Abstract
Recent research on the SARS-CoV-2 pandemic has exploded around the furin-cleavable polybasic insert PRRAR↓S, found within the spike protein. The insert and the receptor-binding domain, (RBD), are vital clues in the Sherlock Holmes-like investigation into the origin of the virus and in its zoonotic crossover. Based on comparative analysis of the whole genome and the sequence features of the insert and the RBD domain, the bat and the pangolin have been proposed as very likely intermediary hosts. In this study, using the various databases, in-house developed tools, sequence comparisons, structure-guided docking, and molecular dynamics simulations, we cautiously present a fresh, theoretical perspective on the SARS-CoV-2 virus activation and its intermediary host. They are a) the SARS-CoV-2 has not yet acquired a fully optimal furin binding site or this seemingly less optimal sequence, PRRARS, has been selected for survival; b) in structural models of furin complexed with peptides, PRRAR↓S binds less well and with distinct differences as compared to the all basic RRKRR↓S; c) these differences may be exploited for the design of virus-specific inhibitors; d) the novel polybasic insert of SARS-CoV-2 may be promiscuous enough to be cleaved by multiple enzymes of the human airway epithelium and tissues which may explain its unexpected broad tropism; e) the RBD domain of the feline coronavirus spike protein carries residues that are responsible for high-affinity binding of the SARS-CoV-2 to the ACE 2 receptor; f) en route zoonotic transfer, the virus may have passed through the domestic cat whose very human-like ACE2 receptor and furin may have played some role in optimizing the traits required for zoonotic transfer.
Keywords: Spike protein, Furin, Host, Proteases, SARS-CoV-2, Feline CoV, RBD
Highlights
-
•
Polybasic insert of the SARS-CoV-2 spike protein is rare among several hundred proteins with a motif ‘RRARS’.
-
•
SARS CoV-2 shares furin-like site and RBD interface residues including hotspot sites, with some of the lethal form of Feline coronavirus spike protein and those from the healthy cats.
-
•
Polybasic sequence PRRARS binds less well to furin in structural models.
-
•
SARS-CoV-2 may have passed through the domestic cat during zoonotic transfer.
1. Introduction
Most viruses that infect humans often cause seasonal flu which results in fever, and other mild symptoms that last for a few days after which natural immunity takes over to clear the viruses. Once in a while some of these viruses become highly virulent, contagious, spread fast, and furious with fatal consequences. Occasionally, as seen with the SARS-CoV-2, H1N1, H5N1, and H9N2, they cross zoonotic barriers with infections rising to epidemic or pandemic proportions [[1], [2], [3], [4]]. Mammals such as the bat, camel, civet act as reservoirs for these viruses where some of the major tricks for the zoonotic jump emerge [5]. For the virus to be able to cross the species boundaries, it has to undergo a couple of modifications, one of which is the ability to recognize and bind to the receptors of the human airway and the second is the capacity to hijack the host proteolytic system that can activate the virus [[6], [7], [8]]. To achieve these the virus engages its surface glycoprotein, be it the Hemagglutinin (HA) of the Influenza or the spike protein of the coronaviruses (CoVs). These glycoproteins carry two distinct domains separated by a linker. A precise proteolytic cut is necessary to separate the receptor binding S1 domain from the fusion competent S2 domain. The disengaged S2 domain then undergoes a series of conformational changes resulting in the fusion of the viral and host cell membranes, ultimately delivering viral genetic contents into the host cell cytoplasm. This is a highly conserved mechanism of viral entry [[9], [10], [11]]. The highly specific proteolytic cleavage that disengages the two domains of the spike protein (envelope proteins) is indispensable for zoonotic transfer. Bats are one of the intermediary hosts for the ‘human’ coronaviruses. Many of these CoVs seem to have the capacity to bind to human receptors. Yet when isolated, these viruses even after binding to the receptor, do not enter the human cells grown in culture, unless pre-treated with trypsin. Exogenous addition of trypsin separates the S1 and S2 domains allowing host-viral fusion. Receptor binding accompanied by a proteolytic cut unlocks the major barrier for cross-species transmission of infection [12].
Enhanced virulence and pathogenicity of viruses as compared to the non-pathogenic forms, is attributed to the acquisition of a sequence rich in arginine and lysine residues [10] creating the polybasic insert such as the R-X-R/K-R motif. Such a motif creates a binding and cleavage sites for furin and furin like enzymes which belong to the class of preprotein convertases or PCs. The primary function of the PCs is to activate other cellular proteins by proteolytic processing. Among these, furin is ubiquitous in its presence and is crucial for most cellular functions. Mammalian proteins such as hormones and enzymes, bacterial toxins such as the Anthrax, envelope proteins such as the Herpes virus and Coronavirus S are processed by furin [13]. Altered levels and aberrant activation of furin can result in diseases such as cancer and neurodegenerative disorders. Viruses have evolved by adapting to the proteolytic environment of the host and despite the differences in the sequences and structures of their envelope proteins, diverse types of viruses show a conserved dependency of furin and PC enzymes for activation. It is for this reason that when a virus acquires a sequence preferred by furin and PC enzymes, it is considered to be more pathogenic as it can enter the cells via the non-endocytic route, it can enhance the capacity for cell-cell fusion and expand the tissue tropism. These mechanisms although not fully understood, are well discussed and illustrated in Refs. [7,14,15].
There are two main proteolytic sites in the coronavirus (CoV) spike proteins – the S1/S2 site situated at the S1 and S2 domain boundary, and the S2′ site located close to the S2 domain. The cleavage at S1/S2 site is considered dispensable for coronavirus entry (viral and host membrane fusion) and is not inhibited by furin inhibitors. However, cleavage at the S1/S2 site is required for the virus to exit from virus-producing cells and enter a new target cell (via cell-cell fusion) and is sensitive to inhibition by furin inhibitors [15]. A polybasic insert is not uncommon in spike proteins. However, at the S1/S2 site, in the majority of the spike proteins related to SARS-CoV-2, a conserved monobasic sequence (R↓S) preferred by trypsin-like enzymes is normally seen. Therefore, when multiple Arg residues similar to those seen with the MERS and other infecious SARS viruses were found at the S1/S2 site of the SARS-CoV-2 spike protein [[16], [21]], it led to a series of speculations on the role of furin in zoonotic transfer, activation of the virus, and the widespread pandemic. The first report on the role of the polybasic insert in experimental model systems showed that the membrane-bound receptor-activated type II serine protease, TMPRSS2, cleaves the SARS-CoV-2 and this cleavage is responsible for viral entry. Later, furin-mediated cleavage of the spike protein at the polybasic site was found necessary for the virus to enter the human lung cells in culture [16,17]. Mutations at this site and the addition of furin inhibitors affect cleavage and entry into the human lung cells in vitro [16].
