Abstract
SARS-CoV-2 antibody development and immunity will be crucial for the further course of the pandemic. Until now, it has been assumed that patients who are infected with SARS-CoV-2 will develop antibodies as has been the case with other coronaviruses, like MERS-CoV and SARS-CoV. In the present study, we analyzed the development of antibodies in 77 patients with an oncologic diagnosis 26 days after positive RT-qPCR testing for SARS-CoV2. RT-qPCR and anti-SARS-CoV2-antibody methods from BGI (MGIEasy Magnetic Beads Virus DNA/RNA Extraction Kit) and Roche (Elecsys Anti-SARS-CoV-2 immunoassay) were used, respectively, according to the manufacturers’ specifications. Surprisingly, antibody development was detected in only 6 of 77 individuals with a confirmed history of COVID-19. Despite multiple testing, the remaining patients did not show measurable antibody concentrations in subsequent tests. These results undermine the previous hypothesis that SARS-CoV2 infections are regularly associated with antibody development and cast doubt on the provided immunity to COVID-19. Understanding the adaptive and humoral response to SARS-CoV2 will play a key role in vaccine development and gaining further knowledge on the pathogenesis.
Keywords: SARS-CoV2 antibody development, COVID-19, Cancer, Immunity
Introduction
SARS-CoV2 and its underlying disease, COVID-19, has spread around the world, so far causing over 61,299,371 confirmed infections and 1,439,784 deaths, according to the WHO Coronavirus Disease (COVID-19) Dashboard as of November 28, 2020. Coronaviruses are a subgroup in a spectrum of viruses that are phenotypically and genotypically diverse and have provoked recent epidemics [1,2]. Coronaviruses are enveloped viruses containing single-stranded positive-sense RNA with a viral genome of about 27-32 kb, which encodes for structural and nonstructural proteins [3], [4], [5]. The novel SARS-CoV2 consists of 4 structural proteins, namely: the spike protein (S), the envelope protein (E), the membrane glycoprotein (M), and the nucleocapsid protein (N) [3,6]. The majority of antibodies that are produced are formed against the nucleocapsid, which are therefore considered to be highly sensitive for antibody testing, even though it has to be noted that there is a sequence of homologies which could lower the sensitivity [3,7]. So far, millions of cases have been registered with positive RT-qPCR result whereas antibody testing has just recently become a factor.
Patients suffering from chronic diseases are generally thought to be at higher risk of developing a severe course of COVID-19, which could lead to intensive care treatment [8]. In contrast, Hempel et al has shown in a recent study that cancer patients treated in oncological outpatient settings, who tested positively for SARS-CoV2 in RT-qPCR, remained mostly asymptomatic virus carriers without an impact on the applied systemic cancer therapy (submitted manuscript). Nevertheless, measures are made to counter and minimize the risk of SARS-CoV2 infection and severe complications. Due to this reason, adjuvant chemotherapies, surgeries, and other compromising therapies were eventually postponed or changed [9].
As the symptoms and course of COVID-19 vary broadly, tests by nasopharyngeal or throat swabs were recently also taken from asymptomatic patients to identify virus carriers. It is estimated that over 50% of the cases are asymptomatic [10], and there is also a risk of false negative results because of poor swab techniques or a sparse amount of virus-RNA. However, an antibody test with high sensitivity and specificity could provide epidemiological information on the actual rate of infection. So far, it is unclear whether the majority of SARS-CoV2 infected patients produce a sufficient quantity of antibodies that sustains immunity. Until now, it has been assumed that antibodies are formed after the viral infection, as it is the case with other coronaviruses, namely MERS-CoV and SARS-CoV [11], [12], [13], [14]. Numerous studies also describe antibody production after infection with SARS-CoV2 [15,16]. Long et al. were able to detect positive rates of IgG and IgM at a median of 13 days after the onset of symptoms. IgG was detected in all patients between 17 and 19 days after the onset of symptoms whereas IgM showed a positive rate in 94.1% after 20–22 days of the beginning of symptoms [16]. The authors recommended that serological testing could be helpful for the diagnosis of asymptomatic virus carriers as well as for questionable cases with negative RT-PCR results [16]. Zhao et al [17] analyzed the samples of 173 patients, detecting the presence of antibodies <40% among patients within 1-week after symptom onset, and showed a rapid increase of up to 94.3% for IgM, and 79.8% for IgG from day 15 after the onset of illness. Xiang et al described antibody development even earlier, on the fourth day after symptom onset. According to the authors they provide strong support for the utility of serological testing in routine diagnostics regarding diagnosis and management [18].
