Abstract
Global warming poses severe threats to agricultural production, including soybean. One of the major mechanisms for organisms to combat heat stress is through heat shock proteins (HSPs) that stabilize protein structures at above‐optimum temperatures, by assisting in the folding of nascent, misfolded, or unfolded proteins. The HSP40 subgroups, or the J‐domain proteins, functions as co‐chaperones. They capture proteins that require folding or refolding and pass them on to HSP70 for processing. In this study, we have identified a type‐I HSP40 gene in soybean, GmDNJ1, with high basal expression under normal growth conditions and also highly inducible under abiotic stresses, especially heat. Gmdnj1‐knockout mutants had diminished growth in normal conditions, and when under heat stress, exhibited more severe browning, reduced chlorophyll contents, higher reactive oxygen species (ROS) contents, and higher induction of heat stress‐responsive transcription factors and ROS‐scavenging enzyme‐encoding genes. Under both normal and heat‐stress conditions, the mutant lines accumulated more aggregated proteins involved in protein catabolism, sugar metabolism, and membrane transportation, in both roots and leaves. In summary, GmDNJ1 plays crucial roles in the overall plant growth and heat tolerance in soybean, probably through the surveillance of misfolded proteins for refolding to maintain the full capacity of cellular functions.
Keywords: co‐chaperone, GmDNJ1, heat shock protein, heat stress, J domain, soybean
1. INTRODUCTION
Global warming has been gaining worldwide attention for decades. According to the National Aeronautics and Space Administration (NOAA National Centers for Environmental Information, 2020), in 2019, the earth's surface temperature was 0.98°C higher than the 20th‐century average and the second highest in the past century. Increasing air temperature could hamper plant growth and development from germination to harvest by damaging the photosystems, generating excessive reactive oxygen species (ROS), destabilizing molecular structures, and finally resulting in crop yield reduction (Wahid et al., 2012).
Heat shock proteins (HSPs) are members of a big protein family and are an important component of abiotic stress tolerance in all organisms. Plant HSPs are divided into five groups based on their molecular sizes, namely HSP100, HSP90, HSP70, HSP60, and small HSPs (Al‐Whaibi, 2011). Among them, HSP70s, which are ATP‐dependent molecular chaperones, are the most conserved in both functions and structures among different organisms (Usman et al., 2017). In order for HSP70s to carry out their chaperone function effectively, they require the involvement of HSP40s and the nucleotide exchange factor (Sharma & Masison, 2009).
In general, HSP40s can be divided into three types based on their functional domains, but all three groups contain a J domain which defines the HSP40. The J domain is responsible for the binding of an HSP40 to its specific HSP70 partner. In addition, both type‐I and II HSP40s contain a glycine/phenylalanine‐rich region followed by a C‐terminal protein‐binding domain, but only type I has four repeats of the CXXCXGXG‐type zinc finger in between the J domain and the C‐terminal domain, having the same structure as the Escherichia coli DnaJ protein. The other HSP40s without these conserved domains are grouped into type III. The grouping is not related to their functions, and HSP40s do not only work as co‐chaperones (Kampinga & Craig, 2010). For example, they can also suppress protein aggregation independent of HSP70s (Cyr & Ramos, 2015).
HSP40s, as co‐chaperones of HSP70s, are responsible for delivering client proteins to HSP70s. In general, to complete a chaperone activity cycle, an HSP40 directs client proteins to its partner HSP70 and activates the HSP70‐ATP hydrolysis to facilitate client protein unfolding. The nucleotide exchange factor then binds to the complex to release the unfolded client proteins and replaces the ADP with a new ATP (Kampinga & Craig, 2010).
Besides the folding of newly synthesized proteins or the unfolding of misfolded proteins, the HSP70/HSP40 complex is also involved in diverse cellular activities. For example, HSP70/HSP40 mediates protein degradation through the ubiquitin–proteasome system (Howarth et al., 2007), clears aggregated protein through autophagy (Higgins et al., 2018), mediates protein trafficking to different organelles (Cheng et al., 2008; Craig, 2018; Prasad et al., 2018; Shi et al., 2009), and participates in mitochondrial DNA and plasmid replication (Sozhamannan & Chattoraj, 1993; Tyc et al., 2015).
In this study, we identified an HSP40 homologue from soybean, GmDNJ1, which was highly expressed under normal growth conditions and was much more strongly induced under abiotic stress treatments, especially heat stress, compared to other J‐domain proteins in the soybean genome. Gmdnj1‐knockout mutants exhibited stunted growth and were more prone to heat stress damage. Through gene expression analyses, physiological and biochemical assays, as well as aggregated protein profiling, we were able to explore the possible modes of action of GmDNJ1. This is the first time a soybean J‐domain protein has been studied with respect to its involvement in both growth and stress tolerance.
2. MATERIALS AND METHODS
2.1. Soybean materials
Glycine max cultivars C01 (Lam et al., 2010) and Williams 82 used in this study were from our laboratory stock. The Gmdnj1 mutant was constructed in the Williams 82 background using the KeyPoint® Breeding (Keygene NV) (Rigola et al., 2009). The successful knockout event of GmDNJ1 (Glyma.12G095700) was identified from heterozygous M1 plants. Subsequent generations were self‐pollinated and selected for individuals that were homozygous for the mutation in GmDNJ1. Seeds of M4 lines (the M5 generation) from two different M3 homozygous individuals (Gmdnj1‐3.2 and Gmdnj1‐3.11) were used for experiments in this study. Mutation at the target gene was verified using Sanger sequencing.
2.2. Localization study of GmDNJ1 in tobacco Bright Yellow‐2 cells
The coding sequence of GmDNJ1 (Glyma.12G095700) was cloned into the V7 binary vector downstream of a 35S promoter (Brears et al., 1993). The coding sequence of the c‐Myc tag was cloned either upstream or downstream of GmDNJ1 to produce either V7‐c‐Myc‐GmDNJ1 or V7‐GmDNJ1‐c‐Myc. These constructs and the V7 empty vector were transformed into tobacco Bright Yellow‐2 (BY‐2) cells separately by Agrobacterium‐mediated transformation using A. tumefaciens strain LBA4404 (An, 1985). Positive transformants were used for subcellular localization studies by immunostaining according to a previous study (Li et al., 2015) with the following modifications. In brief, 4‐day‐old BY‐2 cells were fixed with 1% paraformaldehyde for 1 hr. After washing twice with phosphate‐buffered saline (PBS), the cells were digested with 0.1% cellulase, 0.01% pectolyase Y23 in PBS for 15 min, followed by three PBS washes. The cells were then blocked with 4% bovine serum albumin fraction V with 0.1% Triton X‐100 in PBS with gentle agitation for 1 hr. After two PBS washes, the cells were incubated with 1:1,000 anti‐Myc monoclonal antibodies (R950‐25, Thermo Fisher Scientific) at 4°C overnight. After that, the cells were incubated with 1:500 goat anti‐mouse IgG H&L (Alexa Fluor® 488 conjugated) (ab150113, Abcam) for 1 hr, with PBS washes before and after the incubation. After staining with 4′,6‐diamidino‐2‐phenylindole (DAPI) for 15 min, the cells were observed under an SP8 Lightning confocal microscope (Leica).
