Abstract
The diffusion of novel SARS-CoV-2 coronavirus over the world generated COVID-19 pandemic event as reported by World Health Organization on March 2020. The huge issue is the high infectivity and the absence of vaccine and customised drugs allowing for hard management of this outbreak, thus a rapid and on site analysis is a need to contain the spread of COVID-19. Herein, we developed an electrochemical immunoassay for rapid and smart detection of SARS-CoV-2 coronavirus in saliva. The electrochemical assay was conceived for Spike (S) protein or Nucleocapsid (N) protein detection using magnetic beads as support of immunological chain and secondary antibody with alkaline phosphatase as immunological label. The enzymatic by-product 1-naphtol was detected using screen-printed electrodes modified with carbon black nanomaterial. The analytical features of the electrochemical immunoassay were evaluated using the standard solution of S and N protein in buffer solution and untreated saliva with a detection limit equal to 19 ng/mL and 8 ng/mL in untreated saliva, respectively for S and N protein. Its effectiveness was assessed using cultured virus in biosafety level 3 and in saliva clinical samples comparing the data using the nasopharyngeal swab specimens tested with Real-Time PCR. The agreement of the data, the low detection limit achieved, the rapid analysis (30 min), the miniaturization, and portability of the instrument combined with the easiness to use and no-invasive sampling, confer to this analytical tool high potentiality for market entry as the first highly sensitive electrochemical immunoassay for SARS-CoV-2 detection in untreated saliva.
Keywords: Immunosensors, Screen-printed electrodes, Carbon black, Magnetic beads, Differential pulse voltammetry
Highlights
-
•
Carbon black-modified screen-printed for smart immunosensor able to detect SARS-CoV-2.
-
•
Magnetic beads as support for rapid and reliable SARS-CoV-2 detection in untreated saliva.
-
•
Saliva clinical samples and cultured SARS-CoV-2 virus analysis.
-
•
Selectivity when tested with influenza pandemic/seasonal H1N1 viruses.
1. Introduction
The pandemic event of novel SARS-CoV-2 infection started on 31 December 2019 when Chinese health authorities reported to World Health Organization (WHO) Country Office a pneumonia of unknown cause detected in the city of Wuhan. Later, on 5 January 2020 WHO published the first Disease Outbreak News on the new virus reporting the risk assessment and advice as well as the status of patients and the public health response on the cluster of pneumonia cases in Wuhan. On 11 February, WHO named COVID-19 (COrona VIrus Disease 2019) the disease due to SARS-CoV-2 and one month later WHO reported COVID-19 as a pandemic outbreak, having impact all over the world.
In Europe, the President of European Commission reported as top priority the safeguarding of the health and well-being of citizens by using any tool available by coordinating all Member States (https://ec.europa.eu/info/live-work-travel-eu/health/coronavirus-response/public-health_en). Several countermeasures are taken to tackle this unexpected pandemic event including european strategy to accelerate the development, manufacturing, and deployment of vaccines against COVID-19 and the establishment of the guidelines for high-quality coronavirus test methodologies which encompasses the analytical tools for detecting SARS-CoV-2 and those for measuring the immune response of the human body to the infection (https://ec.europa.eu/info/live-work-travel-eu/health/coronavirus-response/public-health_en).
As highlighted by EU, one of the main pillars is related to analysis, indeed the fast and accurate detection represents a huge task for the management of COVID-19, being the key point for identifying the infected people, especially asymptomatic subjects, enabling the correct countermeasure in timely fashion.
To evaluate the immune response of human body to the infection, different serological tests have been developed and are present in the market. For instance, qSARS-CoV-2 IgG/IgM Rapid Test is manufactured by Cellex and it is a lateral flow chromatographic immunoassay to detect IgM and IgG antibodies against the SARS-CoV-2 (https://www.cellexcovid.com). However, this kit producer does not report which epitopes of virus proteins able to bind IgM and IgG present of the patient's blood are used to fabricate the device. COVID-19 IgM/IgG System, manufactured by Chembio Diagnostic Systems, reports that the detection of antibodies is carried out using Nucleocapsid protein as target (https://chembio.com/dpp-covid-19-igm-igg-system-ous-2/). VITROS Immunodiagnostic Products Anti-SARS-CoV-2 Total Reagent Pack, manufactured by Ortho-Clinical Diagnostics, Inc., is an ELISA test using Spike protein as target protein (Ward et al., 2020).
Regarding the type of the above-mentioned kits, the EU reported in 2020/C 122 I/01 that “The effectiveness of antibody tests in early COVID-19 diagnosis is very limited because antibodies become detectable in the patient's blood only several days after infection. This depends on the one hand on the individual's immune system and on the other hand on the sensitivity of the technique employed. In addition, antibodies persist for some time after the infection has cleared. They do not give a definite answer on the presence or absence of the SARS-CoV-2 virus and thus they are not suitable to assess if the tested individual may be contagious for others” (EUROPEAN COMMISSION COMMUNICATION FROM THE COMMISSION Guidelines on COVID-19 in vitro diagnostic tests and their performance (2020/C 122 I/01)). However, the utility of these tests relies on their effectiveness to perform large-scale sero-epidemiological population surveys to evaluate and guide the management when the pandemic is under control, nevertheless they cannot exclude that the patient having developed an immunitary defence is still contagious.
Regarding the detection of the virus, the quantification of SARS-CoV-2 at low level is the most challenging issue in the rapid diagnosis of COVID-19, because the detection of SARS-CoV-2 at the start of infection is the main and successful approach for an effective COVID-19 management to limit the spread of this viral contagious.
For detecting SARS-CoV-2, the approach recommended by the WHO and the European Centre for Disease Prevention and Control (ECDC) is related to the quantification of virus genetic material by reverse transcription polymerase chain reaction in nasal or throat secretions (i.e. swabs). The currently available kits based on real-time PCR technology offers the highest sensitivity to detect the presence of SARS-CoV-2 virus. However, in case of samples with low viral load, the repetition of the test may be required for delivering a definitive diagnosis (Cui and Zhou, 2020). Actually, it is considered as positive for SARS-CoV-2 RNA the specimen with a limit of 38/40 cycle threshold (CT) (corresponding to 5610 virus copies/mL) (Wyllie et al., 2020; Wang et al., 2020). The main drawbacks of this method include the long analysis time i.e. at least 3 h, laboratory set-up, and skilled personnel.
In addition to PCR for SARS-CoV-2 RNA detection, ELISA for antibody analysis have been developed, but both these methods are characterised by some limitations (Lassaunière et al., 2020; Liu et al., 2020). As widely reported in literature, immunosensors are characterised by sensitive, cost-effective, and accurate detection (Khristunova et al., 2019, 2020), and the detection of antigens such as viral proteins, which is the basis to develop smart and reliable biosensing systems, can be an added value for the management of SARS-CoV-2 outbreak (Morales-Narváez and Dincer, 2020; Mahapatra and Chandra, 2020).
However, all the developed methods are tests based on serum analysis, while our method uses saliva as specimen, which is a non-invasive sample and which accuracy has been demonstrated, through PCR methods, to be comparable with nasopharyngeal sampling method (Alizagar et al., 2020; Azzi et al., 2020).
In detail, SARS-CoV-2 is a coronavirus with size ranges 50–200 nm diameter and consists of positive-sense single-stranded ribonucleic acid (RNA), characterized by an envelope with superficial glycoproteins including transmembrane Spike (S), Envelope (E), and Membrane (M) proteins. S protein is cleaved by a host cell furin-like protease into two separate polypeptides S1 and S2 and it is characterized by an affinity with the human angiotensin-converting enzyme 2 (hACE2) used as a receptor to infect human cells (Verdecchia et al., 2020; Xia et al., 2020).