Until recently, there was a lack of a widespread availability of valid test kits making antibody testing in routine clinical care challenging. In May 2020, an Elecsys antibody-test was released by Roche Diagnostics to detect anti-SARS-CoV2 immunoglobulins, with the ability to bind the viral nucleocapsid antigen [19]. According to the manufacturer, the sensitivity 14 days after a positive SARS-CoV2 test is up to100% and the specificity 99.91%, respectively [19]. Currently, there are no studies available to confirm these numbers. Moreover, studies describing antibody production in oncologic patients after SARS-CoV-2 infection are lacking. The aim of our study was to observe the course of antibody development and analyze the seroprevalence of antibodies against SARS-CoV2 in oncologic patients with a history of COVID-19.
Material and methods
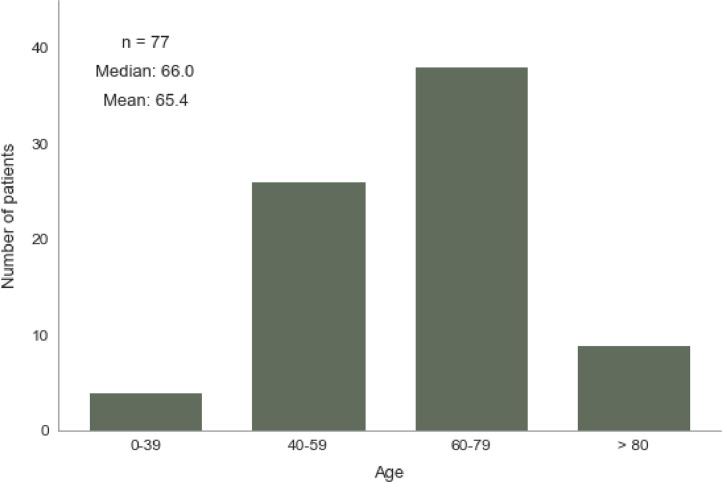
From April 15, 2020, all patients visiting one of the 7 participating outpatient clinics were tested for SARS-CoV2 infection by collecting throat swab from upper respiratory tract and RT-qPCR, regardless of symptoms. A total of 77 oncology patients who tested positive for SARS-CoV2 by RT-qPCR were enrolled in the analysis of anti-SARS-CoV2-antibodies. Clinical characteristics and demographics of the enrolled patients are shown in Table 1 . The distribution of age of the enrolled patients is shown in Fig. 5. For RNA isolation, the MGIEasy Magnetic Beads Virus DNA/RNA Extraction Kit was used on MGI SP-960 instruments. The extracted RNA was analyzed by RT-qPCR using the BGI Real-time fluorescent RT-PCR kit for detecting 2019-nCoV2. RT-qPCR and signal interpretation were performed on Applied Bioscience ABI 7500 Fast machines according to the instruction manual. The target sequences in RT-qPCR were ORF1ab for SARS-CoV2 and human GADPH, which served as an internal reference for effective RNA isolation. Positive and negative controls were included on each plate. The average number of days between a patient's positive RT-qPCR result and the first subsequent negative result was 14 days (SD 7,9).
Table 1.
Studies describing antibody production after infection with SARS-CoV2.
| Title | Authors | Number of patients | Statements |
|---|---|---|---|
| Antibody responses to SARS-CoV2 in patients with COVID-19 | Long et al [16] | n = 285 (63 follow-up measurements for SARS-CoV2 positive hospitalized tested patients) |
|
| Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019 | Zhao et al [17] | n = 173 |
|
| Antibody detection and dynamic characteristics in patients with COVID-19 | Xiang et al [18] | n = 109 (85 with confirmed diagnosis of SARS-CoV2 and 24 patients with suspected diagnosis of SARS-CoV2 infection) |
|
Fig. 5.