Based on the above reports and analogous studies on other infectious viruses, furin, and furin-like enzyme cleavage seem to be one of the major events that resulted in the zoonotic transfer, the infectivity of the SARS-CoV-2 in pandemic proportions, and lethality. However, equally compelling evidence points to other possibilities. For example, the feline enteric coronavirus (FECV), responsible for a milder and localized form of enteric infection in the infected cat, carried a highly optimized furin cleavage site at S1/S2 [18]. In contrast, in the spike protein of the feline infectious peritonitis virus (FIPV), that caused systemic infection and death of the infected cat, the polybasic insert was either completely lost or found mutated. The catalytic efficiency of furin against these various peptide sequences, was measured and the optimal sequences were found enriched in FECV. The FIPV spike protein carried mutations at all crucial positions in the polybasic region and many of these mutations adversely affected cleavage by furin [18]. It was hypothesized that the ability to be cleaved by enzymes of the macrophages such as the Cathepsins, MMPs, and PSCK1 enabled the virus to expand its tissue tropism.
Even in influenza and HIV viruses with an established role for the furin mediated proteolysis in the fusion events and infectivity [10], contrary observations have been reported. For example, in H5N1 influenza, one of the only two events that led to lethality was the acquisition of tandem furin cleavage sites between the HA1 and HA2 proteins. However, the infectivity of the virus was found to be rapidly attenuated, preventing widespread infection. The reasons for these were not clear. In the case of HIV-1, a less optimal polybasic site within the gp160 was found conserved in several isolates [19]. Upon introduction of a furin optimal sequence in gp160, the infectivity of the engineered virus was found attenuated compared to HIV with the suboptimal cleavage sequence. From the foregoing discussion, it is clear that associating proteolytic cleavage with infectivity, zoonotic transfer, virulence, tissue tropism, and survival advantage is complicated and can be extremely challenging to investigate [20].
The second ‘gain of function’ that seems to have allowed the SARS-CoV-2 to make the zoonotic transfer is the enhanced affinity of the RBD domain to the ACE2 receptor [3,21]. The affnity of the RBD domain of SARS-CoV and SARS-CoV-2 and their full-length spike proteins to the human ACE2 receptor has been compared. The RBD domain of SARS-CoV-2 has a better affinity for the ACE2 receptor than the SARS-CoV RBD, although the fold differences were dependent on the type of experiments performed [22,23]. The affinity of the full-length SARS-CoV-2 spike protein to ACE2 was similar to SARS-CoV spike protein at 37 °C and 38 °C but was lesser than that of the SARS-CoV spike protein at lower or higher temperatures [24]. These observations make it difficult to precisely define the role of binding affinity in the enhanced virulence of SARS-CoV-2. Besides affinity, structural and unique conformationl changes in the SARS-CoV-2 spike protein are implicated in the enhanced virulence of this virus. Meanwhile, there have been studies exploring the conservation of the proteolytic site and the RBD domain/spike protein interaction with the ACE2 receptor to predict the likely intermediate host(s). So far, the bat [25] and the pangolin are considered the most likely players in the adaptation of the virus [[25], [26], [27], [28]]. The rampant and large-scale recombination between different lineages of coronaviruses and mutations/insertions/deletions at the S1/S2 site are considered as the major reasons for ambiguous determinations. In addition to such molecular mechanisms required to define the intermediary host, a more pragmatic view is as follows: for the originating virus to have acquired both the polybasic insert and the high-affinity RBD at the same time, it would have had to encounter a host with high population density [21]. In this context, the domestic cat has been speculated to be one of the intermediary hosts of the SARS-CoV-2 [29].
It is against this background that we set out to understand the novel polybasic insert in the context of what is known about it in naturally occurring furin substrates and its abundance in human and viral sequences. As a surrogate to experimental methods, we used information from databases, structure-guided docking, and Molecular Dynamics Simulations (MD), to compare the binding affinity of the polybasic insert with the fully optimized furin cleavage sequence known in the literature. Since the rapid evolution of a mild feline corona virus to its lethal form was accompanied by critical changes in the polybasic insert, we asked whether the direction of evolution is encrypted in the acquired novel polybasic insert in SARS-CoV-2. As the domestic cat could be a potential intermediary host, it was pertinent to compare the receptor-binding domains of the two spike proteins to identify any conserved hot spot residues. The outcome of this investigation was rather striking and not reported so far. They not only provide a compelling new perspective but are also very important in the ongoing efforts to understand the pathogenicity of the virus and the means to control it.
2. Results
2.1. The polybasic insert of the SARS-CoV-2 spike protein is rare among several hundred proteins with a motif ‘RRAR↓S’
The presence of the novel insert ‘PRRA’ in the spike protein of SARS-CoV-2 just before the RS, a monobasic sequence found conserved at the S1/S2 boundary in many coronaviruses, creates a polybasic sequence PRRAR↓S. This site can be cleaved by furin and furin-like enzymes. We referred to various literature reports on the polybasic inserts in viruses and natural substrates of furin. Proline was rare at any position, and Ala at the P2 position occurs at low frequency in the furin ‘logos’ derived from 1000s of sequences [30]. One would expect that it would be beneficial for the virus if it shared the polybasic site with human proteins that can be cleaved by furin and furin-like enzymes. Therefore, we decided to obtain an unbiased count of the abundance of PRRARS and its relationship to known natural substrates of furin. To achieve this, a comprehensive analysis of polybasic inserts within non-redundant databases was necessary. We scanned the human databases (Taxon 9606) and found that the exact sequence PRRARS was present only in two human proteins: AAB17869.1, Hermansky-Pudlak syndrome protein (HPS1), and AAF79955.1, RhoGEF. There are 17 isoforms of HPS1; incidentally, RhoGEF in the curated UniProt data does not carry this motif (Supplemental Table 1).