E protein is found in small quantities embedded within the host-derived viral envelope membrane while M protein is the structural protein with two different conformations to promote membrane curvature as well as bind to the nucleocapsid (J Alsaadi et al., 2019; Bhowmik et al., 2020; Coutard et al., 2020).
SARS-CoV-2 is also characterised by highly immunogenic phosphoprotein namely the nucleocapsid (N) protein which binds the viral genome in beads on a string type conformation. N protein of coronavirus is often used as a marker in diagnostic assays (Fehr and Perlman, 2015). Other authors also indicate N antigen of SARS-CoV-2 for reliable diagnosis of severe acute respiratory syndrome in patients (Di et al., 2005). More recently, Diao et al. reported N protein-based assay for accurate and rapid diagnosis of COVID-19, measuring N protein in urine as well as nasofaringeal swab within 10 min with fluorescence immunochromatographic assay. After comparing their N protein detection with data obtained on the same samples with the nucleic acids test, authors concluded that their N antigen detection method may guarantee early diagnosis in hospitals, moreover it may be used in large-scale screening in community (Diao et al., 2020).
The N and S2 proteins share a highly conserved structure with SARS-CoV, while S1 subunit is less conserved and more highly specific to SARS-CoV-2, thus these proteins can be used as antigens for rapid assay development (Ward et al., 2020). Seo et al. developed a field-effect transistor–based device for detecting SARS-CoV-2 by using as target analyte SARS-CoV-2 S protein and the antibody for the S protein as biocomponent immobilised on the graphene sheets coating on field-effect transistor (Seo et al., 2020). This biosensor was tested in nasopharyngeal swab specimens from COVID-19 patients and cultured virus, observing a detection limit equal to 100 fg/mL and 1.6 × 101 PFU/mL, respectively. In the market, for antigen detection a kit namely COVID-19 antigen Respi-Strip is available, which consists in a lateral flow assay for qualitative detection of virus in a nasopharyngeal or nasal sample with a LOD value of 5 × 103 PFU/mL (https://www.intermed.be/en/professional-products/laboratory-diagnostics/serology-amp-virology/rapid-test/coris-covid-19-ag-respi-strip.html).
These biosensing tools are conceived to be easily used in respect of PCR, however they require nasopharyngeal swab specimen, thus the sampling needs to be carried out by skilled personnel.
Taking into account that the seasonal influenza virus is present in salivary liquid, To et al. (2020) investigated and reported that also SARS-CoV-2 is present in the saliva of 91,7% of patients, demonstrating that saliva is a promising non-invasive specimen for diagnosis, monitoring, and infection control in patients with COVID-19. Indeed, simple sputum samples are processed with in-house 1-step real-time reverse transcription–quantitative polymerase chain reaction (RT-qPCR) assay targeting the S gene of SARS-CoV-2 (Chan et al., 2020). In addition, another study reported that between nasopharyngeal and saliva samples, saliva yielded greater sensitivity with the huge advantage of self-sample collection of saliva with less variability in respect to nasoparyngeal samples (Wyllie et al., 2020). Among the different biosensors, the electrochemical biosensors based on the use of screen-printed electrochemical cell, i.e. screen-printed electrodes (SPEs), have attracted a huge attention for the miniaturization of both the electrochemical sensor as well as the reader (Caratelli et al., 2020), the sensitivity, the capability to work in complex matrices, and the easiness to use (Arduini et al., 2016). In addition, SPEs are easily modified with nanomaterials namely gold nanoparticles, graphene, and carbon black during the mass-production to improve the electroanalytical performances and increasing the sensitivity of biosensors (Antuña-Jiménez et al., 2020; Arduini et al., 2020; Cinti and Arduini, 2017).
The magnetic beads-based electrochemical assay is an antibody-based assay in which magnetic beads (MBs) are used as support for the immunological chain with the following advantages i) the possibility to load high amount of capture antibody thanks to the high surface-to-volume ratio, allowing for an augmented sensitivity; ii) the use of magnetic field to move MBs in a selected volume eliminating with a simple washing step any matrix during the measurement; iii) the pre-concentration of MBs on the working electrode surface using a miniaturized customized magnetic tool for increasing the enzymatic by-product in close contact to the working electrode surface and thus the sensitive of electrochemical measurement (Pedrero et al., 2012).
In this study, in order to have a miniaturized electroanalytical device, we developed the first sensitive electrochemical biosensor for SARS-CoV-2 detection in saliva using a MBs-based electrochemical assay and carbon black-based SPE as sensor combined with PALM SENS portable potentiostat as reader. Both SARS-CoV-2 proteins namely S protein and N protein were used as target analyte developing a sandwich assay with immobilised antibodies for S or N proteins on MBs. The binding was evaluated by using secondary antibody labelled with alkaline phosphatase enzyme. The developed MBs-based assay was tested in buffer solution, saliva samples, and cultured SARS-CoV-2 virus, as well as in saliva clinical samples, comparing the data with the ones obtained by RT-PCR using nasopharyngeal swab specimen.
2. Materials and methods
2.1. Reagents, solutions and samples
The target SARS-CoV Spike (subunit S1) protein (0.5 mg/mL), the Monoloclonal antibody anti SARS-CoV (produced in mouse, 1 mg/mL) and the Polyclonal antibody SARS-CoV Spike (produced in rabbit, 1 mg/mL) which recognizes the S protein, were purchased from Sinobiological (Germany); the target SARS Coronavirus 2019 Spike Recombinant protein (1 mg/mL) and the Polyclonal Antibody SARS-CoV-2 to Spike S1 (produced in rabbit, 0.1 mg) were from ProSci (USA); the target SARS-CoV-2 Nucleocapside protein was from Acrobiosystem (USA, 0.6 mg/mL); Monoclonal antibody to SARS-CoV-2 (produced in mouse, 3.4 mg/mL) and Polyclonal antibody to SARS-CoV-2 (produced in rabbit, 7.8 mg/mL), which recognized N protein, were purchased from Biorbyt (UK); high affinity Anti-Rabbit IgG Antibody Alkaline Phosphatase-labelled (PAb-AP, 1 mg/mL) that binds rabbit antibodies and anti-mouse IgG-AP (MAb-AP, 1 mg/mL) that binds mouse antibodies were from Vector Laboratories (USA); the bovine serum albumin (BSA), used as blocking agent for covered MBs or plates to reduce the aspecific adsorptions, the electrochemical AP substrates 1-Naphthyl phosphate disodium salt, the optical AP substrate 4-Nitrophenyl phosphate disodium salt (PNPP), diethanolamine, tween 20, sodium azide and all other reagents were obtained from Sigma (USA). Maxisorp™ surface, 96well polystyrene microtitre plates, were purchased from Nunc (Roskilde, Denmark). Dynabeads® Pan Mouse IgG pre-coated with anti-Mouse IgG able to bind monoclonal antibody (supplied as a suspension containing 4 x 108 beads/mL in phosphate buffered saline, pH 7.4) was from Life Technologies (USA). 25 μL of MBs are able to bind ~1 μg of antibody. A rotary shaker and a magnetic rack/particle concentrator were from Dynal Biotech (USA). Phosphate saline buffer (PBS) = 0.0015 M KH2PO4, 0.0081 M Na2HPO4, 0.137 M NaCl, 0,0027 M KCl, pH 7.4; Washing Buffer = PBS+0.05 % Tween 20; Storage buffer = PBS + 0.02% NaN3, and DEA buffer = DEA–MgCl2–KCl buffer = 0.97 M DEA+1 mM MgCl2+0.1 M KCl pH 9.6 were used as buffer solution.