2.3. In‐vitro luciferase refolding assay
The full‐length coding sequence of GmDNJ1 was cloned between EcoRI and XhoI downstream of and in‐frame with the GST‐tag coding sequence. The recombinant protein was expressed in E. coli strain BL21 (DE3). The recombinant protein was purified using GST SpinTrap (27‐4570‐03, GE Healthcare, Chicago, IL, USA) according to the manufacturer's protocol. Purified proteins were quantified with Protein Assay Kit II (500‐0002, Bio‐Rad). Luciferase refolding assay was performed as described (Zmijewski et al., 2004). Luciferase (E1701, Promega) was denatured at 42°C for 10 min before the assay. Then the denatured luciferase was preincubated in luciferase refolding buffer (40 mM Tris‐HCl [pH 7.4], 50 mM KCl, 5 mM dithiothreitol [DTT], 10 mM MgCl2, and 10% glycerol) with different combinations of the chaperone proteins at 25°C for 10 min. The concentrations of Dnak, DnaJ, GrpE, (ADI‐SPP‐630‐J, ALX‐201‐144‐C025, ADI‐SPP‐650‐F, respectively, Stressgen, Victoria, Canada), purified GST‐GmDNJ1, and luciferase were 0.1 µM, 0.2 µM, 0.5 µM, 0.2 µM, and 80 nM, respectively. GST‐GmDNJ1 was replaced with either 0.2 µM GST or 0.5 mg/ml BSA in the negative‐control assays. The luciferase was renatured at 25°C for 20 min after the addition of 5 mM ATP. Luciferase activity was determined using the Bright‐Glo Luciferase Assay System (E2610, Promega) and a Lumi‐Imager (2012847, Roche).
2.4. Abiotic stress treatments of wild‐type soybean seedlings for gene expression analyses
Seeds of C01 were germinated in vermiculite. Seedlings were transferred to a hydroponic system with half‐strength modified Hoagland's solution at 1 week after germination. Seedlings were grown at 28°C until the first trifoliate had fully opened before the abiotic stress treatments began. For NaHCO3 and polyethylene glycol (PEG) treatments, the half‐strength Hoagland's solution was replaced with fresh half‐strength Hoagland's solution with 50 mM NaHCO3 (pH 8.5) or 5% PEG6000 and treated for 24 hr. Plants grown in fresh half‐strength Hoagland's solution only were used as controls. For paraquat treatment, 1 ml 10 mM paraquat (PQ) was sprayed on the leaves of the plants. Water‐sprayed plants were used as controls. For heat treatment, soybean seedlings in a hydroponic system were relocated to a growth chamber set to 42°C with a light–dark cycle of 16/8 hr. The seedlings were treated for 4 hr in PQ and heat treatments. After treatments, the trifoliate and roots were collected separately and snap‐frozen in liquid nitrogen for subsequent RNA extraction.
2.5. Heat treatment of soybean Gmdnj1 mutant lines
Seeds of Williams 82 or Gmdnj1 mutants were germinated in vermiculite at 28°C in a growth chamber in the dark for 3 days. The seedlings were then grown until 2 weeks old with fully expanded first trifoliate in the growth chamber at 28°C with a light–dark cycle of 16:8 hr. For heat treatment, plants were moved to another growth chamber 4 hr after light‐on with a thermal cycle of 45/28°C following the 16/8 hr light–dark cycle. Untreated plants were maintained at 28°C in the original growth chamber. Primary leaves and roots were harvested at 0 and 6 hr after the start of the heat treatment for gene expression analyses. Samples for reactive oxygen species (ROS) content analyses were harvested 6 hr after heat treatment. Samples for determining chlorophyll contents, fresh weights, and dry weights were harvested at 4 days after treatment.
2.6. Gene expression analysis
RNA was extracted using a modified phenol:chloroform:isoamylalcohol protocol (Ausubel et al., 1995). RNA was treated with DNase I (18068015, Invitrogen) according to the manufacturer's protocol. DNase I‐treated RNA was diluted 10‐fold before reverse transcription quantitative polymerase chain reaction (RT‐qPCR). RT‐qPCR was carried out using One Step TB Green® PrimeScript™ RT‐PCR Kit II (Perfect Real Time) (RR086A, Takara) on a CFX96 Touch Real‐Time PCR detection system (Bio‐Rad). Each 20 µl reaction contained 15 ng cDNA, 1X One Step TB Green RT‐PCR Buffer 4, 0.4 µl PrimeScript 1 step Enzyme Mix 2, 0.2 µM of each primer. Reverse transcription was carried out at 42°C for 5 min and 95°C for 10 s. Amplification was then performed under a thermal cycle of 95°C for 3 min followed by 40 cycles of 95°C for 10 s and 60°C for 30 s. The specificity of primers was determined with melt curve analysis following the RT‐qPCR. The relative expression of target genes was calculated with the 2−ΔΔCT method (Livak & Schmittgen, 2001) and was normalized to α‐tubulin. Primer information can be found in Table S1.
2.7. Measurement of chlorophyll content
Chlorophyll content was measured according to previous publications (Moran, 1982; Moran & Porath, 1980). In brief, a leaf disc of around 0.03 g was excised from the primary leaf, and the actual weight of the leaf disc was determined. Chlorophyll was extracted with 0.8 ml N,N‐dimethylformamide in a 1.5‐ml microcentrifuge tube at −20°C overnight. The absorbance of the extract at 664, 647, and 603 nm was measured with a Synergy H1 Hybrid multimode plate reader (BioTek Instruments Inc.). The amount of chlorophyll in the extracts was calculated with the following equations:
FW was the actual fresh weight of the leaf disc.
2.8. Reactive oxygen species (ROS) measurement
The ROS content in the treated plant was measured as previously described (Jambunathan, 2010; Valkonen & Kuusi, 1997), with the following modifications. About 10 mg of finely ground tissue sample was suspended with 1 ml of 10 mM Tris‐HCl (pH 7.2) in a 1.5‐ml microcentrifuge tube. The extract was then centrifuged at 12,000 × g for 20 min at 4°C. The protein concentration in the cleared supernatant was measured as A280. Ten microliters of cleared extract was mixed with 89 µl of 10 mM Tris‐HCl and 1 µl of 1 mM H2DCFDA. Reaction without H2DCFDA was used as background and reaction with 10 µl 50 mM Vitamin C was used as control. The H2DCFDA was excited at 480 nm and the emission at 526 nm was measured using a Synergy H1 Hybrid multimode plate reader. The relative ROS concentration was calculated as the emission of the sample minus the emission of the background and then normalized to the protein concentration.
2.9. Aggregated protein extraction and analysis
Primary leaves and roots were collected at 6 and 24 hr after heat treatment, respectively, from the Gmdnj1 mutants and Williams 82 for aggregated protein extraction (Planas‐Marquès et al., 2016). In brief, 200 mg of ground tissue was resuspended in 4 ml fractionation buffer (20 mM tris‐HCl [pH 8], 1 mM EDTA [pH 8], and 0.33 M sucrose) by vortex in a 15‐mL centrifuge tube. The homogenized solution was filtered through Miracloth (475855, Merck Millipore), followed by centrifugation at 2,000× g for 5 min. The supernatant was further centrifuged at 6,000 × g for 10 min, and the subsequent supernatant at 100,000 × g for 90 min at 4°C. The final pellet was resuspended in 3 ml fractionation buffer with 0.3% Triton X‐100 and incubated at 4°C for 1 hr with gentle shaking. The solubilized protein was centrifuged at 50,000 × g for 60 min at 4°C. The pellet containing the aggregated proteins was thoroughly resuspended in 8 M urea by sonication. The protein concentration was quantified with a 2‐D quant kit (GE80‐6483‐56, GE Healthcare, Chicago, IL, USA). Fifteen milligrams of protein were treated with 5 mM DTT at 37°C for 30 min followed by 15 mM iodoacetamide at room temperature for 30 min. The sample was diluted with three volumes of 50 mM sodium bicarbonate. Trypsin (V5111, Promega) equivalent to one tenth (w/w) of the protein amount was added for digestion at 37°C overnight.