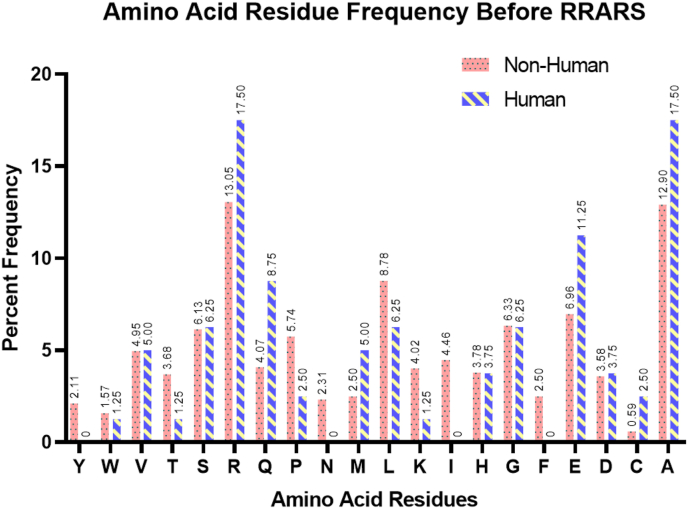
The minimum cleavage motif for furin and furin-like enzymes is a tetrapeptide sequence defined by P4P3P2P1 residues. Therefore, we changed the query sequence to RRARS (which includes the conserved Serine at the P1’) and searched the human-only database, viral non-redundant database, and the non-human/non-viral databases for an exact match (Supplemental Table 1). We then plotted the frequency of residues that would occur in the P5 position (i.e., if the protein were a furin substrate; Fig. 1). Proline occurred at a frequency of ~2.5% in human proteins (2 proteins out of 241 sequences with an exact match to RRARS), 5.74% in all other non-human databases, and at a frequency of 3.79% within the viral database (taxon ID: 10239). We also surveyed the all-viral database ViPR for the presence of RRARS and found only two matches, the M protein of NL63-related Bat coronavirus, and an Alpha coronavirus from a feline coronavirus (Supplemental Table 2). The number of proteins searched and the hits obtained are compiled in Supplemental Table 3.
Fig. 1.
Frequency of distribution of the ‘putative P5 residue’ in proteins that may be substrates of furin and furin-like enzymes as they carry the RRARS motif. Proteins carrying the perfect match for this cleavage sequence RRARS were obtained using the program BLAST.
2.2. SARS CoV-2 furin-like site shares features of the milder and lethal forms of feline CoV spike protein
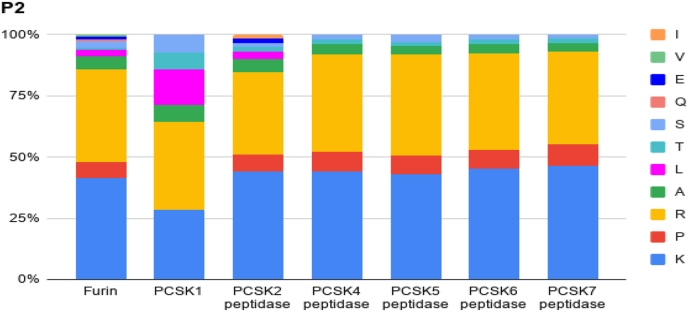
The presence of a furin cleavage site RRARR↓S in the feline spike protein of the less infectious strain FECV and the presence of PRRAR↓S in the SARS-CoV-2 prompted us to analyse the sequences more carefully [18]. RRARR↓S is one of the bonafide furin substrates (changes at P3 are tolerated by furin) with the sequence satisfying all major rules for the position-specific amino acids, although Lys is marginally better than Arg at P2. A detailed catalog of amino acids preferred by furin indicates that Pro is disfavored at the P5 position [30] and yet the novel SARS-CoV-2 polybasic insert (PRRAR↓S) has a Pro at P5 (Pro is also seen at the P5 position of the polybasic sequence in MERS). Furthermore, furin has a stringent requirement for a basic residue (R/K) at the P2 position [19]. This prompted us to look for the frequency of P2 Ala (present in PRRAR↓S) in cleavage sites of furin and furin-like enzymes. As seen from Fig. 2, Ala is found at very low frequency at this position, present at only 0.03–0.04%. Peptides corresponding to the polybasic insert of the spike protein from FECV and the corresponding mutations in FIPV were tested in a furin cleavage assay [18]. P1 Arg in FECV when mutated to a Gly, Met or Thr results in loss of activity. His, Leu or Ser at P2 (FIPV) instead of the Arg in the canonical sequence (FECV) compromised cleavage efficiency; Pro at this position (FIPV) enhanced activity. Ala is not seen at the P2 position in the sequences compared and has not been tested. In all of the peptides (regardless of the isolate), an Arg or Lys was present at the P5 position. At this position in Cov-2 spike protein the polybasic insert carries a Proline residue. Thus the polybasic insert of CoV-2 spike protein is closer to that of the spike protein of the milder feline coronavirus but carries crucial substitutions that are either uncommon or are disfavored in classical Furin substrates. This may compromise binding or cleavage efficiency as seen with the sequences in FIPV.
Fig. 2.
Frequency of amino acid residues found at the P2 position of all the known cleavage sequences of furin and furin-like enzymes. The cleavage sequences of the form P4P3P2P1P1′P2′P3′P4′ archived in the MEROPS database were extracted. Each amino acid present at the P2 position was counted and normalized to the total observed sites. Note that the basic residues R/K contribute to about 80% of the P2 residues.