Saliva and Nasopharyngeal swab samples were collected with the express consent, free and with informed agreement, to such collection and use, of the person from whom this material was taken, according to the current legislation.
2.2. Screen-printed electrode production and modification
SPEs were produced onto a transparent and flexible polyester support by employing 245 DEK (Weymouth, UK) serigraphic printer. A three-electrode cell was realised by using a graphite-based ink (Electrodag 423 SS) for the working electrode (surface area equal to 0.07 cm2) and counter-electrodes, and a silver-based ink (Electrodag 6033 SS) for the pseudo-reference electrode. The SPEs were modified with 6 μL of CB N220 dispersion (1 mg/mL in N,N-dimethylformamide:distilled H2O 1:1 v/v), by a three-step consecutive drop-casting procedure of 2 μL on the working electrode. The optimization of modification procedure and morphological characterisation of CB-modified SPEs by the scanning electron microscope were reported in our previous article (Arduini et al., 2012).
2.3. ELISA procedure
This test was performed to verify the capability of the antibodies to bind target proteins. 100 μL of both S proteins (from Sinobiological and from ProSci) 2 ng/mL prepared in PBS buffer were added in each well of a microtitre-plate and left overnight at 4 °C. Then, the coated plate was blocked with 100 μL of 3% (w/v) BSA in PBS and incubated for 1 h at 37 °C. Binding curves for each anti-SARS-CoV antibody were constructed by adding into the wells 100 μL of different antibody dilutions prepared in PBS and ranging from 2 μg/mL to 0.5 μg/mL (we have tested all the antibody purchased), for 1 h of incubation at 37 °C. The labelling step involves the addition of 100 μL of anti-rabbit IgG-AP 2 μg/mL and an incubation for 1 h at 37 °C (when we test MAb antibody, the labelling step involves anti-mouse IgG-AP 2 μg/mL). Between each step, a three-cycle washing procedure, using 150 μL of PBS +0.05% Tween 20, was performed. Finally, 100 μL of PNPP solution (2 mg/mL in DEA buffer, pH 9.6) were used and the absorbance was read (at 415 nm) after 30 min of incubation. To evaluate the efficiency of MBs to tether antibodies, a competitive ELISA was performed: 100 μL of 10 μg/mL of MAb anti-SARS-CoV-2 (Sinobiological) prepared in PBS buffer were added in each well of microtitre-plate and left overnight at 4 °C. Then, the coated plate was blocked with 100 μL of 3% (w/v) BSA in PBS and incubated for 1 h at 37 °C. Competition curves for anti-SARS-CoV antibody were constructed by adding into the wells 100 μL containing different antibody concentrations prepared in PBS ranging from 0.005 to 20 μg/mL + anti-mouse IgG-AP 0.5 μg/mL in an incubation for 1 h at 37 °C. Between each step, a three-cycle washing procedure, using 150 μL of PBS +0.05% Tween 20, was performed. Finally, 100 μL of PNPP solution (2 mg/mL in DEA buffer, pH 9.6) were used and the absorbance was read (at 415 nm) after 15 min of incubation.
2.4. Electrochemical immunoassay
The electrochemical MBs-based assay involves three sequential procedures: I) a preliminary blocking-coating procedure of the Dynabeads® Pan Mouse IgG (to store them at 4 °C until use for several months); II) an immunoassay procedure (in which the classical sequential incubations for the immuno-recognition events are merged in a single incubation of 30 min); III) an electrochemical detection using SPEs.
-
I)
Blocking-coating procedure: 250 μL of MBs 4 x 108 beads/mL was pipetted into 2 mL tube and washed twice with 1 mL of PBS pH 7.4 and the MBs were blocked incubating them in 1 mL of PBS pH 7.4 + 3% BSA for 30 min at room temperature (RT) with slow tilt rotation (using Dynal sample mixer). Then the supernatant was discarded and 500 μL of PBS, containing 10 μg MAb anti SARS-CoV-2, were added to the beads and incubated for 30 min at RT with slow tilt rotation. The blocked and coated MBs were washed twice with 1 mL of PBS. Finally, they were resuspended in 250 μL of PBS +0.02% NaN3, and stored for up to several months at 4 °C.
Between each washing step, the Eppendorf tube containing the magnetic beads was placed in the Dynal MPC and after 30 s the supernatant was discarded.
-
II)
Immunoassay procedure:
-
1.
shake and transfer 10 μL of the coated and blocked beads suspension (stored at 4 °C) into 2 mL tube (in number required for the samples to analyze);
-
2.
wash 2 times with 1 mL PBS, every time shaking and discarding the supernatant by placing for 1 min a magnet outside the tube in a lateral position;
-
3.
add consecutively 200 μL of PAb anti SARS-CoV-2 1 μg/mL in PBS (produced in rabbit) + 200 μL of PAb-AP anti rabbit IgG 1 μg/mL in PBS + 300 μL of sample;
-
4.
incubate for 30 min at RT;
-
5.
wash 2 times with 1 mL PBS +0.05% Tween 20 and one time with PBS, every time shaking and discarding the supernatant by using the magnet as above,
-
III)
Electrochemical measurements: At the end of the immunoassay procedure, the beads (with their immunological chain) were resuspended in 100 μL of DEA buffer and 20 μL of these suspension (three replicates for each sample) were drop cast on the working electrode and magnetically concentrated onto the surface through the magnet positioned just under the working electrode. The immune complex was then revealed by adding 70 μl of 1- naphthyl phosphate (5 mg/mL, prepared in DEA–MgCl2–KCl buffer pH 9.6) into each well. Waiting 2 min for the enzymatic reaction, the electroactive enzymatic product (1-naphthol) is measured by applying differential pulse voltammetry (DPV) with the following parameters, based on our previous work (Fabiani et al., 2019): Ebegin = −0.2 V; Eend = 0.4 V; Estep = 0.016 V; Epulse = 0.05 V; tpulse = 0.06 s; scan rate = 0.016 V/s.
The electrochemical measurements were performed by a portable potentiostat, PalmSens3 (Palm Instrument, The Netherlands), connected to a personal computer.
2.5. SARS-CoV-2 virus propagation
SARS-CoV-2 was passaged once in Vero cells to generate a virus master stock used to generate a virus working stock. The virus was propagated in Vero cells cultured in minimum essential medium (MEM) containing 2% (w/v) fetal bovine serum (Euroclone S.p.A.). After infection, virus stock was collected by centrifuging the culture supernatants of infected Vero cells at 600 g for 5 min. The clarified supernatant was supplemented to 20% with fetal bovine serum (w/v), frozen and kept at −80 °C until use. The concentration of infectious virus was determined by plaque-forming titre assay. Virus propagation, virus isolation or neutralization assays of SARS-CoV-2 needs to be conducted in a bio-safety Level-3 facility according to WHO laboratory biosafety guidance (https://www.who.int/publications/i/item/laboratory-biosafety-guidance-related-to-coronavirus-disease-(covid-19https://www.who.int/publications/i/item/laboratory-biosafety-guidance-related-to-coronavirus-disease-(covid-19)).