Age distribution of SARS-CoV-2 antibody positive tested patients.
After the confirmation of the SARS-CoV2 infection, blood was drawn from the patients according to the individual therapy algorithm, within a median time interval of 26 days (SD 13,6) after a positive test result in RT-qPCR. The samples were taken at different intervals, expecting the presence of antibodies at least at day 14 after SARS-CoV2 detection.
For measurements of anti-SARS-CoV2 antibodies (IgM and IgG), the Elecsys anti-SARS-CoV2 immunoassay from Roche was used on a Cobas e801 according to the vendor's instructions. The assay targets a recombinant protein representing the nucleocapsid (N) antigen for the determination of antibodies against SARS-CoV2.
Differences of clinical characteristics of the patients between the 2 subgroups (positive and negative anti-SARS-CoV2 antibody results) were tested for statistical significance using the Chi-square test. Due to multiple testing, test results were adjusted using the Bonferroni method. Adjusted P-values <.05 were regarded as statistically significant.
The study was approved by the Ethics Committee of the Bavarian Chamber Of Physician (BLÄK) with the ethic committee´s approval No. 20037.
Results
Out of 77 patients with a positive SARS-CoV2 RT-qPCR result enrolled in the study, only 6 patients developed measurable antibodies for SARS-CoV2 after 14 days or longer, whereas 71 of the tested patients were below the assay's cut-off value, even after multiple testing later in the course.
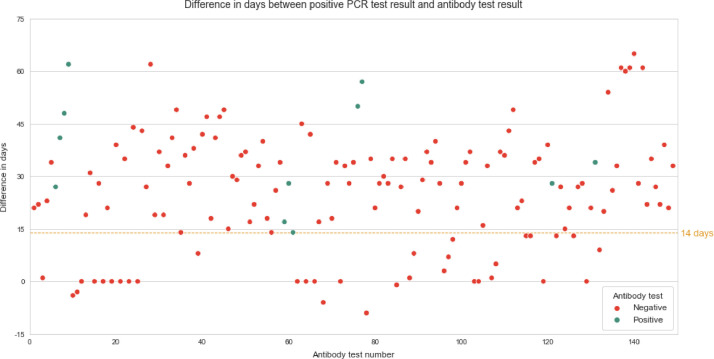
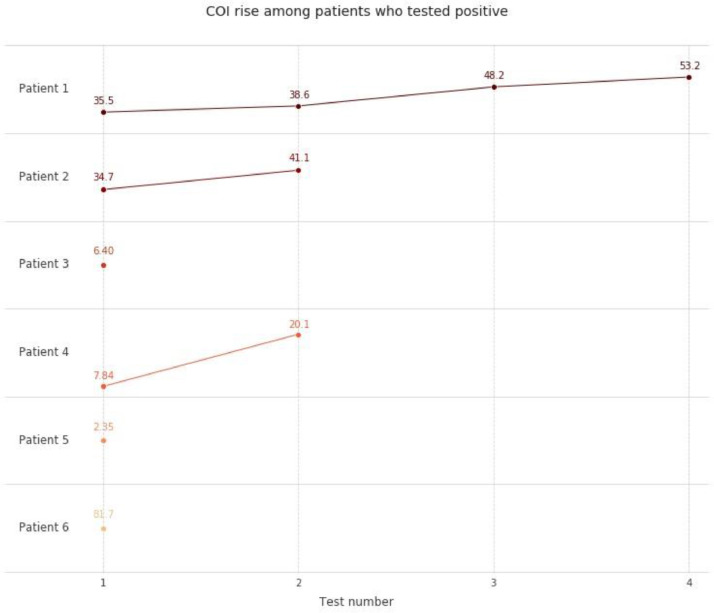
The first antibody test was performed on average 26 (SD 13,61) days after a positive SARS-CoV2 RT-qPCR result (Fig. 1 ). A second measurement was performed in 45 patients after 35 days (median, SD 9,9). Thirty patients received a third measurement at day 41 (mean, SD 10,8). 13 patients were tested multiple times (<6). The patients who tested negative for antibodies in the first sample did not show any increase in antibody concentration signal in subsequent tests. However, in three out of 6 patients who tested positive for antibodies, an increase in concentration signal was observed over the course of the study (Fig. 2 ). Three patients who developed antibodies showed mild symptoms including shortness of breath and common cold symptoms. These patients had slightly elevated temperature (median 37,5°C). Two of the 6 patients with positive SARS-CoV2 antibody results experienced severe forms of COVID-19. One of the patients who tested positive, developed pneumonia (CURB-65 score of 2) and had to be hospitalized, but was not admitted to the ICU and did not require assisted ventilation according to a low CURB-65 index. This patient suffered from an active tumor disease and was receiving immunotherapy with lenalidomide (Revlimid) at the time of testing. Due to the critical medical condition of the patient, tumor therapy had to be aborted. Eventually this patient tested negative on SARS-CoV2 PCR (7 days after the first positive RT-qPCR result) with complete remission of pneumonia and continued immunotherapy. One additional patient suffering from Acute Respiratory Distress Syndrome (ARDS) was hospitalized and had to be treated in the ICU using extracorporeal membrane oxygenation. This patient had not received systemic oncologic therapy in the previous 6 months. After 5 days of treatment in the ICU, using extracorporeal membrane oxygenation followed by a 2-week stay in hospital, condition of patient was stabilized.
Fig. 1.
Difference in days between positive RT-qPCR test result and first anitbody test result.
Fig. 2.
Rise of cut off index among SARS-CoV-2 antibody positive tested patients.
In the antibody positive group 4 patients suffered from hematological or lymphatic malignancies compared to 15 patients in the antibody negative group (Table 1). The second most common malignancies were tumors of the urinary tract with 2/6 in the positive tested group. Within the cohort of patients with negative antibody tests most solid cancer types were breast tumors 12/71, tumors of digestive organs 9/71, and tumors of male genital organs 7/71 (Table 1).
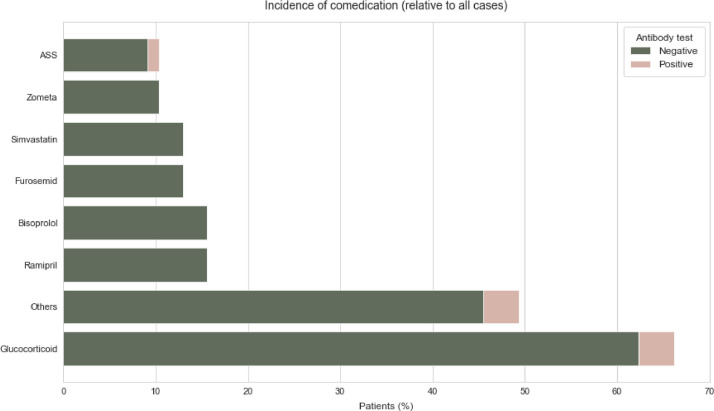
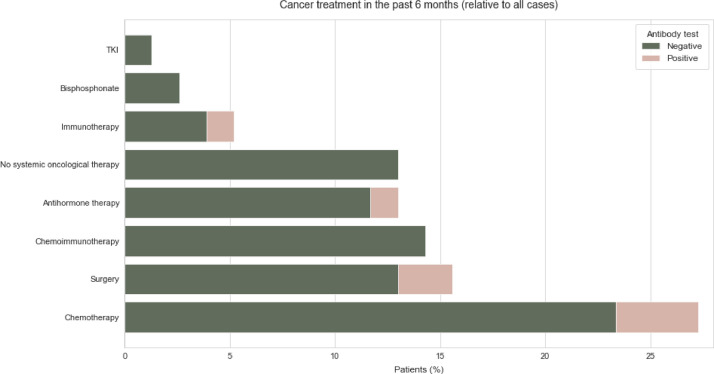
The most common comorbidities in both the individuals who tested positive for antibody and those who tested negative were hypertension and diabetes. Therapies carried out up to 6 months before the positive RT-qPCR result were taken into account in the evaluation. Glucocorticoids were applied in 3 of the 6 antibody positive patients compared to 48 patients in the group without detectable antibodies. In the group who tested negative for antibodies, 12 patients were treated with Bisoprolol and 12 with Ramipril (Fig. 3 ). The most common applied systemic cancer therapies in the group who tested positive for antibody were chemotherapy in three patients, antihormonal therapy in 1 patient and immunotherapy in 1 patient (Fig. 4 ). Within the group who tested negative for antibody 18 received chemotherapy, 11 received chemoimmunotherapy, and 9 received antihormone therapy (Fig. 4).