2.3. PRRARS binds less well to furin as compared to RRKRRS
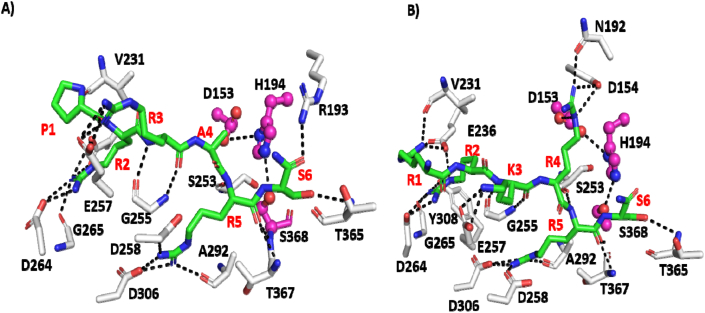
To better understand the interaction of the less abundant sequence PRRARS with furin, a model of the furin-PRRAR↓S was generated and compared with RRKRR↓S, an optimized furin sequence. Both the models were subjected to Molecular Dynamics Simulations. During MD simulations, both the peptides remained stably bound and formed several hydrogen bond interactions with furin. To further understand the differences between the two sequences that may influence binding affinity, binding energy calculations were carried out. The polybasic sequence (RRKRRS) had a considerably improved binding energy, ~ 30 kcal/mol more, as compared to the sequence PRRAR↓S from the SARS-CoV-2 protein. All the six residues in the RRKRR↓S complex are involved in hydrogen bond interactions with furin, whereas in the case of the PRRAR↓S, the proline is not involved in any interactions, and the interactions with alanine are restricted to its backbone atoms (Fig. 3). While the peptide with residues P5 Pro and P2 Ala in PRRARS remain bound during the 100 ns simulation, it is the loss of several crucial hydrogen bonds made by P5R and P2R at the binding pocket that results in the big difference in binding energy. When 'pseudo' viruses carrying the wild-type PRRARS or the fully optimized mutant RRRKRS sequence in their spike proteins were compared for their ability to be cleaved by furin during entry, no major differences were observed. Poor binding substrates can be turned over by enzymes as efficiently or better than the optimal binding sequences. However, when these viruses were compared for their ability to form syncytia (cell-cell fusion), the mutant virus (optimal furin sequence RRRKRS), was found to be far more effective than the virus with the wild-type sequence (PRRARS) [16]. The ability to induce cell-cell fusion is dependent on furin cleavage within the secretory pathway and furin is credited to play a more important role in disease transmission via cell-cell fusion [15,31]. Differences in the observed phenotype are probably due to better binding of the optimized sequence, RRRKRS, to furin and its efficient cleavage as compared to PRRARS, which seems to bind less well to the furin active site (Fig. 3).
Fig. 3.
Snapshots of conformations sampled during the MD simulations of (A) furin-PRRARS (B) furin–RRKRRS complexes. The furin protein is shown as a cartoon (grey) and the bound peptides are shown as a cartoon (green), with the peptide-protein-interacting residues and hydrogen-bond interactions highlighted in sticks and dashed lines, respectively. The three catalytic residues (His194, Asp153, and Ser368) are colored pink. P1'S is labelled as S6 (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.4. Can other proteases cleave the polybasic sequence PRRARS?
The presence of what seems to be a less optimal furin cleavage site in SARS-CoV-2 prompted us to ask if the sequence could be cleaved by other enzymes (besides the already reported TMPRSS2). We used an in-house computational method called the PNSAS, Prediction of Natural Substrates from Artificial Substrates, developed to predict proteolytic sites within the human proteome [32]. The simple algorithm takes tripeptides of the type P1↓P2P3 or tetrapeptides of the kind P1’↓P1P2P3 which includes the scissile bond and scans for the presence of the motif within the intended substrate. If a match is found, the algorithm looks for a crystal structure of the protein in the PDB and if coordinates are found for at least 75% of the protein sequence (75% sequence coverage), the surface accessibility is calculated. Subsequently, the accessibility is compared with a completely disordered region and relative accessibility is calculated. Yet another filter is applied for subcellular co-localization upon which the enzyme-substrate relationship is predicted. Prediction from this algorithm was tested on an enzyme and its novel substrate and was authenticated in a breast cancer cell line [33]. The prediction from this algorithm was also validated by two other independent studies [34,35]. We used the same principles here and integrated different databases for accessibility calculations.
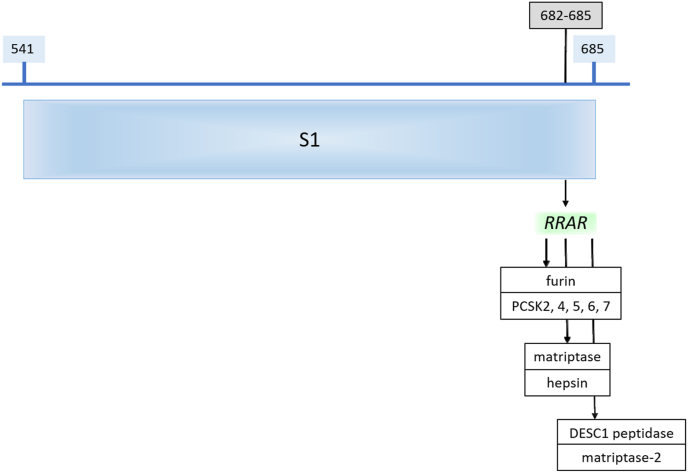
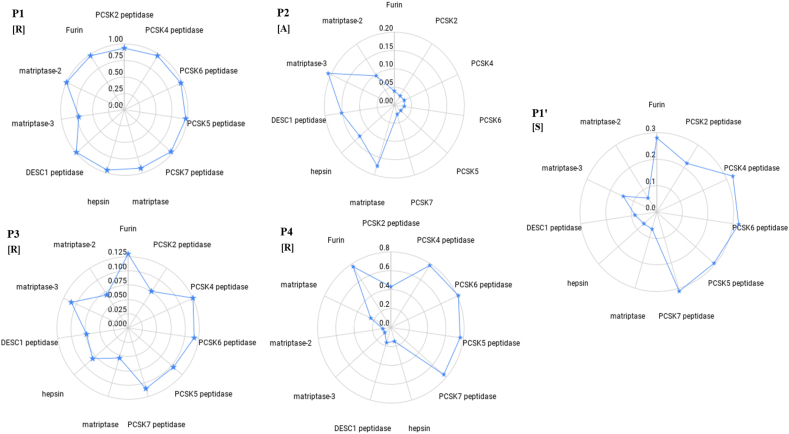
We scanned the dictionary of the cleavage sites of serine proteases for a match with RRAR↓S and RRAR. With the restricted and stringent pentapeptide motif, not surprisingly, we precisely identified furin and the preprotein convertases PCs 2,4,5,6 and 7 as the only possible enzymes that would cut the polybasic sequence RRAR↓S (Fig. 4). However, when the tetrapeptide RRAR motif was used to increase the possible number of matches besides these enzymes, human airway proteases, matriptase 1,2, and 3, hepsin, and Desc1 aligned with the RRAR (Fig. 4).