2.6. SARS-CoV-2 quantification by real-time PCR
Nasopharyngeal swabs collected and tested from Italian Scientific dept. of Army Medical Center for SARS-CoV-2. Viral RNA was extracted from 300 μL of swab's medium using a Maxwell RSC Viral Total Nucleic Acid Purification Kit on Maxwell RSC Instrument (Promega, USA). Total nucleic acid was eluted in a final volume of 50 μL of nuclease-free water. Then, one step Real-Time PCR was performed using Novel Coranavirus (2019-nCoV) Nucleic Acid Diagnostic Kit (Sansure Biotech Inc., China) on LC480 instrument (Roche Diagnostics, Germany). This test utilizes 2019-CoV ORF1ab and N protein genes as target regions and an internal control, which monitors the presence of PCR inhibitors. Reaction mixture contained 13 μL of 2019-nCoV-PCR Mix, 2 μL 2019-nCoV-PCR and 5 μL of extract viral RNA to reach to final volume of 20 μL. The cycling program was as follows: Reverse transcription at 50 °C for 30 min; cDNA denaturation at 95 °C for 1 min and 45 cycles of PCR at 95 °C for 15 s and 60 °C for 30 s (signal acquisition). FAM, ROX and Cy5 channels were selected for ORF1ab, N and the internal control detection, respectively.
2.7. Electrochemical measurement of SARS- CoV-2 in biosafety level 3 environment
To date it is impossible to relate the protein concentration to the total viral load, so, to evaluate the sensitivity of our assay on native virus, we tested SARS-CoV-2 directly in electrochemical MBs-based assay in Biosafety Level 3 laboratory. Starting from a SARS-CoV-2 aliquot of 6.5 x 105 PFU/mL, we prepared five 1:10 v/v serial dilutions in PBS and incubated for 30 min 300 μL of each dilution (and a negative control) with 10 μL of MBs (precoated with MAb anti SARS-CoV and washed), 200 μL of PAb anti-SARS-CoV 1 μg/mL in PBS and 200 μL of PAb-AP 1 μg/mL in PBS. The test was performed on N protein and S protein in parallel using antibodies that recognize protein N and S, respectively. After the washing step, we proceeded with the electrochemical measurement. All the apparatus used were covered in plastic to avoid any contamination.
2.8. 2009 H1N1 influenza pandemic and seasonal H1N1 influenza virus propagation for selectivity test
2009 H1N1 influenza pandemic (2009 pH1N1) and seasonal H1N1 influenza virus master stocks were passaged in Madin-Darby canine kidney (MDCK) to generate virus working stocks. The viruses were propagated in MDCK cultured in minimum essential medium (MEM) containing 2% (w/v) fetal bovine serum (Euroclone S.p.A.), 2 μg/mL trypsin-TPCK (Merck, Germany). After infection, virus stocks were collected by centrifuging the culture supernatants of infected MDCK cells at 600 g for 5 min. The clarified supernatants were supplemented to 20% with fetal bovine serum (w/v), frozen and kept at −80 °C until use. The concentration of infectious viruses was determined by the 50% tissue culture infectious dose (TCID50) by 10-fold serial titration in MDCK cells.
3. Results and discussions
3.1. Electrochemical MBs-based set-up
The high sensitivity of electrochemical detection combined with the miniaturization is the key of this work to reach the overriding goal for a cost-effective and reliable high sensitivity detection of SARS-CoV-2 using a no-invasive and attractive biological fluid namely saliva.
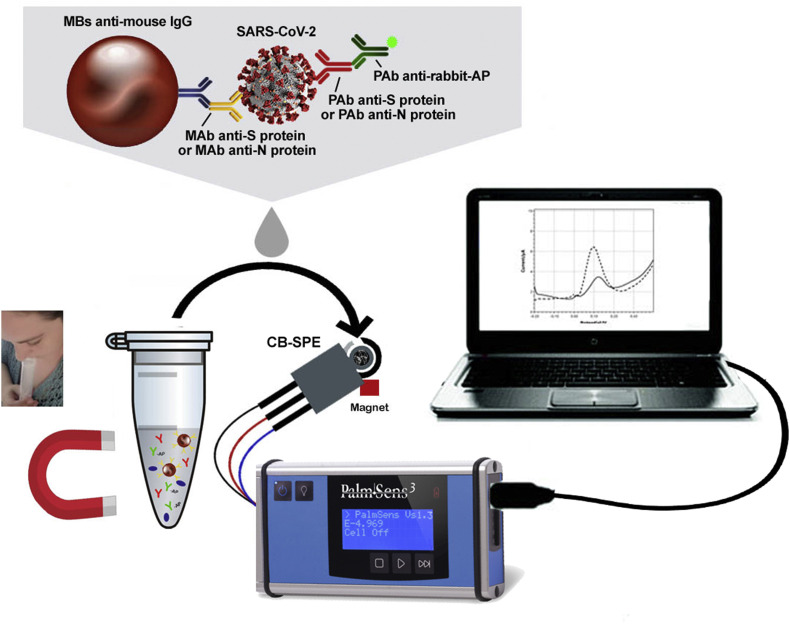
To accomplish this task, we have exploited MBs as support for immunological chain with their outstanding features as well as SPE modified with the nanomaterial carbon black (CB) to reach improved sensitivity as demonstrated in our previous papers for several electroactive compounds (Arduini et al., 2010, 2011, 2012, 2020; Talarico et al., 2015) (Fig. 1 ).
Fig. 1.
The MBs-based assay for SARS-CoV-2 detection in untreated saliva.
For making a simple and easy analysis, saliva is used by simply adding the sputum in the tube previously loaded with the reagents needed for the measurement, without requiring no-extra task to the end-users.
After the selected minutes for immunological chain construction, washing steps are required followed by the addition of MBs on the working electrode surface and the enzymatic substrate i.e. 1- naphthyl phosphate.
To develop a highly performant analytical tool in terms of sensitivity and selectivity, we start to optimize the assay using specific antibody for S1 subunit of S protein, taking into account that several S proteins are present on each viral particles.
3.2. Electrochemical MBs-assay optimization for spike protein detection
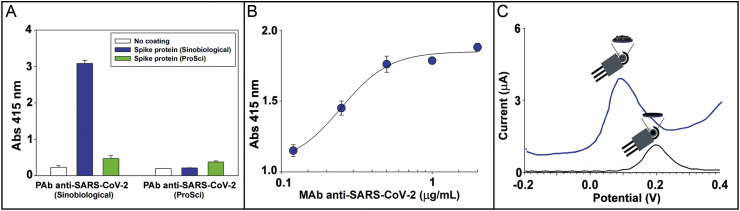
The use of the high sensitive antibodies is one of the main task to develop high sensitive and selective analytical device, thus for the selection of antibodies, spectrophotometric ELISA was carried out to assess the reactivity of MAb and two different PAb (Sinobiological, Germany and ProSci, USA) towards two different S proteins namely SARS Coronavirus 2019 Spike Recombinant protein (1000–1200 aa) and Recombinant Spike protein SARS-CoV Spike protein, S1 subunit. For all optical measurements, the secondary antibody labelled with alkaline phosphate enzyme was added subsequently using 4-nitrophenyl phosphate as enzymatic substrate and monitoring the binding through the formation of enzymatic by-product 4-nitrophenol at 415 nm. In Fig. 2 A the response of ELISA was reported testing the two PAb and two S proteins and demonstrating the better affinity between PAb belonging from Sinobiological, Germany and Recombinant Spike protein SARS-CoV, S1 subunit from Sinobiological, Germany, thus these reagents were selected for further studies. Subsequently, MAb was tested towards this S protein in ELISA and the binding curve was reported in Fig. 2B, demonstrating high affinity.
Fig. 2.