Digested peptides were cleaned up with a Pierce™ C18 Spin Column (89873, Thermo Fisher Scientific) followed by a Pierce™ Detergent Removal Spin Column (87777, Thermo Fisher Scientific). Samples were analyzed on an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) coupled with the LC Ultimate 3000 RSLCnano system equipped with a C‐18µ‐precolumn (300‐µm i.d. × 5 mm) and an Acclaim Pepmap RSLC nanoViper C‐18 column (75 µm × 25 cm). Mobile phase A (1.9% acetonitrile and 0.1% formic acid) and mobile phase B (98% acetonitrile and 0.1% formic acid) were used in the liquid chromatography (LC). The LC mobile phase gradient profile was set as follows: 50°C chamber with 0.3 µl min−1 flow rate, 100% mobile phase A for 5 min, increase mobile phase B to 6% from 0% from the 5 min mark to the 8 min mark, increase mobile phase B to 18% in the following 40 min, then increase mobile phase B to 30% in the following 10 min, then increase mobile phase B to 80% in the following 7 min, and finally re‐equilibrate with 100% mobile phase A for 10 min. Raw data files generated by Xcalibur software (Thermo Fisher Scientific, Waltham, MA, USA) were analyzed using Proteome Discoverer v2.3 (Thermo Fisher Scientific). The MS/MS spectra were searched against a protein database of Williams 82 (Wm82.a2) with 74733 entries using the SEQUEST HT engine. Searches were configured with dynamic modifications for carbamidomethylation (+57.021 Da) of cysteine, oxidation of methionine (+15.995 Da), N‐terminal protein acetylation (+42.011 Da), precursor mass tolerance of 10 ppm, fragment mass tolerance of 0.02 Da, and trypsin cleavage (max two missed cleavages). False discovery and identifications were validated by Percolator, and only peptides with q‐value ≤ 0.05 were accepted. Samples were compared using the label‐free quantification (LFQ) method. Three biological replicates were performed independently from sample collection to protein identification using LC‐MS/MS. Only proteins appearing in at least two replicates with a twofold enrichment (adjusted p‐value ≤ .05) in both mutants compared to wild‐type Williams 82 were used for Gene Ontology (GO) enrichment analysis (Morales et al., 2013).
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez‐Riverol et al., 2019) partner repository with the dataset identifier PXD019799 and 10.6019/PXD019799. All other relevant data were described in the main text and the Supporting Information files.
3. RESULTS
3.1. GmDNJ1 is a typical type‐I HSP40 localized in both the cytoplasm and the nucleus
From our previous transcriptomic study of salt‐treated soybean samples (Liu et al., 2019), we identified one highly expressed and salt‐inducible gene encoding a J‐domain protein, Glyma.12G095700 (Figure S1), which we named GmDNJ1 in this study. GmDNJ1 encodes a typical type‐I HSP40 with an N‐terminal J domain, a glycine/phenylalanine‐rich region, four repeats of the CXXCXGXG‐type zinc finger, and a C‐terminal protein‐binding domain (Figure 1a). A phylogenetic tree was constructed using GmDNJ1 along with other well‐characterized HSP40s and it put GmDNJ1 into a clade with other reported type‐I HSP40s and the DnaJ protein of E. coli (Figure 1b), which were distinctively separated from the type‐II and type‐III HSP40s. The predicted 3D structure of GmDNJ1 consists of two β‐barrel structures (Figure S2), which is the signature of a broad substrate binder. Furthermore, two nuclear localization signal sequences (NLSs) and one nuclear exporting signal (NES) were found in GmDNJ1 (Figure 1a). Immunostaining detected c‐Myc‐tagged GmDNAJ1 in both the cytoplasm and the nucleus in transgenic tobacco BY‐2 cells overexpressing the fusion proteins regardless whether the tag was located in the N terminus or the C terminus of GmDNJ1 (Figure 1c). To further confirm the biochemical functions of GmDNJ1 as an HSP40, a luciferase recovery assay was done (Figure 1d). Heat‐denatured luciferase without the addition of heat shock proteins showed the lowest activity after renaturation. When the denatured luciferase was incubated with the complete E. coli chaperone system composing of DnaK, DnaJ and GrpE, a significant increase in luciferase activity was detected, and this was used as the positive control. When the E. coli DnaJ was replaced with GST‐tagged GmDNJ1, the luciferase activity was comparable to that with the complete E. coli chaperone system, suggesting that GmDNJ1 is a bona fide HSP40. As negative controls, the DnaJ was either omitted or replaced with GST or BSA. Without a functional HSP40, the luciferase activity dropped to only 40%–50% of the activity with a complete HSP system.
FIGURE 1.
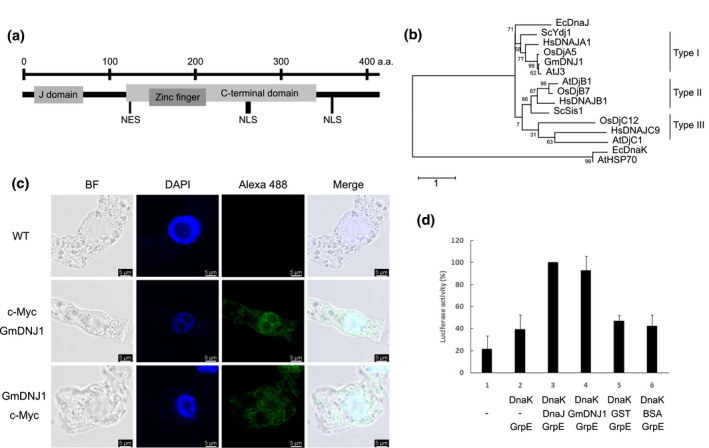
GmDNJ1 is a typical type‐I HSP40. (a) A cartoon depicting the protein structure of GmDNJ1. The various domains were determined by Pfam (El‐Gebali et al., 2019). The nuclear localization signal (NLS) and nuclear export signal (NES) were determined by LOCALIZER (Sperschneider et al., 2017) and NetNES 1.1 Server (la Cour et al., 2004), respectively. (b) Phylogenetic tree of characterized HSP40s. The tree was built with MEGA X (Kumar et al., 2018) using the Maximum Likelihood method and the JTT matrix‐based model with 1,000 bootstrap replications for phylogeny test. Bootstrap values were labeled as percentages on the branches. The accession numbers of the HSP40 protein sequences included in this tree are GmDNJ1: NP_001238341.1; AtJ3: NP_189997.1; AtDjB1: NP_195759.1; AtDjC1: NP_187752.2; HsDNAJA1: NP_001530.1; HsDNAJB1: NP_006136.1; HsDNAJC9: NP_056005.1; ScYdj1: NP_014335.1; ScSis1: NP_014391.1; EcDnaJ: NP_414556.1; OsDjA5: XP_015632121.1; OsDjB7: XP_015637533.1; OsDjC12: EEE55258.1; AtHSP70: NP_001328002.1; and EcDnaK: WP_102804582.1. (c) Subcellular localization study of GmDNJ1. A total of >50 cells were observed for each line with similar results. (d) Luciferase refolding assay for testing GmDNJ1 co‐chaperone activity. The luciferase activity in each reaction was normalized to that containing E. coli DnaJ–DnaK‐GrpE in combination (lane 3), which was set at 100%. Each bar represents the average of at least three technical replicates with the error bar representing standard error
3.2. Expression of GmDNJ1 was inducible by multiple abiotic stresses
Seedlings of soybean cultivar C01 were subjected to salt stress (NaCl), osmotic stress (PEG), alkaline stress (NaHCO3), oxidative stress (PQ), and heat stress (42°C) treatments (Figure 2 and Figure S3). In additional to GmDNJ1, we have also tested the expression of four other stress‐responsive genes. GmRD22 was known to be responsive to salt and osmotic stresses (Wang et al., 2012). GmCHX20a was known to be responsive to short‐term NaCl stress (Jia et al., 2020). Hsf2A is a heat‐inducible transcription factor (Charng et al., 2007). GsCHX19.3 is a salt and alkaline stresses inducible gene (Jia et al., 2017). The expression of the known stress‐responsive genes indicated that all the treatments were effective. Only GmDNJ1 and Hsf2A were significantly induced by all four kinds of abiotic stresses in at least one of the tissues. Under salt, PEG, NaHCO3, or PQ treatment, the fold of induction of GmDNJ1 was similar in both root and leaves while the induction of GmDNJ1 was more prominent in roots under heat stress. Furthermore, compared to the other abiotic stress treatments, the folds of induction of GmDNJ1 in both roots and leaves were exceptionally higher, by an order of magnitude, when under heat treatment (Figure 2 and Figure S3).