Fig. 4.
PCs (PSCK) and trypsin like enzymes that can cleave the S1/S2 boundary and activate the virus. A more flexible tetra peptide query set derived from cleavage sites of Serine proteases deposited in MEROPS was used to scan the SARS-CoV-2 spike protein. The logic was based on our program PNSAS created to identify proteolytic sites on natural substrates. The sequence matched sites, accessible on the surface of the S1 domain are indicated.
To obtain an idea of the suitability of the RRARS to bind to the active site of these enzymes, we calculated the number of times P4R, P3R, P2A, P1R, and P1′S occurred in the cleavage sites (MEROPS) of the three Trypsin like enzymes (Fig. 5). As expected, these enzymes have a very strong preference for P1 Arg. Interestingly, they have a greater preference for P2 Ala than do the furin-like enzymes. However, they accommodate P1′ Ser and P4 Arg at a lower frequency compared to the furin-like enzymes. Assembling these various aspects, we hypothesize that the SARS-CoV-2 polybasic insert has retained some of the classic and primary requirements for cleavage by furin and the furin-like enzymes i.e., P4 R, P1 R, and P1′S residues. This makes Site 1 a better furin substrate as compared to all other coronavirus spike proteins that cause human infections. However, the P2 Ala and P5 Pro may compromise the efficiency of the furin and furin-like enzymes unless compensated by other mechanisms. The sequence can also be cleaved by other trypsin-like enzymes mentioned above, albeit less efficiently. It is important to note that these enzymes are present in the human airway epithelium, the entry point of the virus.
Fig. 5.
Frequency plot of P4R, P3R, P2A, P1R, and P1′S within the known cleavage sites of the trypsin-like enzymes. The cleavage sequences of the form P4P3P2P1P1′ archived in the MEROPS database for the trypsin-like enzymes (X-axis) were extracted. Each amino acid present at these positions were counted. The number of times the individual amino acids of the polybasic insert RRARS occur at the respective position were normalized to the total observed sites.
2.5. RBD of feline CoV spike protein with an intact/mutated but not deleted polybasic site, shows conservation of interface residues
If the two traits, receptor binding and the acquisition of the furin site, were to have evolved together, then the intermediate host should be highly populous [21]. Because of our foregoing analysis on the furin site, we asked if the RBD domain of the feline CoV spike protein has features that can help in binding to the human ACE2 receptor? The feline ACE2 receptor is 85.2% identical to the human ACE2 receptor (Supplemental File 2). While the type I FCoV spike proteins were expected to bind to the aminopeptidase receptor, experiments designed to test this in cell culture model systems indicated otherwise [36]. The RBD of the feline spike protein has not been analysed for its ability to bind to human ACE2. We compared the two spike protein sequences and find that despite the low sequence homology and distant evolutionary relationship to the novel SARS-CoV-2 spike protein (Fig. 6 and Supplemental File 3), the RBD domain of the feline coronavirus carries at least six residues that are identical to those at the interface of SARS-CoV-2 RBD-ACE2 receptor complex (Fig. 7). Notable among them are K417 (not found in SARS-CoV) which forms a salt bridge with the Asp 30. The feline ACE2 has glutamate at the corresponding position. Tyr 505, which forms an extensive hydrogen bond network with Lys 343, is conserved in the feline ACE2 receptor (Fig. 7). Not surprisingly, the amino acids of the human ACE2 involved in the interaction with SARS-CoV-2 RBD are conserved in the feline ACE2 receptor (Supplemental File 2 and Fig. 7). These observations render strong support for the domestic cat as one of the possible intermediary hosts that may have nurtured the SARS-CoV-2 in its zoonotic jump. In this context, it is pertinent that the domestic cat is one among the animals that was found to support the replication of SARS-CoV-2 [29].
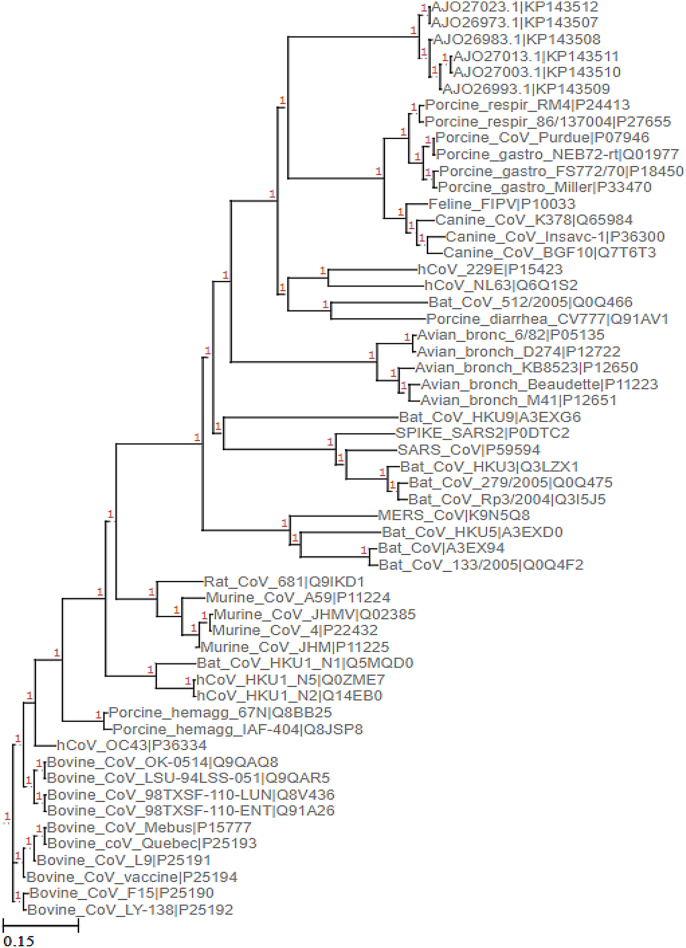
Fig. 6.
Phylogenetic map of coronaviruses based on the sequence homology at the S2 domain of the Spike protein. Curated UniProt sequences of Coronavirus were clubbed with the Feline spike protein sequences reported in Ref. [9] and the phylogenetic tree was constructed according to Multiple Sequence Alignment (please refer to methods for details).