A) ELISA response for two different PAb anti-SARS-CoV-2 1 μg/mL (Sinobiological and ProSci) towards two different Spike proteins coated at 2 ng/mL. B) Binding curve of colorimetric ELISA for MAb anti-SARS-CoV-2 ranging from 0.12 – 2 μg/mL. Coating of Spike protein: 2 ng/mL. C) Electrochemical response using the MBs-based assay using CB-based modified electrode (blue line) and bare electrode (black line). The mean value (n = 3) with the corresponding standard deviation was reported for each measurement. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
For the electrochemical measurement, the secondary antibody labelled with alkaline phosphate enzyme was used with 1-naphthyl phosphate as enzymatic substrate, monitoring the formation of enzymatic by-product 1-naphtol at around 0.1 V by using differential pulse voltammetry. The use of CB-based SPE has allowed to increase the sensitivity as reported in Fig. 2C. Indeed, the enzymatic by-product was detected with increased sensitivity of ca. 4 folds at lower potential 0.1 V using CB-modified electrode (blue line) in respect to 0.2 V using unmodified electrode (black line). To evaluate how many MAbs were tethered on beads, we have measured the solution after the incubation of 125 μL of MBs with 5 μg of MAb by means of spectrophotometric ELISA, with the aim to estimate the amount of MAb unbounded. In detail, the amount of MAb tethered on beads was calculated by interpolating the data (red point) within the competition curve reported in Fig. S1, funding that 125 μL of MBs capture 2.6 μg of antibody, thus each MB tethers ~2 × 105 antibodies. This number is also consistent with the spherical surface of one bead (~63.6 μm2) and the dimension of one Ab (~10 nm) (Reth, 2013).
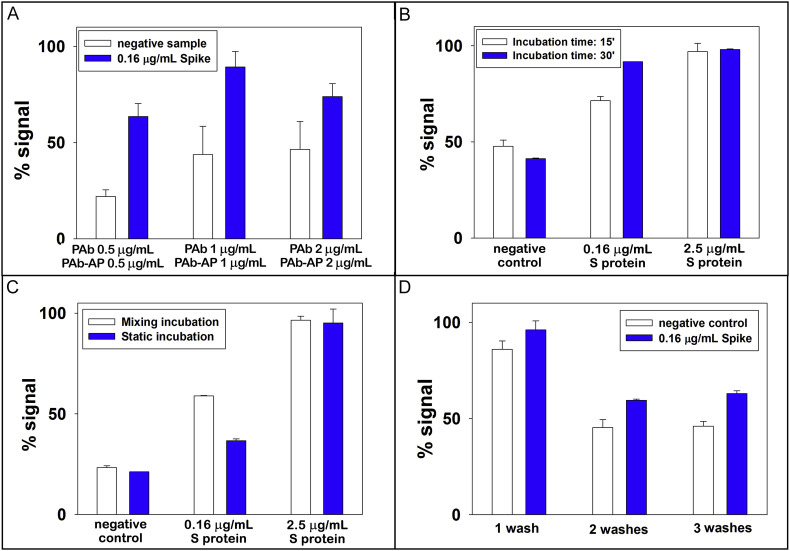
The construction of immunological chain requires the optimization of the concentration of PAb i.e. the antibody free in solution, the time of reaction between antigen and antibodies, the mode of reaction (i.e. tilt rotation or static condition), and number of washing steps.
PAb antibody is free in solution, thus it affects the response of the assay being the element that completes the sandwich immunological format. We tested amounts equal to 0.5, 1, and 2 μg/mL observing the highest signal using 1 μg/mL, thus this value was selected for further studies (Fig. 3 A). This experiment showed that 0.5 μg/mL of antibody was not a sufficient concentration to bind all the proteins present, while 2 μg/mL of antibody probably creates a steric hindrance that did not allow the correct binding (in the figure the positive signal is lower than the one obtained with 1 μg/mL of antibody). In Fig. 3B the effect of incubation time was evaluated demonstrating that 15 min were not sufficient for a good sensitivity, thus 30 min were selected. Because the assay involves a single incubation step, a minimum time of 30 min is required to correctly create the antibody-antigen-antibody bonds. Within the aim to develop a device to be easily customised for the end-user, the effect of stirring was evaluated. As depicted in Fig. 3C, the lower concentration i.e. 0.16 μg/mL demonstrated high signal in stirring condition, with a signal significantly different from the blank signal. However, in the case of the higher concentration 2.5 μg/mL, the same sensitivity was observed. This happens because the probability of binding at high concentrations is greater than the one at low concentrations, being the assay carried out in static condition. Taking into account that the device has been designed for an easy miniaturization, the measurement without stirring was selected for the rest of work. To reduce the tasks required by end-users for the analysis, the effect of number of washing steps needed to avoid the aspecific absorption on the MBs was evaluated, choosing two washing steps for the construction of the calibration curve (Fig. 3D). Under the optimized condition, the effect of pH on the assay was evaluated testing DEA buffer in a pH range comprised between 8.5 and 10, observing the highest sensitivity at pH = 9.6, thus this value was selected (Fig. S2).
Fig. 3.
A) Study of the signal response for evaluating PAb concentration. 10 μL of MBs, 200 μL of PAb anti SARS-CoV-2 0.5, 1, and 2 μg/mL (in PBS) + 200 μL of PAb-AP anti rabbit IgG 0.5, 1, and 2 μg/mL (in PBS) + 300 μL of tested sample, incubation time 30 min without stirring, 3 washing steps, testing a negative control and 0.16 μg/mL of S protein. B) Study of the signal response for evaluating the effect of incubation time. 10 μL of MBs, 200 μL of PAb anti SARS-CoV-2 1 μg/mL (in PBS) + 200 μL of PAb-AP anti rabbit IgG 1 μg/mL (in PBS) + 300 μL of tested sample, incubation time (15/30 min) without stirring, 3 washing steps, testing a negative control, 0.16 and 2.5 μg/mL of S protein. C) Study of the signal response as function of mixing during the incubation step. 10 μL of MBs, 200 μL of PAb anti SARS-CoV-2 1 μg/mL (in PBS) + 200 μL of PAb-AP anti rabbit IgG 1 μg/mL (in PBS) + 300 μL of tested sample, incubation time 30 min with and without stirring, 3 washing steps, testing a negative control and 0.16 and 2.5 μg/mL of S protein. D) Study of the signal response for evaluating the effect of number of washing steps. 10 μL of MBs, 200 μL of PAb anti SARS-CoV-2 1 μg/mL (in PBS) + 200 μL of PAb-AP anti rabbit IgG 1 μg/mL (in PBS) + 300 μL of tested sample, incubation time 30 min without stirring, 1/2/3 washing steps, testing a negative control and 0.16 μg/mL of S protein. The mean value (n = 3) with the corresponding standard deviation was reported for each measurement.
