer, H. , Memelink, J. , & Goossens, A. (2015). The bHLH transcription factor BIS1 controls the iridoid branch of the monoterpenoid indole alkaloid pathway in Catharanthus roseus . Proceedings of the National Academy of Sciences of the United States of America, 112, 8130–8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack, C. , & Strnad, M. (2019). Jasmonates are signals in the biosynthesis of secondary metabolites — Pathways, transcription factors and applied aspects — A brief review. New Biotechnology, 48, 1–11. [DOI] [PubMed] [Google Scholar]
- Wei, T. , & Simko, V. (2017) R package “corrplot”: Visualization of a Correlation Matrix (Version 0.84). https://github.com/taiyun/corrplot
- Xu, W. , Dubos, C. , & Lepiniec, L. (2015). Transcriptional control of flavonoid biosynthesis by MYB‐bHLH‐WDR complexes. Trends in Plant Science, 20, 176–185. [DOI] [PubMed] [Google Scholar]
- Yamamura, C. , Mizutani, E. , Okada, K. , Nakagawa, H. , Fukushima, S. , Tanaka, A. , Maeda, S. , Kamakura, T. , Yamane, H. , Takatsuji, H. , & Mori, M. (2015). Diterpenoid phytoalexin factor, a bHLH transcription factor, plays a central role in the biosynthesis of diterpenoid phytoalexins in rice. The Plant Journal, 84, 1100–1113. [DOI] [PubMed] [Google Scholar]
- Yang, F. , Wang, Q. , Schmitz, G. , Muller, D. , & Theres, K. (2012). The bHLH protein ROX acts in concert with RAX1 and LAS to modulate axillary meristem formation in Arabidopsis. The Plant Journal, 71, 61–70. [DOI] [PubMed] [Google Scholar]
- Yin, Y. , Vafeados, D. , Tao, Y. , Yoshida, S. , Asami, T. , & Chory, J. (2005). A new class of transcription factors mediates brassinosteroid‐regulated gene expression in Arabidopsis . Cell, 120, 249–259. 10.1016/j.cell.2004.11.044 [DOI] [PubMed] [Google Scholar]
- Yuan, L. (2020). Clustered ERF transcription factors: Not all created equal. Plant and Cell Physiology, 61, 1025–1027. 10.1093/pcp/pcaa067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. B. , Bokowiec, M. T. , Rushton, P. J. , Han, S. C. , & Timko, M. P. (2012). Tobacco transcription factors NtMYC2a and NtMYC2b form nuclear complexes with the NtJAZ1 repressor and regulate multiple jasmonate‐inducible steps in nicotine biosynthesis. Molecular Plant, 5, 73–84. [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Hedhili, S. , Montiel, G. , Zhang, Y. , Chatel, G. , Pre, M. , Gantet, P. , & Memelink, J. (2011). The basic helix‐loop‐helix transcription factor CrMYC2 controls the jasmonate‐responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus. The Plant Journal, 67, 61–71. 10.1111/j.1365-313X.2011.04575.x [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Koes, R. , Shang, H. , Fu, Z. , Wang, L. , Dong, X. , Zhang, J. , Passeri, V. , Li, Y. , Jiang, H. , Gao, J. , Li, Y. , Wang, H. , & Quattrocchio, F. M. (2019). Identification and functional analysis of three new anthocyanin R2R3‐MYB genes in Petunia. Plant Direct, 3, e00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1‐S4‐Table S1