Fig. 7.
Interface of the SARS-CoV-2 RBD and the ACE2 receptor complex compared with the sequence of the feline ACE2 and feline CoV ‘RBD domain’. The crystal structure (PDB ID: 6M0J) was used to obtain the interaction map in the 2-dimensional format using PDBsum. Using the pairwise sequence alignment (in Supplementary File 2 – for ACE2 and Supplementary File 3 – for Spike proteins), we juxtaposed the corresponding residues from feline ACE2 and feline CoV CoV RBD domain. Tyr 505, Lys417 of the RBD domain are absolutely conserved and Asn 501 is replaced by Gln. The strain in which this residue has undergone mutation to Tyr has been identified. The charged residues Asp30, Lys31, Lys353, Asp355 of ACE2 are absolutely conserved.
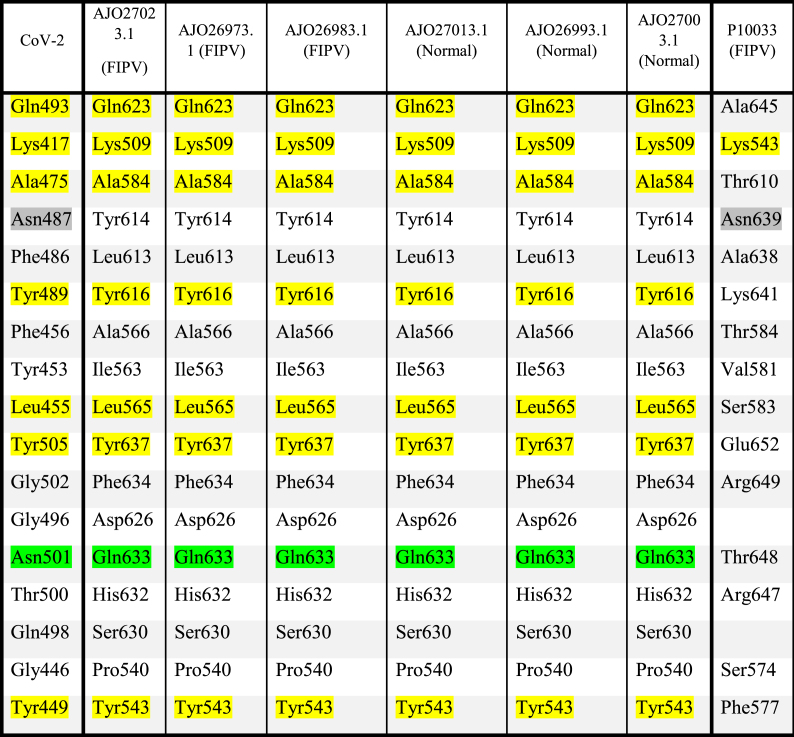
In the phylogenetic tree, the spike proteins of the feline coronavirus with sequences corresponding to the AJO series formed a separate and distant clade (Fig. 6). Among the six isolates, three were from cats infected with FIPV and the other three were from the healthy ones. The furin cleavage site was mutated in all of the three FIPV isolates (as reported in the paper). In the same comparison, a FIPV spike protein (UniProt ID P10033) with a completely deleted polybasic insert, clustered closer to the SARS-CoV-2. The RBD sequence of the feline spike protein (AJO270131.1) used for comparison with the RBD of SARS-CoV-2 (Fig. 7) was derived from the coronavirus isolated from a healthy cat with an intact furin cleavage site. The differences between the polybasic site in the the spike protein of FIPV P10033 and the sequences of the six isolates (Fig. 8), prompted us to compare their RBD regions. The sequences are identical in all of the six isolates (not shown) and they share the same interface residues (Table 1). However, in the spike protein P10033, only two amino acids of the RBD were conserved, one common with the six isolates and SARS-CoV-2 RBD and the other with the SARS-CoV-2 RBD only. That is, in the sequence where the polybasic insert is lost, the RBD domain does not carry all of the conserved residues at the interface! We do not fully understand the implications of these results, but these observations seem to emphasize the difficulties seen with sequence comparisons when large scale deletions and mutations accumulate. In any event, the changes in the polybasic insert in the spike protein [18] and the changes in the RBD (as noted here) during the evolution of the feline coronavirus from the benign to the aggressive form within the infected cats is quite remarkable, to say the least.
Fig. 8.
The polybasic insert in different isolates of feline corona viruses. The AJO27013.1, AJO26993.1, AJO27003.1 are from spike proteins of SARS isolated from healthy cats. AJO27023.1, AJO26973.1, and AJO26983.1 are from infected cats and belong to the FIPV isolates associated with systemic infection and lethality. RRARR↓S at the S1/S2 boundary found in isolates from healthy cats is one of the well-optimized furin cleavage sequences. This is mutated to a sequence that cannot be cleaved by Furin or has been lost in the FIPV strains.
Table 1.
Comparison of residues in the RBD region of the feline CoV spike protein with the interface residues of the SARS-CoV-2-ACE2 receptor complex. Individual pairwise alignment of each of the six isolates with SARS-CoV-2 spike protein was performed using the Needleman-Wunsch algorithm. All six isolates have the same RBD residues. Residues that form the interface of SARS-CoV-2 RBD with the ACE2 receptor as reported in PDB 6MOJ (Fig. 6), were compared with all six isolates and the FIPV isolate reported in Uniprot. The conserved residues are highlighted in yellow., the synonymous mutation Asn to Gln in green and those conserved only with P10033 FIPV in grey.