3.3. Analytical features of electrochemical MBs-based assay for spike protein detection in standard solution and saliva
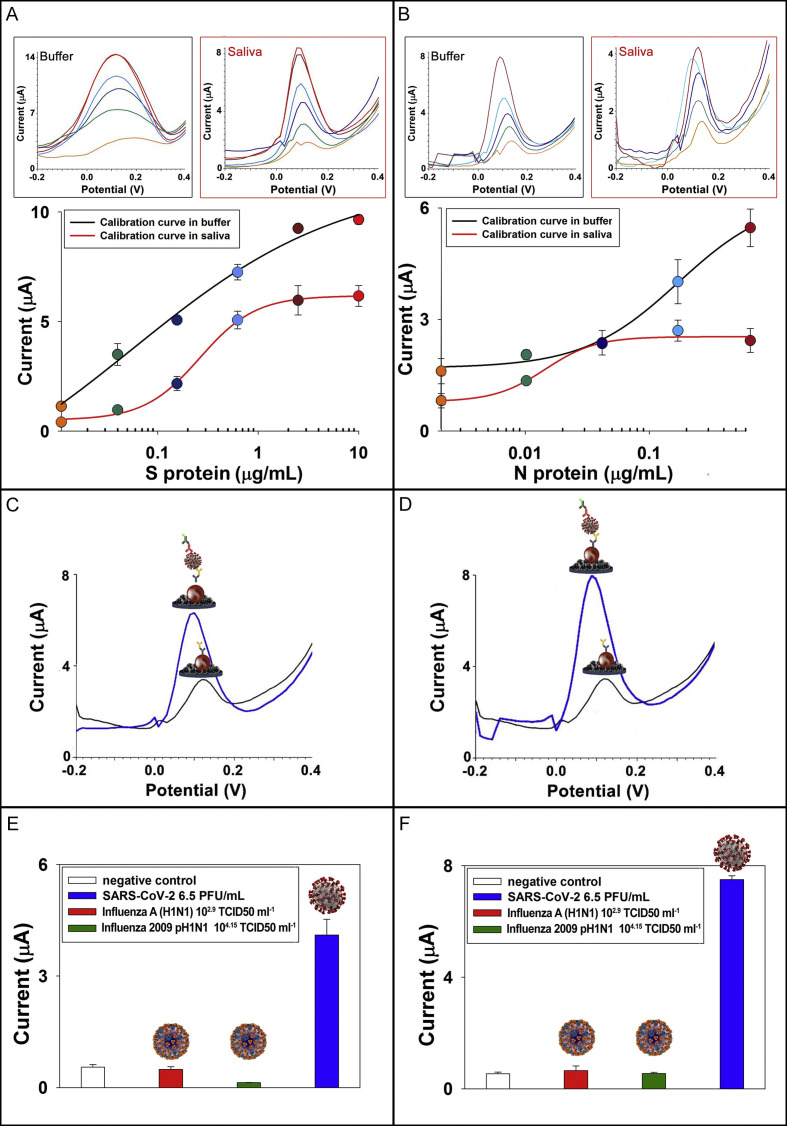
To assess the analytical features of the developed assay for measuring S protein in standard solutions, different concentrations of S protein diluted in 0.015 M phosphate buffer +0.137 M NaCl +0.0027 M KCl pH = 7.4 ranging from 0.04 to 10 μg/mL were tested, observing a sigmodal behavior (Fig. 4 A) described by non-linear four-parameter logistic calibration plot as follows:
| (1) |
where a and d are the asymptotic maximum and minimum values, c is the value of x at the inflection point and b is the slope, obtaining a detection limit equal to 14 ng/mL, calculated as blank signal + 3 standard deviation (SD). The matrix effect due to the analysis in saliva without any pre-treatment was evaluated by constructing the calibration curve in untreated saliva (Fig. 4A), observing the same sigmodal behavior with lower intensity of peak current due the matrix effect. The calibration curve in saliva was described by non-linear four-parameter logistic calibration plot with detection limit equal to 19 ng/mL.
Fig. 4.
A) Electrochemical calibration curve (inset the voltammograms) using the optimized parameters for S protein detection in buffer (black line) and in untreated saliva (red line) (n = 3). B) Electrochemical calibration curve (inset the voltammograms) for N protein detection in buffer (black line) and untreated saliva (red line) (n = 3). C) Voltammograms obtained without (black line) and with (blue line) cultured SARS-CoV-2 virus at concentration 6.5 PFU/mL using MBs-based immunoassay for S protein. D) Voltammograms obtained without (black line) and with (blue line) cultured SARS-CoV-2 virus at concentration 6.5 x 104 PFU/mL using MBs-based immunoassay for N protein. E) Exclusivity test on seasonal influenza virus A (H1N1) (102.9 TCID50 mL−1) and Influenza 2009 pH1N1 virus (104.15 TCID50 mL−1) in comparison to SARS-CoV-2 (6.5 PFU/mL) using assay for S protein (n = 3). F) Exclusivity test on seasonal influenza virus A (H1N1) (102.9 TCID50 mL−1) and 2009 influenza virus pH1N1 (104.15 TCID50 mL−1) in comparison to SARS-CoV-2 (6.5 x 104 PFU/mL) using assay for N protein (n = 3). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. Analytical features of electrochemical MBs-based assay for N detection in standard solution and saliva
To assess the analytical features of the developed assay for measuring N protein in standard solutions, different concentrations of N protein diluted in 0.015 M phosphate buffer +0.137 M NaCl +0.0027 M KCl pH = 7.4 ranging from 0.01 to 0.6 μg/mL were tested (Fig. 4B), using the previously optimized parameters, observing a sigmoidal behavior described by non-linear four-parameter logistic calibration plot Eq. (1) observing a detection limit of 4 ng/mL. The matrix effect due to the analysis in saliva without any pre-treatment was evaluated by construction the calibration curve in untreated saliva, observing well-defined sigmoidal behavior with lower intensity of peak current due the matrix effect with detection limit equal to 8 ng/mL.
3.5. Measurement of SARS-CoV-2 virus
The structural studies on SARS-CoV-2 reported in literature until now are not able to furnish unequivocal information related to the number of S proteins present on each virus (Ke et al., 2020; Kiss et al., 2020), thus it is difficult to calculate the number of viruses detected using the calibration curve for S protein. To evaluate the response of our assay for native virus, we tested the native virus ranging from 6.5 x 105 to 6.5 PFU/mL in Biosafety Level 3 environment. In Fig. 4 C/D, the voltammograms show the signal related to virus concentration in respect to the signal of negative control using the electrochemical MBs-assay for S protein and N protein, respectively.
As depicted in Fig. 4C, when the test is carried out using antibodies directed against S protein, the sensitivity of the assay is excellent (can detect 6.5 PFU/mL), in respect to the assay N protein (Fig. 4D). Instead, in case of N protein-based assay the sensitivity is lower (can be detect 6.5 x 103 PFU/mL), because the amount of N protein is significantly lower than the amount of S protein in SARS-CoV-2 virus.
3.6. Exclusivity study
To assess the selectivity of the proposed MBs-based assay, an exclusivity test was performed analyzing seasonal influenza virus A (H1N1) (102.9 TCID50 mL−1) and 2009 influenza virus pH1N1 (104.15 TCID50 mL−1), comparing the results with the SARS-CoV-2 previously analyzed (section 3.5). These tests were carried out using the assay directed against the S protein (Fig. 4E) and the assay directed against the N protein (Fig. 4F). In both cases, the results show a 100% of exclusivity.
3.7. Measurement of SARS-CoV-2 in clinical samples
To assess the effectiveness of the MBs-based assay using clinical samples, saliva, and nasopharyngeal swabs were tested using MBs-assay and Real-Time PCR, respectively. In detail, we tested saliva samples using fresh samples and freezed samples, observing a reduction of signal in case of freezed sample for both positive and negative patients, thus we decided to set 1.8 μA as threshold in case of unfreezed samples, while in case of freezed sample the threshold selected was 1 μA. We suggested the use of fresh saliva sampled after drinking a glass of water, with the aim of easy sampling without any treatment to match the requirement of the point of care system. As depicted in Table 1 , we have observed an agreement in 22/24 samples, which is a very satisfactory result, taking into account the low viral load detected as well as the analytical features of biosensors reported in literature (Table 2 ).
Table 1.