FIGURE 2.
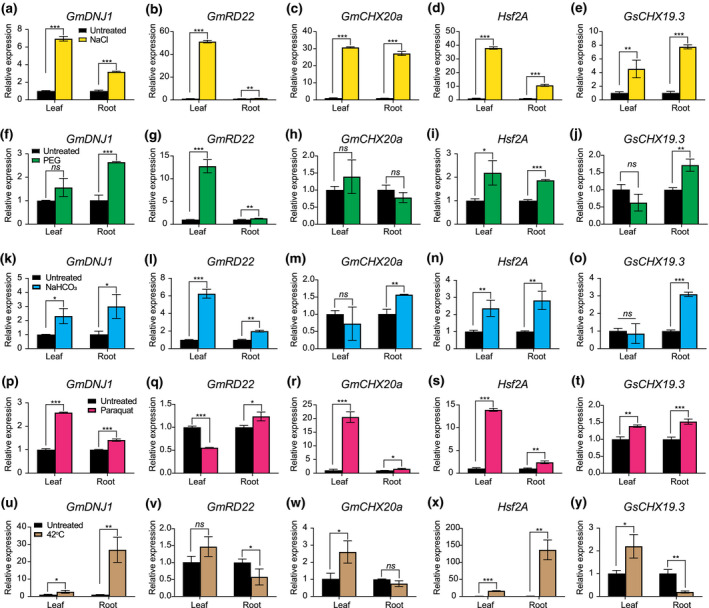
Expressions of GmDNJ1 were induced under abiotic stress treatments. First‐trifoliate seedlings of G. max cultivar C01 were treated with (a–e) 9% NaCl for 4 hr, (f–j) 5% PEG for 24 hr to induce osmotic stress, (k–o) 50 mM NaHCO3 at pH 8.5 for 24 hr, (p–t) 10 mM paraquat for 4 hr to induce oxidative stress, and (u‐y) heat stress at 42°C for 4 hr. Expressions of GmDNJ1 and other stress‐responsive genes in leaves and roots were analyzed by RT‐qPCR. The expressions of GmDNJ1 in the treated tissues were normalized to those in the respective untreated tissues. α‐tubulin was used as the housekeeping gene for normalizing RNA input. Relative gene expression was calculated by the 2‐ΔΔCT method. The error bar represents the standard deviation of three technical repeats. Two‐tailed student's t test was adopted to compare the expressions between untreated and treated samples. *, **, and ***indicates a significant difference at p < .05, p < .01, and p < .001, respectively. ns means that there was no statistically significant difference. A biological repeat of this experiment can be found in Figure S3
3.3. Characterization of GmDNJ1‐knockout mutant under heat treatment
An EMS mutant of GmDNJ1 in the G. max cultivar Williams 82 background was identified through the KeyPoint® Breeding (Rigola et al., 2009) and denoted as Gmdnj1. Gmdnj1 harbors a C‐to‐T mutation in the GmDNJ1 coding sequence which introduces a stop codon (Cag/Tag: Q72*) immediately following the J domain leading to premature termination. To minimize the effect of residual background mutations, seeds of two M5 generation pools (Gmdnj1‐3.2 and Gmdnj1‐3.11) from two different M3 individuals, homozygous for the mutation, were used for the subsequent experiments. As the expression of GmDNJ1 was the most responsive to heat treatment, the mutant was characterized using this treatment. Since the previous gene expression study was done using the cultivar C01, and although the protein sequences of GmDNJ1 from the two cultivars are the same, to ensure GmDNJ1 behaves similarly in Williams 82, the expression of GmDNJ1 under heat treatment was evaluated in wild‐type Williams 82 as a quality control (Figure S4). GmDNJ1 was also strongly induced in Williams 82 under heat stress and the fold of induction was also higher in the root than in the leaf.
Upon prolonged heat treatment, both the Gmdnj1‐3.2 and Gmdnj1‐3.11 mutant lines showed more severe browning compared to their wild‐type parent, Williams 82 (Figure 3a), indicating that the mutant is more sensitive to the heat stress. This was consistent with the lower chlorophyll contents in the mutants after heat treatment (Figure 3b), suggesting that the loss of a functional GmDNJ1 could sensitize the plant toward heat stress and may in turn reduce the chlorophyll content.
FIGURE 3.
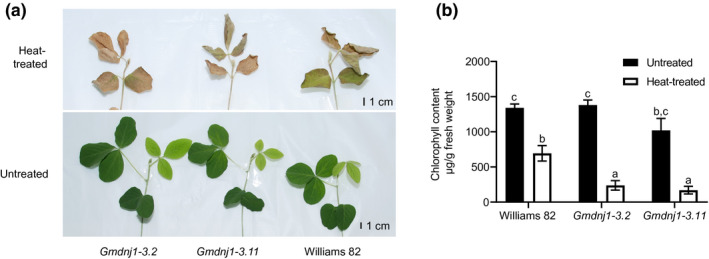
Growth performance and chlorophyll contents of Gmdnj1 mutant lines. (a) Photographs showing 2‐week‐old Gmdnj1 mutant plants treated at 45/28°C (heat‐treated) and 28/28°C (untreated) following the 16/8 hr light–dark cycle for 4 days. (b) Chlorophyll contents of the mutant lines with or without heat treatment. Data were assessed with one‐way ANOVA followed by Tukey's post hoc test. Different letters above the bars indicate means that were significantly different at p < .05. N ≥ 4. Errors bars: SEM. The experiment was performed twice with similar results. Replicate of the experiment can be found in Figure S5
Other physiological parameters were subsequently assessed. Under normal conditions, the differences in growth between wild‐type Williams 82 and the Gmdnj1 mutants were more pronounced in the roots than in the shoots, especially in terms of dry weights (Figure 4). Upon heat treatment, both the wild‐type Williams 82 and the Gmdnj1 mutant had significant reductions in the fresh weights of both roots and shoots, probably due to higher rates of evapotranspiration at the elevated temperature (Figure 4). However, the dry weights of both roots and shoots did not change significantly upon heat stress in both the mutant lines and the wild type, which may be the result of a relatively short treatment duration.
FIGURE 4.
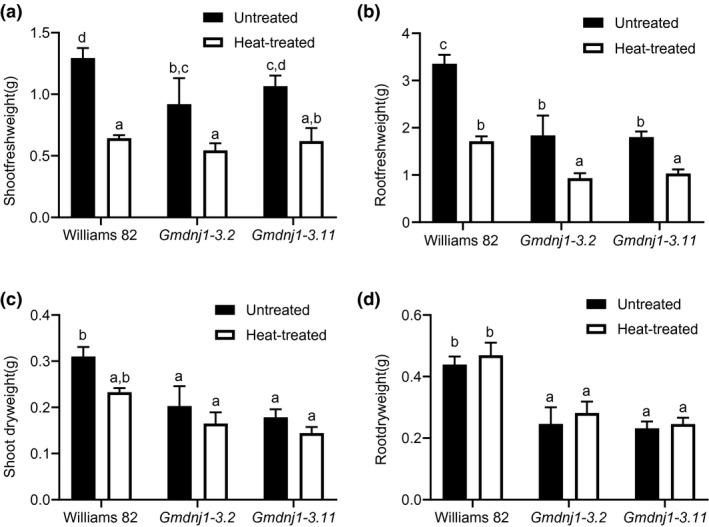
Fresh weights and dry weights of Gmdnj1 mutant lines under normal and heat treatment conditions. (a) Shoot fresh weights, (b) root fresh weights, (c) shoot dry weights, and (d) root dry weights of wild‐type Williams 82 and two Gmdnj1 mutant lines after 4 days of heat treatment at 45/28°C under the 16/8 hr light–dark cycle. Data were assessed with one‐way ANOVA followed by Tukey's post hoc test. Different letters above the bars indicate means that were significantly different at p < .05. N ≥ 4. Errors bars: SEM. Replicate of the experiment can be found in Figure S6
3.4. Knocking out GmDNJ1 led to elevation of total reactive oxygen species (ROS) content under heat treatment
Heat treatment could lead to ROS bursts which could be both beneficial and detrimental to the plant. In the root, only Gmdnj1‐3.2 showed significantly higher ROS content than wild‐type Williams 82 under normal conditions (Figure 5a). The difference between wild type and mutants were more prominent in the root than in the shoot upon heat treatment, with both Gmdnj1‐3.2 and Gmdnj1‐3.11 having higher ROS contents than the wild‐type Williams 82 (Figure 5b).