Fig. 3.
Incidence of comedication of SARS-CoV-2 anitbody positive tested and SARS-CoV-2 negative tested patients relative to all cases.
Fig. 4.
Cancer treatment in the past 6 month of SARS-CoV-2 anitbody positive tested and SARS-CoV-2 negative tested patients relative to all cases.
Discussion
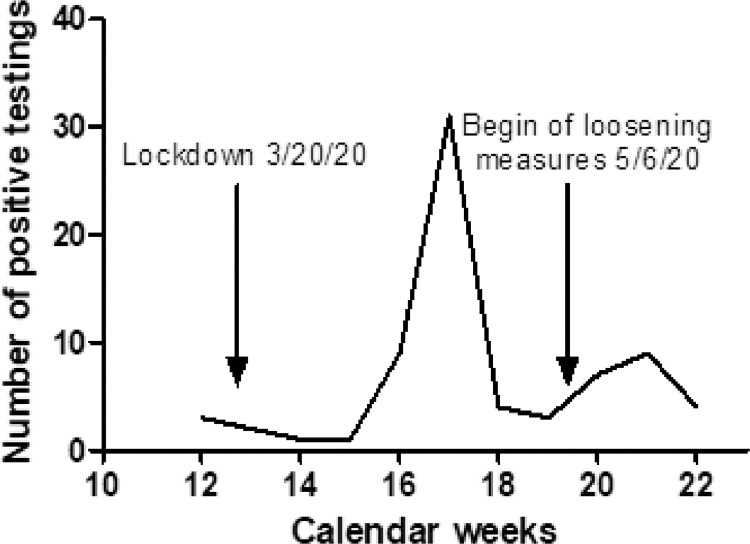
Thus far, COVID-19 has globally led to more than 1,439,784 deaths and to enormous socioeconomic damage due to shutdowns worldwide [20]. Importantly, our data suggest that an infection with SARS-CoV2 is not automatically accompanied by the development of antibodies. After the peak of positive SARS-CoV2 PCRs in Bavaria between April 15 and March 30, there should have been a peak phase of antibody development in those patients (Fig. 6 ). Our results clearly indicate that far from all patients develop antibodies, as shown by multiple testing at regular intervals. This is particularly interesting in view of the therapies administered during this period. Most of the patients tested positive in RT-qPCR received the first negative RT-qPCR result 13 days (in median, SD 8,1) after confirmation of the positive test. The timeframe of positive PCR detection of the virus was therefore only a few days. The reason for the short-time interval of positive RT-qPCR results might be a low virus load to which patients were exposed. Based on patient surveys we presume that the patients consistently adhered to the requirements of social distancing, and may therefore have only been exposed to low virus innoculums. Since we assume that patients with a low viral load may not be infectious, the viral load determination could be used to enable selective isolation measures, which would make a decisive socioeconomic contribution. The patient cohort of oncologic patients is particularly suitable for this purpose, as they are predominantly asymptomatic SARS-CoV2 carriers (manuscript submitted).
Fig. 6.
Course of the RT-qPCR tests for SARS-CoV-2.
A further explanation for the mild cases could be special oncological therapies that inhibit virus replication and thus have a positive effect on the course of the infection. So far it is unclear why many infected patients with tumors remain asymptomatic or show only mild symptoms, whereas previously healthy individuals can develop a fatal infection. This illustrates the importance of determining the viral load in addition to RT-qPCR test. The observed lower incidence of COVID-19 disease in oncology patients offers a completely new perspective on the possible underlying pathomechanisms of the disease.