Clinical samples analyzed using MBs-based immunoassay and RT-PCR. * freezed samples.
| Sample | Spike protein MBs-assay | N protein MBs-assay | RT-PCR | CT number |
|---|---|---|---|---|
| Patient#1 | + | + | + | 38 (for both ORF and N) |
| Patient#2 | + | – | + | 38 for ORF, negative for N |
| Patient#3 | – | – | – | NO CT |
| Patient#4 | – | – | – | NO CT |
| Patient#5* | + | + | + | 37 for ORF and 33 for N |
| Patient#6* | + | + | + | 35 for ORF and 33 for N |
| Patient#7* | + | + | + | 31 for ORF and 28 for N |
| Patient#8* | + | No tested for low volume of sample | + | 31 for ORF and 27 for N |
| Patient#9* | + | + | – | NO CT |
| Patient#10* | – | – | – | NO CT |
| Patient#11* | – | – | – | NO CT |
| Patient#12* | – | – | – | NO CT |
| Patient#13* | – | No tested for low volume of sample | – | NO CT |
| Patient#14* | – | No tested for low volume of sample | – | NO CT |
| Patient#15* | – | No tested for low volume of sample | – | NO CT |
| Patient#16 | + | – | – | NO CT |
| Patient#17 | + | + | + | 31 for ORF and 30 for N |
| Patient#18 | – | – | – | NO CT |
| Patient#19 | – | – | – | NO CT |
| Patient#20 | – | – | – | NO CT |
| Patient#21 | – | – | – | NO CT |
| Patient#22 | – | – | – | NO CT |
| Patient#23 | – | – | – | NO CT |
| Patient#24 | – | – | – | NO CT |
Table 2.
Overview of biosensors for SARS-CoV-2 detection.
| BIOSENSOR | METHOD | ANALYTE | SAMPLE | ANALYTICAL FEATURES |
REF. |
|---|---|---|---|---|---|
| CARMEN-Cas13a | Microwell-array system for color-coded droplets of CRISPR detection reagents (fluorescent signal) | Amplified nucleic acid | Plasma, nasal or throat swabs | Attomolar sensitivity | Ackerman et al., 2020 |
| CRISPR-Chip | Label-free electrical detection | Unamplified nucleic acid | Buccal swab | – | https://cardeabio.com/crispr-chip/ |
| CRISPR-powered assays | RT-PCR CRISPR-Cas12a fluorescent reporter assay | Amplified nucleic acid | Nasal swab | LOD: 2 copy /μL | Huang et al., 2020 |
| Dual-Functional Plasmonic Photothermal Biosensors | Biosensor combining the plasmonic photothermal (PPT) effect and localized surface plasmon resonance (LSPR) sensing transduction | Unamplified nucleic acid | – | LOD: 0.22 pM | Qiu et al., 2020 |
| CONVAT | Optical biosensor | Unamplified nucleic acid | Nasal or saliva swabs | – | https://optics.org/news/11/4/13 |
| FET biosensor | Label-free, real-time electrical detection with graphene-based FET functionalized with antibody | Spike protein | Nasopharyngeal swabs | LOD: 100 fg /mL | Seo, G. et al., 2020 |
| Portable, Ultra-Rapid and Ultra-Sensitive Cell-Based Biosensor | Bioeectric recognition assay based on mammalian Vero cells, which were engineered by electroinserting the human chimeric spike S1 antibody | Spike protein | LOD: 1 fg /mL | Mavrikou et al., 2020 | |
| Toroidal plasmonic metasensors | Miniaturized plasmonic immunosensor based on toroidal electrodynamics concept | Spike protein Miniaturized plasmonic immunosensor based on toroidal electrodynamics concept |
LOD: 4.2 fmol | Ahmadivand et al., 2020 | |
| Fluorescence immunochromato-graphic assay | Fluorescence immunochromatographic assay in blind analysis | Nucleocapsid protein | Nasopharyngeal swab, Urine |
Sensitivity: 68%, Specificity: 100%, Accuracy: 72% |
Diao, B. et al., 2020 |
| Electrochemical immunosensor | Magnetic bead-based immunosensor combined with carbon black-modified screen-printed electrode | Spike and Nucleocapsid protein | Untreated saliva | 19 ng/mL and 8 ng/mL in untreated saliva, respectively for S and N protein | This work |
4. Conclusions
The COVID-19 pandemic event declared by World Health Organization on March 2020 has required great effort by the scientific community, including the research activity for the development of rapid assay for SARS-CoV-2. Herein, we developed a smart immunosensor for SARS-CoV-2 detection in saliva by combining the use of MBs as support for immunological chain and carbon black-based SPEs for sensitive and reliable detection. This sensor configuration demonstrated the capability to detect S and N proteins in untreated saliva with a detection limit equal to 19 ng/mL and 8 ng/mL, respectively as well as SARS-CoV-2 in saliva clinical samples and cultured SARS-CoV-2, without any cross-reactivity when tested with seasonal H1N1 influenza virus and 2009 pH1N1 influenza pandemic. The satisfactory analytical features found in terms of sensitivity, accuracy, and selectivity with the time of analysis (30 min), easiness to use, and the requirement of portable instrumentation boost this biosensor to acquire a relevant position in SARS-CoV-2 device scenario, taking into account also the easy sampling of saliva.
CRediT authorship contribution statement
Laura Fabiani: Experimental investigation, Methodology, Data curation, Formal analysis, Writing - review & editing. Marco Saroglia: Conceptualization, Writing - review & editing. Giuseppe Galatà: Conceptualization, Writing - review & editing. Riccardo De Santis: Experimental investigation, Methodology, Data curation, Formal analysis, Writing - review & editing. Silvia Fillo: Experimental investigation, Methodology, Data curation, Formal analysis, Writing - review & editing. Vincenzo Luca: Experimental investigation, Methodology, Formal analysis. Giovanni Faggioni: Data curation, Formal analysis, Writing - review & editing. Nino D'Amore: Data curation, Formal analysis, Writing - review & editing. Elisa Regalbuto: Experimental investigation, Methodology, Data curation, Formal analysis. Piero Salvatori: Experimental investigation, Methodology, Data curation, Formal analysis. Genciana Terova: Data curation, Formal analysis, Writing - review & editing. Danila Moscone: Data curation, Formal analysis, Writing - review & editing. Florigio Lista: Formal analysis, Writing - review & editing. Fabiana Arduini: Conceptualization, Data curation, Formal analysis, Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2020.112686.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Ackerman C.M., Myhrvold C., Thakku S.G., Freije C.A., Metsky H.C., Yang D.K., Ye S., Boehm C.K., Kosoko-Thoroddsen T.-S., Kehe J., Nguyen T., Carter A., Kulesa A., Barnes J., Dugan V., Hung D., Blainey P., Sabeti P. Nature. 2020;582:277–282. doi: 10.1038/s41586-020-2279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadivand A., Gerislioglu B., Ramezani Z., Kaushik A., Manickam P., Ghoreishi S.A. 2020. Preprint arXiv:2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alizargar J., Sh M.E., Aghamohammadi M., Hatefi S. J. Formos. Med. Assoc. 2020;119:1234–1235. doi: 10.1016/j.jfma.2020.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antuña-Jiménez D., González-García M.B., Hernández-Santos D., Fanjul-Bolado P. Biosensors. 2020;10:9. doi: 10.3390/bios10020009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arduini F., Amine A., Majorani C., Di Giorgio F., De Felicis D., Cataldo F., Moscone D., Palleschi G. Electrochem. Commun. 2010;12:346–350. [Google Scholar]
- Arduini F., Cinti S., Mazzaracchio V., Scognamiglio V., Amine A., Moscone D. Biosens. Bioelectron. 2020;156:112033. doi: 10.1016/j.bios.2020.112033. [DOI] [PubMed] [Google Scholar]
- Arduini F., Di Nardo F., Amine A., Micheli L., Palleschi G., Moscone D. Electroanalysis. 2012;24:743–751. [Google Scholar]
- Arduini F., Majorani C., Amine A., Moscone D., Palleschi G. Electrochim. Acta. 2011;56:4209–4215. [Google Scholar]
- Arduini F., Micheli L., Moscone D., Palleschi G., Piermarini S., Ricci F., Volpe G. Trac. Trends Anal. Chem. 2016;79:114–126. [Google Scholar]
- Azzi L., Carcano G., Gianfagna F., Grossi P., Dalla Gasperina D., Genoni A., Fasano M., Sessa F., Tettamanti T., Carinci F., Maurino V., Rossi A., Tagliabue A., Baj A. J. Infect. 2020;81:45–50. doi: 10.1016/j.jinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmik D., Nandi R., Jagadeesan R., Kumar N., Prakash A., Kumar D. Infect. Genet. Evol. 2020;84:104451. doi: 10.1016/j.meegid.2020.104451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caratelli V., Ciampaglia A., Guiducci J., Sancesario G., Moscone D., Arduini F. Biosens. Bioelectron. 2020;165:112411. doi: 10.1016/j.bios.2020.112411. [DOI] [PubMed] [Google Scholar]
- Chan J.F.W., Yip C.C.Y., To K.K.W., Tang T.H.C., Wong S.C.Y., Leung K.H., Fung A.Y.-F., Ng A.C.-K., Zou Z., Tsoi H.-W., Choi G., Tam A.R., Cheng V., Chan K.-H., Tsang O., Yuen K.-Y. J. Clin. Microbiol. 2020;58:e00310–e00320. doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinti S., Arduini F. Biosens. Bioelectron. 2017;89:107–122. doi: 10.1016/j.bios.2016.07.005. [DOI] [PubMed] [Google Scholar]
- Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. Antivir. Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui F., Zhou H.S. Biosens. Bioelectron. 2020;165:112349. doi: 10.1016/j.bios.2020.112349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di B., Hao W., Gao Y., Wang M., Qiu L.W., Wen K., Zhou D., Wu X., Lu E., Liao Z., Mei Y., Zheng B., Che X. Clin. Diagn. Lab. Immunol. 2005;12:135–140. doi: 10.1128/CDLI.12.1.135-140.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao B., Wen K., Chen J., Liu Y., Yuan Z., Han C., Chen J., Pan Y., Chen L., Dan Y., Wang J., Chen Y., Deng G., Zhou H., Wu Y. Preprint medRxiv. 2020. [DOI]
- COMMUNICATION from the COMMISSION Guidelines on COVID-19 in Vitro Diagnostic Tests and Their Performance (2020/C 122 I/01).