FIGURE 5.
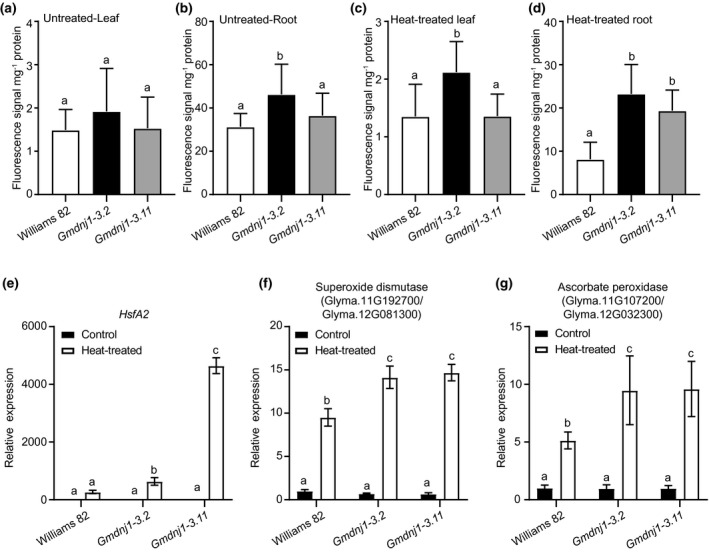
ROS contents and expressions of genes encoding ROS‐scavenging enzymes in Gmdnj1 mutant lines. The ROS contents in leaf (a) and root (b) of untreated plants, and in leaf (c) and root (d) of heat‐treated plants were compared by measuring the H2DCFCA fluorescence per unit protein in the extract. Expressions of HsfA2 (e), heat shock element containing superoxide dismutase‐encoding genes (f), and heat shock element containing ascorbate peroxidase‐encoding genes (g) in root of Gmdnj1 mutant were monitored. The data in a–d were analyzed with one‐way ANOVA followed by LSD test. Different letters above the bars indicate means that are significantly different at p < .05. N ≥ 4. Error bar: SEM. The data in (e–g) were analyzed with one‐way ANOVA followed by Tukey's test. Different letters above the bars indicate means that are significantly different at p < .05. N ≥ 3. Error bar: standard deviation. Replicate of the experiment can be found in Figure S7
Next, we examined the expression of an important heat‐inducible transcription factor, HsfA2 (Charng et al., 2007) and ROS‐scavenging enzyme‐encoding genes, which carry heat shock elements on their promoters. As expected, HsfA2 was highly induced by heat treatment only in the Gmdnj1 mutant lines (Figure 5c), suggesting that the mutant lines were more sensitive to the heat stress. In turn, the expressions of genes encoding superoxide dismutase and ascorbate peroxidase, which are potential targets of HsfA2, were also significantly more highly induced under heat treatment in the two mutant lines than in the wild type (Figure 5d,e).
3.5. Knocking out GmDNJ1 led to aggregation of proteins of diverse functions under heat stress
Knocking out a major HSP40 may lead to the impairment of the protein refolding system, and in turn lead to a change in the aggregated protein profile. Therefore, the aggregated protein profiles of the Gmdnj1 mutant lines were compared against that of the wild‐type Williams 82 under both untreated and heat‐treated conditions. Aggregated proteins with higher abundance (twofold difference, p‐value < .05, present in at least two replicates) in both mutant lines compared to Williams 82 in untreated and heat‐treated samples were identified for Gene Ontology (GO) analysis. In total, 35, 26, 158, and 54 aggregated proteins were found fulfilling these criteria in untreated root, heat‐treated root, untreated leaf, and heat‐treated leaf, respectively (Data Set S1). These proteins were annotated with 726 GO terms, which were distributed among diverse biological processes (Data Set S2). Nineteen of the GO terms were common to all four sets of samples (Table 1), and they refer to the proteins involved in protein catabolic process (GO: 0042176), sugar metabolism (GO:0006090, GO: 0019320), and membrane transportation (GO: 0015576, GO: 0015591, GO: 0015717, GO: 0035436, GO: 0042176, GO: 0070838). These processes are important as they are involved in protein turnover, energy production, and transportation across cell membrane. Furthermore, proteins annotated with GO: 0000302 (response to reactive oxygen species) were found in the aggregated protein profiles of both heat‐treated and untreated leaves of the Gmdnj1 mutants (Data Set S2). The fact that these proteins had a higher tendency to be misfolded or unfolded in the Gmdnj1 mutants implies that the biological processes they are involved in are more likely to be impaired in the absence of a functional GmDNJ1.
TABLE 1.
Gene ontology (GO) terms common to the aggregated proteins identified from the leaf and root with and without heat treatment
| GO ID | GO Description | Number of genes a | |||
|---|---|---|---|---|---|
| Untreated Root | Heat‐treated Root | Untreated Leaf | Heat‐treated Leaf | ||
| GO: 0000287 | Magnesium ion binding | 3 | 1 | 4 | 6 |
| GO: 0004033 | Aldo‐keto reductase (NADP) activity | 2 | 1 | 7 | 3 |
| GO: 0004849 | Uridine kinase activity | 2 | 1 | 7 | 3 |
| GO: 0006090 | Pyruvate metabolic process | 4 | 1 | 16 | 3 |
| GO: 0008135 | Translation factor activity, nucleic acid binding | 1 | 1 | 1 | 3 |
| GO: 0015576 | Sorbitol transmembrane transporter activity | 1 | 1 | 1 | 1 |
| GO: 0015591 | D‐ribose transmembrane transporter activity | 2 | 1 | 5 | 2 |
| GO: 0015717 | Triose phosphate transport | 2 | 1 | 9 | 8 |
| GO: 0016682 | Oxidoreductase activity, acting on diphenols and related substances as donors, oxygen as acceptor | 3 | 1 | 6 | 4 |
| GO: 0016874 | Ligase activity | 2 | 1 | 4 | 4 |
| GO: 0019320 | Hexose catabolic process | 4 | 1 | 22 | 10 |
| GO: 0019632 | Shikimate metabolic process | 2 | 2 | 6 | 1 |
| GO: 0030234 | Enzyme regulator activity | 2 | 1 | 9 | 5 |
| GO: 0030976 | Thiamine pyrophosphate binding | 2 | 2 | 11 | 5 |
| GO: 0035436 | Triose phosphate transmembrane transport | 2 | 3 | 7 | 4 |
| GO: 0042176 | Regulation of protein catabolic process | 3 | 2 | 16 | 3 |
| GO: 0046482 | Para‐aminobenzoic acid metabolic process | 2 | 1 | 1 | 1 |
| GO: 0047504 | (−)‐Menthol dehydrogenase activity | 1 | 1 | 2 | 1 |
| GO: 0070838 | Divalent metal ion transport | 2 | 1 | 4 | 2 |
Each gene may have more than one GO term.