The limitations of the study are the sample size of only 77 patients, even if they have been followed up over a relatively long period of time. In addition, only one test (Roche) was used to test antibody development, even though the test has a sensitivity of up to 100% and a specificity of 99,1% according to the manufacture´s specification. Furthermore, our cohort consisted of only oncology patients, including immunosuppressed patients and thus represents a special cohort. However, focus on nonhospitalized cases of COVID-19 is a strength and represent real-world data of outpatient oncology medical care.
Conclusion
For the further management of the pandemic and the socioeconomic impact on society, a strategy that allows selective isolation measures is particularly important. So far, it has been assumed that patients suffering from COVID-19 develop antibodies that provide immunity and are thus protected from a reinfection with SARS-CoV2. This also forms the basis of the assumption that rapid vaccine development will lead to rapid control of the pandemic.
Our study indicates that only a part of SARS-CoV2 infected patients develop anti-SARS-CoV2 antibodies. Thus, it has to be noted that RT-qPCR only shows a test result at a certain point in time, whereas antibody tests can provide information about an infection that has occurred in the past. Moreover, it could be that antibody tests detect patients who were infected earlier, without being tested by RT-qPCR. However, further investigations are needed to determine which patients infected by SARS-CoV2 develop antibodies and if this provides immunity. Antibody tests cannot replace the RT-qPCR but could provide further information on immunity. According to our assumption, a negative test cannot rule out an infection that has already occurred. A positive antibody development, however, indicates that the patient has been infected. Comprehensive testing of the population could provide important information on the number of infected persons. In our opinion, antibody tests should be widely available, but in combination with RT-qPCR, solely due to our data which demonstrates that some infected individuals do not develop antibodies. In so far as our understanding goes, on how the mechanism works but determine who develops antibodies and who does not, both tests should be comprehensive. This is particularly important, as it is assumed that people who have suffered from the infection will automatically become immune. Even though our data shows that this is not the case and that these patients could be reinfected. This could prove to be a special challenge for those countries that pursue the strategy of herd immunity. Due to the novelty of SARS-SoV2, there are still no long-term studies on answering the question whether people who have experienced the disease are protected from new infections, therefore it is important to follow an antibody development through long-term studies to find out how long they provide immunity to COVID-19. This underlines the urgent need to validate the antibody detection approaches to support diagnosis, vaccine development, and safety.
Author contributions
Declaration of authorship: Louisa Hempel, Jakob Molnar, Zeljka Trepotec conceived and designed the study; Sebastian Robert, Julia Veloso, Josef Scheiber and Louisa Hempel acquired the data; all authors discussed the results and contributed to the final manuscript; Louisa Hempel carried out the experiment with the help from Sofie Englisch and Philip Weinzierl, Cordula Schick, Beate Gandorfer, Jakob Molnar and Zeljka Trepotec. Valeria Milani, Kathrin Schweneker, Bastian Fleischmann, Axel Kleespies and Dirk Hempel fabricated the sample. Kristina Riedmann helped supervise the project.
All listed authors confirm that they have read the revised version of the manuscript and agree with all changes.
Conflict of Interest
Louisa Hempel, Jakob Molnar, Sebastian Robert, Julia Veloso, Zeljka Trepotec, Sofie English, Philip Weinzierl, Valeria Milani, Cordula Schick, Kathrin Schweneker, Bastian Fleischmann, Josef Scheiber, Axel Kleespies, Kristina Riedmann, Dirk Hempel and Armin Piehler have no conflict of interest.
Human Rights statements and informed consent
All procedures were in accordance with the ethical standard of the responsible committee of the Bavarian Chamber Of Physician (BLÄK) with the ethic committee’s approval No. 20037 and with the Helsinki Declatation of 1964 and it’s later amendments.