3. Discussion
Recently, the authors of a seminal ‘Letters to Nature’ put forth several possibilities to explain the origin of the novel SARS-CoV-2 [21]. One among them is the possible identification of a SARS-CoV-2 like-animal virus with a partial or fully optimized polybasic site. This prompted us to examine the novel insert PRRAR↓S in the spike protein of SARS-CoV-2 from an informatics and structural perspective using data bases and some of our in-house tools. This exact sequence is one of the rare sequences in the databases and even rarer in human proteins (with only two matches). Our analysis also shows that the polybasic sequence in SARS-CoV-2 spike protein deviates from the well known optimized sequences for furin recognition and cleavage. Ala at position P2 and Pro at position P5 are not seen among common furin substrates. We modelled the structures of the complexes of these peptides docked into the active sites of furin and find that these peptide residues do not engage optimally with the active site, and the binding energy of the SARS-CoV-2 polybasic peptide sequence is far less than that of the fully optimized polybasic peptide. Knowledge of the other substrates of furin may enable generating inhibitors of furin that outcompete the SARS-CoV-2 spike protein binding and yet are not strong enough to outcompete the natural substrates. However, it is possible that residues beyond P1'S or P5, other allosteric sites or the secondary exosite recognition by furin-like enzymes may compensate for the rather lower binding energy (as predicted here) at the active site. If this is the case, then detailed comparative structure-activity relationship studies are required both using the peptide sequences and the full-length proteins. Such investigations may help in finding discriminative sites on the SARS-CoV-2 spike protein that can be used to develop inhibitors that may prevent infection or disease transmission.
In the experiments designed to test the role of furin in viral entry and cell-cell fusion [16] some interesting observations were made: a) the spike protein with the optimized furin site was found less infectious for reasons unknown; b) at the same time the virus with the optimized furin sequence RRRKRS caused large syncytia formation resulting in enhanced cell-cell fusion as compared to the virus with the novel insert PRRARS in its spike protein. The second observation suggests that during biogenesis, in the virus producing cells, the spike protein with the classical polybasic sequence, RRRKR↓S, is probably cleaved better by the endogenous furin along the secretory pathway than the spike protein with PRRAR↓S. Based on our analysis PRRAR↓S is likely to bind to furin less well. The authors make a note of this differences between the two viruses and suggest that such optimization of the furin sequence may result in enhanced pathogenicity, which agrees well with the analysis presented here. As reported in other viruses, is is also possible that such optimisation may result in a virus with attenuated infectivity. In vivo the events and corresponding consequences may be different. For example, viral or bacterial infections can induce transcription factors such as hypoxia-inducible factor 1 (HIF-1), which enhances the expression of furin [38]. By virtue of more enzyme molecules, a suboptimal basic insert such as the PRRAR↓S eventually could be cleaved; thus, greater number of protease molecules could compensate for the poor binding.
In addition to optimal active site geometry and extensive interactions with the substrate, other factors such as post-translational modifications may influence proteolytic cleavage. Three O-linked glycosylations are potentially possible in and around the S1 site and these seem to have been ‘inherited’ along with the new insert [21]. The pattern of glycosylation and their branching is dictated by enzymes in the secretory pathway which have very different distributions depending on the cell type. Glycosylation can render the site inaccessible to a protease (or an antibody). Therefore, when and where the cleavage site gets exposed or whether additional deglycosylating enzymes need to be hijacked by the virus are some of the relevant questions that remain to be answered regarding the proteolytic activation of the novel SARS-CoV-2. If these sites were to mutate, they would escape surveillance by the glycosylation machinery and antibody, and can then be actively cleaved by the protease. Such a mechanism has been hypothesized for an influenza virus with an Asn to Asp mutation at the polybasic site [38].
In addition to furin, our in-house algorithm identified some of the human airway trypsin-like epithelial proteases, DESC1, hepsin, and the matriptase 1,2,3, as the possible candidates that can cleave the polybasic site. As seen from their sequence preference at P1' position (MEROPS), it is possible that mutations such as the P1'Ser to P1'Lys may render the site more favorable to these enzymes. None of these enzymes have been associated with SARS infection so far. However, all these enzymes are known to cleave HA of the influenza virus and this cleavage is associated with infection and virulence [39]. Matriptase, in particular, supports multiple rounds of replication of H9N2 in the human airway epithelium [40]. Notably, the expression and secretion of matriptase in multiple tissues make it a dangerous ally in breach of organ tropism, virus spread, and pathogenicity of the H9N2. Matriptase bound to the membrane and within the endosomes is known to activate the Influenza A virion (11, 73). Thus, matriptase, by virtue of such subcellular distribution, could potentially be responsible for the novel SARS-CoV-2 viral-host membrane fusion during entry and cell-cell fusion during egress.
What may be the relevance of the observations in the context of human infection? Even though furin and furin-like enzymes are mandatory for normal functions, they have been proposed as potential targets in respiratory viral infections. The proposed strategy is to use the inhibitors transiently and pack them in vehicles that can be applied in the nasal area [41]. Our investigations based on detailed sequence comparison, uniqueness of the insert, and the identification of residues that deter binding of the novel SARS polybasic insert at the furin active site, suggest that it may be possible to find discriminators against the SARS spike protein vis-a-vis the human proteins that are processed by furin (and furin-like enzymes). If these differences hold true in quantitative experimental studies designed to compare the novel insert PRRARS with the optimized sequence such as the RRKRRS, both in peptides and in the full-length proteins, then the results presented here provide a small window of opportunity to exploitthe differences in binding affinity of the host proteins versus the SARS-CoV-2 spike protein. A more alarming and dismal possibility is that the novel SARS-CoV-2 is en route to evolving into an even more virulent strain by acquiring a fully optimized furin site or sites for other enzymes. Or as reported with other viruses, such a fully optimized furin site may have attenuated infectivity and the strain with a less than optimal binding sequence but with a distinct survival advantage may have remained. Its lethality in certain geographical conditions may be due to the host factors such as the immune response and the proteolytic environment. It seems that a detailed characterization of the proteases that can cleave the spike protein and their efficiency correlated with protease expression/activity in patients is needed to clarify these possibilities. Undoubtedly these are not easy experiments to perform but to fully understand the evolution of the current SARS-Cov2 and any SARS of the future, such studies seem inevitable. In the absence of such experimental evidence, we acknowledge that our analysis remains predominantly speculative in nature.
In addition to the proteolytic cleavage, the most needed adaptation for the virus to cross species barriers, is to bind to the host receptors with high affinity. The conservation of hot spot residues and other interacting residues at the RBD domain of the feline coronavirus and the SARS-CoV-2 spike proteins provides one of the first supporting evidence for the possibility that the dual virulence factors namely the furin cleavage site and the RBD domain could have matured in the same intermediary host. The domestic cat, with its populous presence at high density, with its very human like ACE2 and furin sequences, may have nurtured the SARS-CoV-2 during its zoonotic adaptations have nurt.