4. DISCUSSION
There are over 200 J domain protein‐encoding genes in the soybean genome. Yet, only a few of them have been characterized (Liu & Whitham, 2013; So et al., 2013; Song et al., 2017; Zong et al., 2020). In this study, we have functionally characterized GmDNJ1, which has a relatively high expression in both normal and stress conditions compared with ~200 other J domain‐encoding genes in the soybean genome (Figure S1). GmDNJ1 is a typical type‐I HSP40 that is localized to both the cytoplasm and the nucleus.
A loss‐of‐function mutation with a stop codon introduced early in the coding sequence was constructed through KeyPoint® Breeding (Rigola et al., 2009). Chlorophyll content is a crucial indicator to show the survival of plant (Hu et al., 2020). Increased leaf browning and reduced chlorophyll content in Gmdnj1 plants (Figure 3) indicated that they are more sensitive to heat stress than the wild type. In addition, Gmdnj1 plants exhibited reduced fresh weights and dry weights compared to the wild‐type Williams 82 in both normal and heat stress conditions (Figure 4). Normally, the studies on plant HSP40s are focused on stress responses. However, there are also a few reports about the regulation of plant growth by HSP40s (Verma et al., 2019). For example, AtDjC17 was reported to be important in root hair development through cell fate determination in a tissue‐specific manner (Petti et al., 2014). AtBIL2, a mitochondrial type‐III HSP40, was hypothesized to maintain ATP generation to enhance growth in both shoots and roots by acting through the brassinosteroid signaling pathway (Bekh‐Ochir et al., 2013). To date, this is the first report of diminished growth due to the knocking out of a J‐domain protein in soybean in normal growth conditions.
A persistently high ROS content was detected in the Gmdnj1 mutant. It is still largely unknown how GmDNJ1 alleviates ROS accumulation. Some previous studies reported that HSP40s could play immediate roles in ROS scavenging. For example, the overexpression of LeCDJ1 in transgenic tomato was reported to enhance the activities of ascorbate peroxidase and superoxide dismutase independent of their transcription levels (Kong et al., 2014). Knocking out AtDjB1 led to the disruption of mitochondrial functions, which decreased ascorbate production and thus increased ROS contents (Zhou et al., 2012).
Even though HSP70/HSP40 are involved in multiple functions, HSP70s are generally limited in number compared to HSP40s in the genome. A single HSP70 protein is normally able to interact with multiple HSP40s, which are believed to determine the target client proteins and the functions of an HSP70. HSP40 proteins are structurally diverse in their substrate‐binding domains to fulfill the diverse functions. The substrate‐binding abilities of HSP40 proteins could be briefly divided into specific binder and broad binder. HSP40s such as GmDNJ1 with a double β‐barrel substrate‐binding domain is usually a broad binder (Craig & Marszalek, 2017), supporting the notion that GmDNJ1 could possibly maintain the full capacity of cellular functions by clearing unfolded protein from both the cytosol and the nucleus. Thus, knocking out GmDNJ1 is expected to lead to an overall impairment in growth.
In fact, when GmDNJ1, a major HSP40, was knocked out, we did observe that a number of proteins were consistently more readily aggregated in both Gmdnj1 mutant lines. In the aggregated protein profiles of the four tissue‐treatment combinations, 262 proteins were found in total, sharing 726 GO terms describing diverse but important functions. These proteins may or may not be the direct clients of GmDNJ1, but their proper folding was somehow impacted due to the absence of a fully functional GmDNJ1. For example, some of the differentially aggregated proteins found in Gmdnj1 mutants were involved in sugar metabolism and membrane transport. Carbon metabolism could be an important factor in heat tolerance in plants, while the transportation of sugar was also reported to be altered under heat stress accompanied by the differential expressions of some sugar transporter genes (Julius et al., 2017). In E. coli, DnaK was reported to interact with glucose metabolism‐related enzymes and the mutation of DnaK and DnaJ altered carbon metabolism, leading to a carbon flux to the TCA cycle and the production of ATP (Angles et al., 2017). A human HSP40 was also found to interact with glucose metabolism‐related enzymes to alter the glycolysis and cell proliferation processes (Huang et al., 2014). Furthermore, in heat‐treated roots, the survival rate increases with more available carbohydrates and higher rates of protein turnover (Huang et al., 2012).
In summary, in this report, we have characterized the mutant of a cytoplasm–nucleus localized type‐I J domain‐encoding gene, GmDNJ1, in soybean. Native GmDNJ1 is highly expressed under normal conditions and highly inducible in abiotic stress conditions. Our reverse genetic study suggested that GmDNJ1 is involved in both growth and heat tolerance. Loss of GmDNJ1 functions led to the persistent aggregation of some proteins involved in important biological processes, which may have led to the negative impacts on growth and heat tolerance.
CONFLICT OF INTEREST
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
AUTHOR CONTRIBUTIONS
HML designed the overall research strategy and coordinated this research. SM and AB identified the GmDNJ1 soybean mutants, and TH purified and checked the mutants. KPL, CHW, and CCC designed the experiments. CHW carried out expression studies. CCC carried out the biochemical study. KPL characterized the mutants. KPL and SSC carried out the proteomic analysis. KPL, MWL, and MJTVE participated in data analysis. KPL, MWL, and HML wrote the first draft of this manuscript.
Supporting information
Fig S1‐S7‐Table S1
Data Set S1
Data Set S2
ACKNOWLEDGMENTS
This work was supported by grants from the Hong Kong Research Grants Council General Research Fund (14122715) and Area of Excellence Scheme (AoE/M‐403/16). The authors thank Miss F.‐L. Wong for soybean materials maintenance, Mr. C.‐N. Leung and Mr. T.‐P. Chan for assisting with Orbitrap analyses, Dr. Z. Xiao for bioinformatic support, Miss Q. Wang in assisting with HSE motif identification, and Dr. H. Wang, Mr. W.‐L. Cheung, and Mr. W.‐S. Yung for critical comments and assistance in this project. Ms. J. Chu copy‐edited this manuscript. Any opinions, findings, conclusions, or recommendations expressed in this publication do not reflect the views of the Government of the Hong Kong Special Administrative Region or the Innovation and Technology Commission.
Li K‐P, Wong C‐H, Cheng C‐C, et al. GmDNJ1, a type‐I heat shock protein 40 (HSP40), is responsible for both Growth and heat tolerance in soybean. Plant Direct. 2021;5:e00298 10.1002/pld3.298
Funding information
This work was supported by grants from the Hong Kong Research Grants Council General Research Fund (14122715) and Area of Excellence Scheme (AoE/M‐403/16).