References
- 1.Akhvlediani T, Jelcic I, Taba P, Pfausler B, Steiner I, Sellner J. What did we learn from the previous coronavirus epidemics and what can we do better: a neuroinfectiological point of view. Eur J Neurol. 2020 doi: 10.1111/ene.14395. Published online0-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med. 2020;26(April) doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schoeman D, Fielding BC, Arias-Reyes C, et al. Journal Pre-proof does the pathogenesis of SAR-CoV-2 virus decrease at high-altitude? Does the pathogenesis of SAR-CoV-2 virus decrease at high-altitude? Corresponding authors. Cell Res. 2020;9(1):278–280. doi: 10.3390/ijerph17082932. [DOI] [Google Scholar]
- 4.Astuti I, Ysrafil Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes Metab Syndr Clin Res Rev. 2020;14(4):407–412. doi: 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miłek J, Blicharz-Domańska K. Coronaviruses in avian species-review with focus on epidemiology and diagnosis in wild birds. J Vet Res. 2018;62(3):249–255. doi: 10.2478/jvetres-2018-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeng W, Liu G, Ma H, et al. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem Biophys Res Commun. 2020;527(3):618–623. doi: 10.1016/j.bbrc.2020.04.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herold T, Jurinovic V, Arnreich C, et al. Elevated levels of interleukin-6 and CRP predict the need for mechanical ventilation in COVID-19. J Allergy Clin Immunol. 2020:2020. doi: 10.1016/j.jaci.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Fang X, Cai Z, et al. Comorbid chronic diseases and acute organ injuries are strongly correlated with disease severity and mortality among COVID-19 patients: a systemic review and meta-analysis. Research. 2020;2020:1–17. doi: 10.34133/2020/2402961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The Lancet Oncology COVID-19: global consequences for oncology. Lancet Oncol. 2020;21(4):467. doi: 10.1016/S1470-2045(20)30175-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang W, Guan W, Chen R, et al. Cancer patients in SARS-CoV-2 infection: a nationwide analysis in China. Lancet Oncol. 2020;21(3):335–337. doi: 10.1016/S1470-2045(20)30096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Channappanavar R, Fett C, Zhao J, Meyerholz DK, Perlman S. Virus-specific memory CD8 T cells provide substantial protection from lethal severe acute respiratory syndrome coronavirus infection. J Virol. 2014;88(19):11034–11044. doi: 10.1128/jvi.01505-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hilgenfeld R, Peiris M. From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses. Antiviral Res. 2013;100(1):286–295. doi: 10.1016/j.antiviral.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Payne DC, Iblan I, Rha B, et al. Persistence of antibodies against middle east respiratory syndrome coronavirus. Emerg Infect Dis. 2016;22(10):1824–1826. doi: 10.3201/eid2210.160706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.B S, K S, K W, W P. Perspectives on monoclonal antibody therapy as potential therapeutic intervention for Coronavirus disease-19 (COVID-19) Asian Pacific J Allergy Immunol. 2020;38:10–18. doi: 10.12932/AP-200220-0773. http://huji-primo.hosted.exlibrisgroup.com/openurl/972HUJI/972HUJI_SP?sid=EMBASE&sid=EMBASE&issn=0125877X&id=doi:10.12932%2FAP-200220-0773&atitle=Perspectives+on+monoclonal+antibody+therapy+as+potential+therapeutic+intervention+for+Coronavirus+disease-19+%28COVID-19%29&stitle=Asian+Pac.+J.+Allergy+Immunol.&title=Asian+Pacific+journal+of+allergy+and+immunology&volume=&issue=&spage=&epage=&aulast=Shanmugaraj&aufirst=Balamurugan&auinit=B.&aufull=Shanmugaraj+B.&coden=&i doi:10.12932/AP-200220-0773 LK - Available at. [DOI] [PubMed] [Google Scholar]
- 15.NMA OKBA, Muller MA, Li W, et al. SARS-CoV-2 specific antibody responses in COVID-19 patients. MedRxiv. 2020 doi: 10.1101/2020.03.18.20038059. Published online2020.03.18.20038059. [DOI] [Google Scholar]
- 16.Long QX, Liu BZ, Deng HJ, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26(6):845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 17.Zhao J, Yuan Q, Wang H, et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa344. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiang F, Wang X, He X, et al. Antibody detection and dynamic characteristics in patients with covid-19. clin infect dis. 2020 doi: 10.1093/cid/ciaa461. published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evaluation of Roche Elecsys Anti-Sars-CoV-2 serology assay for the detection of anti-SARS-CoV-2 antibodies.
- 20.Unders anding he Numbers : Provisional Dea h Coun s and COVID-19 How it wo ks.:19.