- Fabiani L., Delibato E., Volpe G., Piermarini S., De Medici D., Palleschi G. Sensor. Actuator. B. 2019;290:318–325. [Google Scholar]
- Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. In: Maier H., Bickerton E., Britton P., editors. Coronaviruses: Methods and Protocols, Methods in Molecular Biology. Springer Science+Business Media; New York: 2015. pp. 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Tian D., Liu Y., Lin Z., Lyon C., Lai W., Fusco D., Drouin A., Yin X., Hu T., Ning B. Biosens. Bioelectron. 2020;164:112316. doi: 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- J Alsaadi E.A., Jones I.M. Future Virol. 2019;14:275–286. doi: 10.2217/fvl-2018-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khristunova E., Barek J., Kratochvil B., Korotkova E., Dorozhko E., Vyskocil V. Bioelectrochemistry. 2020;135:121136. doi: 10.1016/j.bioelechem.2020.107576. [DOI] [PubMed] [Google Scholar]
- Khristunova Y., Korotkova E., Kratochvil B., Barek J., Dorozhko E., Vyskocil V., Plotnikov E., Voronova O., Sidelnikov V. Sensors. 2019;19:2103. doi: 10.3390/s19092103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Z., Oton J., Qu K., Cortese M., Zila V., McKeane L., Nakane T., Zivanov J., Neufeldt C.J., Cerikan B., Lu J.M., Peukes J., Xiong X., Krausslich H.G., Scheres S.H.W., Bartenschlager R., Briggs J.A.G. 2020. Nature; pp. 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss B., Kis Z., Palyi B., Kellermayer M. bioRxiv. 2020. [DOI]
- Lassaunière R., Frische A., Harboe Z.B., Nielsen A.C., Fomsgaard A., Krogfelt K.A., Jørgensen C.S. Medrxiv. 2020. [DOI]
- Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W., Wang Q., Tan L., Wu W., Tang S., Xiong Z., Zheng S. J. Clin. Microbiol. 2020;58:6. doi: 10.1128/JCM.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahapatra S., Chandra P. Biosens. Bioelectron. 2020;165:112361. doi: 10.1016/j.bios.2020.112361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrikou S., Moschopoulou G., Tsekouras V., Kintzios S. Sensors. 2020;20:3121. doi: 10.3390/s20113121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Narváez E., Dincer C. Biosens. Bioelectron. 2020;163:112274. doi: 10.1016/j.bios.2020.112274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrero M., Campuzano S., Pingarrón J.M. Electroanalysis. 2012;24:470–482. doi: 10.1002/elan.201100528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. ACS Nano. 2020;14:5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- Reth M. Nat. Immunol. 2013;14:765–767. doi: 10.1038/ni.2621. [DOI] [PubMed] [Google Scholar]
- Seo G., Lee G., Kim M.J., Baek S.H., Choi M., Ku K.B., Lee C.S., Jun S., Park D., Kim H.G., Kim S.J., Lee J.O., Kim B.T., Park E.C., Kim S. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- Talarico D., Arduini F., Constantino A., Del Carlo M., Compagnone D., Moscone D., Palleschi G. Electrochem. Commun. 2015;60:78–82. [Google Scholar]
- To K.K., Tsang O.T., Chik-Yan Yip C., Chan K.H., Wu T.C., Chan J., Leung W.S., Chik T.S., Choi C.Y., Kandamby D.H., Lung D.C., Tam A.R., Poon R.W., Fung A.Y., Hung I.F., Cheng V.C., Chan J.F., Yuen K.Y. Clin. Infect. Dis. 2020 2020; ciaa149. [Google Scholar]
- Verdecchia P., Cavallini C., Spanevello A., Angeli F. Eur. J. Intern. Med. 2020;76:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. J. Am. Med. Assoc. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S., Lindsley A., Courter J., Assa’ad A. J. Allergy Clin. Immunol. 2020;146:23–34. doi: 10.1016/j.jaci.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P., Geng B., Muenker C., Moore A.J., Volges C.B.F., Petrone M.E., Ott I., Lu P., Lu-Culligan A., Klein J., Venkataraman A., Earnes R., Simoov M., Datta R., Handoko R., Naushad N., Sewanan L.R., Valdez J., White E.B., Lapidus S., Kalinich C., Jiang X., Kin D., Kudo E., Linehan M., Mao T., Moriyama M., Oh E.J., Park A., Silva J., Song E., Takahashi T., Taura M., Weizman O.-E., Wong P., Yang Y., Bermejo S., Odio C., Omer S., Dela Cruz C., Farhadian S., Martinello R.A., Iwasaki A., Grubaugh N., Ko A.I. N. Engl. J. Med. 2020 doi: 10.1056/NEJMc2016359. (in press) [DOI] [Google Scholar]
- Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S., Qin C., Sun F., Shi Z., Zhu Y., Jiang S., Lu L. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- https://ec.europa.eu/info/live-work-travel-eu/health/coronavirus-response/public-health_en
- https://cardeabio.com/crispr-chip/
- https://optics.org/news/11/4/13
- https://www.intermed.be/en/professional-products/laboratory-diagnostics/serology-amp-virology/rapid-test/coris-covid-19-ag-respi-strip.html
- https://www.cellexcovid.com
- https://chembio.com/dpp-covid-19-igm-igg-system-ous-2/
- https://www.who.int/publications/i/item/laboratory-biosafety-guidance-related-to-coronavirus-disease-(covid-19https://www.who.int/publications/i/item/laboratory-biosafety-guidance-related-to-coronavirus-disease-(covid-19))-
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.