REFERENCES
- Al‐Whaibi, M. H. (2011). Plant heat‐shock proteins: A mini review. Journal of King Saud University ‐ Science, 23, 139–150. 10.1016/j.jksus.2010.06.022 [DOI] [Google Scholar]
- An, G. (1985). High efficiency transformation of cultured tobacco cells. Plant Physiology, 79, 568–570. 10.1104/pp.79.2.568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anglès, F. , Castanié‐Cornet, M.‐P. , Slama, N. , Dinclaux, M. , Cirinesi, A.‐M. , Portais, J.‐C. , Létisse, F. , & Genevaux, P. (2017). Multilevel interaction of the DnaK/DnaJ(HSP70/HSP40) stress‐responsive chaperone machine with the central metabolism. Scientific Reports, 7, 41341 10.1038/srep41341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F. , Brent, R. , Kingston, R. , Moore, D. , & Seidman, J. (1995). Current protocols in molecular biology. John Wiley and Sons. [Google Scholar]
- Bekh‐Ochir, D. , Shimada, S. , Yamagami, A. , Kanda, S. , Ogawa, K. , Nakazawa, M. , Matsui, M. , Sakuta, M. , Osada, H. , Asami, T. , & Nakano, T. (2013). A novel mitochondrial DnaJ/Hsp40 family protein BIL2 promotes plant growth and resistance against environmental stress in brassinosteroid signaling. Planta, 237, 1509–1525. 10.1007/s00425-013-1859-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brears, T. , Liu, C. , Knight, T. J. , & Coruzzi, G. M. (1993). Ectopic overexpression of asparagine synthetase in transgenic tobacco. Plant Physiology, 103, 1285–1290. 10.1104/pp.103.4.1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charng, Y.‐Y. , Liu, H.‐C. , Liu, N.‐Y. , Chi, W.‐T. , Wang, C.‐N. , Chang, S.‐H. , & Wang, T.‐T. (2007). A heat‐inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiology, 143, 251–262. 10.1104/pp.106.091322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, X. G. , Belshan, M. , & Ratner, L. (2008). Hsp40 facilitates nuclear import of the human immunodeficiency virus type 2 Vpx‐mediated preintegration complex. Journal of Virology, 82, 1229–1237. 10.1128/JVI.00540-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig, E. A. (2018). Hsp70 at the membrane: Driving protein translocation. BMC Biology, 16, 11 10.1186/s12915-017-0474-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig, E. A. , & Marszalek, J. (2017). How do J‐proteins get Hsp70 to do so many different things? Trends in Biochemical Sciences, 42, 355–368. 10.1016/j.tibs.2017.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyr, D. M. , & Ramos, C. H. (2015). Specification of Hsp70 Function by Type I and Type II Hsp40 In Blatch G. L., & Edkins A. L. (Eds.), The networking of chaperones by co‐chaperones: Control of cellular protein homeostasis (pp. 91–102). Springer International Publishing. [Google Scholar]
- El‐Gebali, S. , Mistry, J. , Bateman, A. , Eddy, S. R. , Luciani, A. , Potter, S. C. , Qureshi, M. , Richardson, L. J. , Salazar, G. A. , Smart, A. , Sonnhammer, E. L. L. , Hirsh, L. , Paladin, L. , Piovesan, D. , Tosatto, S. C. E. , & Finn, R. D. (2019). The Pfam protein families database in 2019. Nucleic Acids Research, 47, D427–D432. 10.1093/nar/gky995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins, R. , Kabbaj, M. H. , Hatcher, A. , & Wang, Y. C. (2018). The absence of specific yeast heat‐shock proteins leads to abnormal aggregation and compromised autophagic clearance of mutant Huntingtin proteins. PLoS One, 13, e0191490 10.1371/journal.pone.0191490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth, J. L. , Kelly, S. , Keasey, M. P. , Glover, C. , Lee, Y.‐B. , Mitrophanous, K. , Chapple, J. P. , Gallo, J. M. , Cheetham, M. E. , & Uney, J. B. (2007). Hsp40 molecules that target to the ubiquitin‐proteasome system decrease inclusion formation in models of polyglutamine disease. Molecular Therapy, 15, 1100–1105. 10.1038/sj.mt.6300163 [DOI] [PubMed] [Google Scholar]
- Hu, S. , Ding, Y. , & Zhu, C. (2020). Sensitivity and responses of chloroplasts to heat stress in plants. Frontiers in Plant Science, 11, 375 10.3389/fpls.2020.00375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, B. R. , Rachmilevitch, S. , & Xu, J. C. (2012). Root carbon and protein metabolism associated with heat tolerance. Journal of Experimental Botany, 63, 3455–3465. 10.1093/jxb/ers003 [DOI] [PubMed] [Google Scholar]
- Huang, L. , Yu, Z. , Zhang, T. , Zhao, X. , & Huang, G. (2014). HSP40 interacts with pyruvate kinase M2 and regulates glycolysis and cell proliferation in tumor cells. PLoS One, 9, e92949 10.1371/journal.pone.0092949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambunathan, N. (2010). Determination and detection of reactive oxygen species (ROS), lipid peroxidation, and electrolyte leakage in plants In Sunkar R. (Ed.), Plant Stress Tolerance. Methods in Molecular Biology (Methods and Protocols) (pp. 291–297). Humana press. [DOI] [PubMed] [Google Scholar]
- Jia, B. W. , Sun, M. Z. , Duanmu, H. Z. , Ding, X. D. , Liu, B. D. , Zhu, Y. M. , & Sun, X. (2017). GsCHX19.3, a member of cation/H+ exchanger superfamily from wild soybean contributes to high salinity and carbonate alkaline tolerance. Scientific Reports, 7(1), 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, Q. , Li, M. W. , Zheng, C. W. , Xu, Y. Y. , Sun, S. , Li, Z. , Wong, F. L. , Song, J. , Lin, W. W. , Li, Q. , & Zhu, Y. (2020). The soybean plasma membrane‐localized cation/H+ exchanger GmCHX20a plays a negative role under salt stress. Physiologia Plantarum. 10.1111/ppl.13250 In press. [DOI] [PubMed] [Google Scholar]
- Julius, B. T. , Leach, K. A. , Tran, T. M. , Mertz, R. A. , & Braun, D. M. (2017). Sugar transporters in plants: New insights and discoveries. Plant and Cell Physiology, 58, 1442–1460. 10.1093/pcp/pcx090 [DOI] [PubMed] [Google Scholar]
- Kampinga, H. H. , & Craig, E. A. (2010). The Hsp70 chaperone machinery: J‐proteins as drivers of functional specificity. Nature Reviews Molecular Cell Biology, 11, 579–592. 10.1038/nrm2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, F. , Deng, Y. , Wang, G. , Wang, J. , Liang, X. , & Meng, Q. (2014). LeCDJ1, a chloroplast DnaJ protein, facilitates heat tolerance in transgenic tomatoes. Journal of Integrative Plant Biology, 56, 63–74. 10.1111/jipb.12119 [DOI] [PubMed] [Google Scholar]
- Kumar, S. , Stecher, G. , Li, M. , Knyaz, C. , & Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35, 1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Cour, T. , Kiemer, L. , Molgaard, A. , Gupta, R. , Skriver, K. , & Brunak, S. (2004). Analysis and prediction of leucine‐rich nuclear export signals. Protein Engineering, Design & Selection, 17, 527–536. 10.1093/protein/gzh062 [DOI] [PubMed] [Google Scholar]
- Lam, H.‐M. , Xu, X. , Liu, X. , Chen, W. , Yang, G. , Wong, F.‐L. , Li, M.‐W. , He, W. , Qin, N. , Wang, B. O. , Li, J. , Jian, M. , Wang, J. , Shao, G. , Wang, J. , Sun, S.‐M. , & Zhang, G. (2010). Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nature Genetics, 42, 1053‐U1041 10.1038/ng.715 [DOI] [PubMed] [Google Scholar]
- Li, M. W. , Zhou, L. , & Lam, H. M. (2015). Paraformaldehyde fixation may lead to misinterpretation of the subcellular localization of plant high mobility group box proteins. PLoS One, 10, e0135033 10.1371/journal.pone.0135033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, A. , Xiao, Z. , Li, M.‐W. , Wong, F.‐L. , Yung, W.‐S. , Ku, Y.‐S. , Wang, Q. , Wang, X. , Xie, M. , Yim, A.‐Y. , Chan, T.‐F. , & Lam, H.‐M. (2019). Transcriptomic reprogramming in soybean seedlings under salt stress. Plant, Cell and Environment, 42, 98–114. 10.1111/pce.13186 [DOI] [PubMed] [Google Scholar]
- Liu, J. Z. , & Whitham, S. A. (2013). Overexpression of a soybean nuclear localized type‐III DnaJ domain‐containing HSP40 reveals its roles in cell death and disease resistance. The Plant Journal, 74, 110–121. 10.1111/tpj.12108 [DOI] [PubMed] [Google Scholar]
- Livak, K. J. , & Schmittgen, T. D. (2001). Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) Method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Morales, A. M. A. P. , O'Rourke, J. A. , van de Mortel, M. , Scheider, K. T. , Bancroft, T. J. , Borém, A. , Nelson, R. T. , Nettleton, D. , Baum, T. J. , Shoemaker, R. C. , Frederick, R. D. , Abdelnoor, R. V. , Pedley, K. F. , Whitham, S. A. , & Graham, M. A. (2013). Transcriptome analyses and virus induced gene silencing identify genes in the Rpp4‐mediated Asian soybean rust resistance pathway. Functional Plant Biology, 40, 1029–1047. 10.1071/FP12296 [DOI] [PubMed] [Google Scholar]
- Moran, R. (1982). Formulas for determination of chlorophyllous pigments extracted with N,N‐dimethylformamide. Plant Physiology, 69, 1376–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran, R. , & Porath, D. (1980). Chlorophyll determination in intact tissues using N,N‐dimethylformamide. Plant Physiology, 65, 478–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NOAA National Centers for Environmental Information . (2020). "State of the Climate: Global Climate Report for Annual 2019".
- Perez‐Riverol, Y. , Csordas, A. , Bai, J. , Bernal‐Llinares, M. , Hewapathirana, S. , Kundu, D. J. , Inuganti, A. , Griss, J. , Mayer, G. , Eisenacher, M. , Pérez, E. , Uszkoreit, J. , Pfeuffer, J. , Sachsenberg, T. , Yılmaz, Ş. , Tiwary, S. , Cox, J. , Audain, E. , Walzer, M. , … Vizcaíno, J. A. (2019). The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Research, 47, D442–D450. 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petti, C. , Nair, M. , & Debolt, S. (2014). The involvement of J‐protein AtDjC17 in root development in Arabidopsis. Frontiers in Plant Science, 5, 532 10.3389/fpls.2014.00532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas‐Marquès, M. , Lema, A. S. , & Coll, N. S. (2016). Detection and quantification of protein aggregates in plants In Lois L. M., & Matthiesen R. (Eds.), Plant proteostasis: methods and protocols (pp. 195–203). Springer, New York. [DOI] [PubMed] [Google Scholar]
- Prasad, R. , Xu, C. C. , & Ng, D. T. W. (2018). Hsp40/70/110 chaperones adapt nuclear protein quality control to serve cytosolic clients. Journal of Cell Biology, 217, 2019–2032. 10.1083/jcb.201706091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigola, D. , Van Oeveren, J. , Janssen, A. , Bonne, A. , Schneiders, H. , Van Der Poel, H. J. A. , van Orsouw, N. J. , Hogers, R. C. , de Both, M. T. , van Eijk, M. J. (2009). High‐throughput detection of induced mutations and natural variation using KeyPoint TM technology. PLoS One, 4, e4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, D. , & Masison, D. C. (2009). Hsp70 structure, function, regulation and influence on yeast prions. Protein and Peptide Letters, 16, 571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, A. , Sun, L. , Banerjee, R. , Tobin, M. , Zhang, Y. , & Grant, B. D. (2009). Regulation of endosomal clathrin and retromer‐mediated endosome to Golgi retrograde transport by the J‐domain protein RME‐8. The EMBO Journal, 28, 3290–3302. 10.1038/emboj.2009.272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So, H. A. , Chung, E. , & Lee, J. H. (2013). Molecular characterization of soybean GmDjp1 encoding a type III J‐protein induced by abiotic stress. Genes & Genomics, 35, 247–256. 10.1007/s13258-013-0078-4 [DOI] [Google Scholar]
- Song, X. , Duanmu, H. , Yu, Y. , Chen, C. , Sun, X. , Zhu, P. , Chen, R. , Duan, X. , Li, H. , Cao, L. , Nisa, Z. U. , Li, Q. , Zhu, Y. , & Ding, X. (2017). GsJ11, identified by genome‐wide analysis, facilitates alkaline tolerance in transgenic plants. Plant Cell, Tissue and Organ Culture (PCTOC), 129, 411–430. 10.1007/s11240-017-1188-5 [DOI] [Google Scholar]
- Sozhamannan, S. , & Chattoraj, D. K. (1993). Heat shock proteins DnaJ, DnaK, and GrpE stimulate P1 plasmid replication by promoting initiator binding to the origin. Journal of Bacteriology, 175, 3546–3555. 10.1128/JB.175.11.3546-3555.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperschneider, J. , Catanzariti, A.‐M. , DeBoer, K. , Petre, B. , Gardiner, D. M. , Singh, K. B. , Dodds, P. N. , & Taylor, J. M. (2017). LOCALIZER: Subcellular localization prediction of both plant and effector proteins in the plant cell. Scientific Reports, 7, 44598 10.1038/srep44598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyc, J. , Klingbeil, M. M. , & Lukes, J. (2015). Mitochondrial heat shock protein machinery hsp70/hsp40 is indispensable for proper mitochondrial DNA maintenance and replication. Mbio, 6, e02425–14. 10.1128/mBio.02425-02414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usman, M. G. , Rafii, M. Y. , Martini, M. Y. , Yusuff, O. A. , Ismail, M. R. , & Miah, G. (2017). Molecular analysis of Hsp70 mechanisms in plants and their function in response to stress. Biotechnology and Genetic Engineering Reviews, 33, 26–39. 10.1080/02648725.2017.1340546 [DOI] [PubMed] [Google Scholar]
- Valkonen, M. , & Kuusi, T. (1997). Spectrophotometric assay for total peroxyl radical‐trapping antioxidant potential in human serum. Journal of Lipid Research, 38, 823–833. [PubMed] [Google Scholar]
- Verma, A. K. , Tamadaddi, C. , Tak, Y. , Lal, S. S. , Cole, S. J. , Hines, J. K. , & Sahi, C. (2019). The expanding world of plant J‐domain proteins. Critical Reviews in Plant Sciences, 38, 382–400. 10.1080/07352689.2019.1693716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahid, A. , Farooq, M. , Hussain, I. , Rasheed, R. , & Galani, S. (2012). Responses and management of heat stress in plants. In Ahmad P., & Prasad M. N. V. (Eds.), Environmental adaptations and stress tolerance of plants in the era of climate change (pp. 135–157). Springer, New York. [Google Scholar]
- Wang, H. , Zhou, L. , Fu, Y. , Cheung, M.‐Y. , Wong, F.‐L. , Phang, T.‐H. , Sun, Z. , & Lam, H.‐M. (2012). Expression of an apoplast‐localized BURP‐domain protein from soybean (GmRD22) enhances tolerance towards abiotic stress. Plant, Cell and Environment, 35, 1932–1947. 10.1111/j.1365-3040.2012.02526.x [DOI] [PubMed] [Google Scholar]
- Zhou, W. , Zhou, T. , Li, M.‐X. , Zhao, C.‐L. , Jia, N. , Wang, X.‐X. , Sun, Y.‐Z. , Li, G.‐L. , Xu, M. , Zhou, R.‐G. , & Li, B. (2012). The Arabidopsis J‐protein AtDjB1 facilitates thermotolerance by protecting cells against heat‐induced oxidative damage. New Phytologist, 194, 364–378. 10.1111/j.1469-8137.2012.04070.x [DOI] [PubMed] [Google Scholar]
- Zmijewski, M. A. , Macario, A. J. L. , & Lipinska, B. (2004). Functional similarities and differences of an archaeal Hsp70(DnaK) stress protein compared with its homologue from the bacterium Escherichia coli . Journal of Molecular Biology, 336, 539–549. 10.1016/j.jmb.2003.12.053 [DOI] [PubMed] [Google Scholar]
- Zong, T. , Yin, J. , Jin, T. , Wang, L. , Luo, M. , Li, K. , & Zhi, H. (2020). A DnaJ protein that interacts with soybean mosaic virus coat protein serves as a key susceptibility factor for viral infection. Virus Research, 281, 197870 10.1016/j.virusres.2020.197870 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1‐S7‐Table S1
Data Set S1
Data Set S2