In summary, our investigations suggest that either the SARS-CoV-2 is in transit on its evolutionary path to gain a fully optimized furin site or it may have promiscuously adapted to the host proteolytic environment for its own better survival. With the selection of residues with lowered binding potential at the P5 and P2 positions, the binding mode seems to have been carefully adapted, posing both a challenge and opportunity for inhibitor design. Our analysis also shows that the polybasic site and the RBD domain of SARS-CoV-2 spike protein, seem to have evolved along the lines of feline coronavirus spike protein providing substantial evidence for the domestic cat as a possible intermediary host during the zoonotic transfer of the SARS-CoV-2 .
4. Methods
4.1. BLAST search to identify proteins carrying the CoV2 polybasic insert
A protein BLAST search for the RRARS motif within the non-redundant database was conducted. This search excluded Homo Sapiens (taxon id: 9606). The maximum allowed number of outputs was 20,000 and we retrieved 15768 hits with 100% identity and 100% coverage for the non-human sequences. From the BLAST results, file accession numbers of all available sequences were extracted and saved into a text file. Then, using batch Entrez, the corresponding sequences for the accession numbers were fetched and downloaded in the FASTA format. In an independent BLAST for the human proteins, 241 sequences with 100% identity and 100% coverage were retrieved through the same pipeline. To increase the coverage and overcome the limitation set by BLAST for searchable sequences (i.e. 20,000 for every search), an independent search was conducted for the virus taxon (taxon ID: 10239). The search fetched 19,998 sequences with a match to RRARS. We also searched the ‘Virus Pathogen’ database [42], which is a depository of sequences exclusive to viruses.
4.2. Filtering of sequences and estimation of amino acid frequency
All FASTA sequences collected by the above method were then run through a python script to cross-check for the RRARS motif, and to identify the residue immediately before the RRARS motif (Putative P5 residue for furin-like enzymes). We further filtered the sequences to remove unknown, unnamed, hypothetical, predicted, and low-quality proteins. To prevent excessive repeats of the motif and skewing of the frequency of the residue at the P5 position, the script included a code to filter the isoforms of the same protein, and only one isoform of each protein was retained. The resulting Excel file was sorted to classify proteins under different organisms of origin (Supplemental Table 1). A frequency distribution graph was created to better visualize the number of times certain residues occurred at the putative P5 position (Fig. 1). We found that residues Arg and Ala are the most frequent. One could consider those proteins with the Arg at ‘P5’ as most likely substrates of furin and furin-like enzymes. The sequences retrieved from the virus taxon were counted for the P5 residues (Supplemental Table 1). An independent search was carried out on the ViPr pathogen database [42] (Supplemental Table 2).
4.3. Construction of the phylogenetic tree
First, the SARS-CoV-2 sequence of spike glycoprotein was queried from NCBI (ID: QHD43416). From the conserved domains via Pfam, the sequence was found to belong to a ‘Corona_S2’ superfamily (PF01601). Corona S2 family is a part of the fusion glycoprotein clan (CL0595) and interacts with the Fusion glycoprotein F0 family (PF00523). InterPro was searched for information on the Corona S2 glycoprotein family (IPR002552). By analysing the collection of proteins related to the family, 5188 (reviewed + unreviewed) Spike proteins were retrieved from UniProt using InterPro. Out of these 5188 proteins, 50 proteins were found to have undergone manual curation (SwissProt) and were included with the SARS-CoV-2 spike sequence. Six feline coronaviruses (FCoV) isolates reported in Ref. [37] were fetched and added to the 50 curated spike proteins from Uniprot. The phylogenetic tree was then built via Multiple Sequence Alignment (MSA) using the Clustal Omega service [43]. The phylogenetic tree obtained from the MSA in Newick format was visualized using the Phylogenetic tree (Newick) viewer (Tool: Tree viewer - Online visualization of phylogenetic trees (Newick) and alignments) [44] and is shown in (Fig. 6).
4.4. Sequence comparison of ACE2 receptor and the RBD domains between FIPV and SARS-CoV-2
The feline and human furin FASTA sequences were fetched from UniProt; (UniProt IDs: M3W594, P09958). Pairwise sequence alignment was performed using the Smith-Waterman algorithm, implemented in the ‘EMBOSS Water’ tool [43]. The result of the sequence comparison among the furin sequences revealed that both the sequences share 96.5% identity, 97.2% similarity, with one gap in the alignment (Supplemental File 1).
The respective FASTA sequences of the feline and human ACE2 receptors, were taken from UniProt; UniProt ID: Q56H28 [ACE2_FELCA], Q9BYF1 [ACE2_HUMAN]. Pairwise sequence alignment was performed using the Smith-Waterman algorithm, implemented in the ‘EMBOSS Water’ tool [43]. The result of the sequence comparison among the ACE2 receptors revealed that both the sequences share 85.2% identity, 92.3% similarity, with no gaps in the alignment (Supplemental File 2).
The FASTA sequences for the respective Spike proteins (containing the RBD domains), of FIPV and SARS-CoV2, were taken from UniProt; UniProt ID: P10033 [SPIKE_FIPV], P0DTC2 [SPIKE_SARS2]. Pairwise sequence alignment was performed using the Smith-Waterman algorithm, implemented in the ‘EMBOSS Water’ tool. The result of the sequence comparison among the Spike proteins revealed that both the sequences share 26.1% identity, 40.5% similarity, with 26.3% gaps in the alignment (Supplemental File 3).
The known interactions between human ACE2 and SARS-CoV-2 RBD were taken from PDBsum (PDB ID: 6M0J) and corresponding pairwise aligned residues, from feline ACE2 and FIPV RBD sequences respectively were mapped onto the diagram (Fig. 6).
The X-ray structure of the furin-peptide co-crystal complex (PDB: 1P8J) was used to generate the model of the furin – PRRARS, furin-RRKRRS complex. Molecular Dynamics simulations were carried out for both the complexes with ff14SB [45] force field using the pmemd. CUDA module from AMBER18 [46]. Hydrogen atoms were added; the N-terminus and the C-terminus of the peptides were capped with the residues ACE and NH2. All the simulation systems were neutralized with appropriate numbers of counter ions. The neutralized system was solvated in an octahedral box with TIP3P [47] water molecules, leaving at least 10 Å between the solute atoms and the borders of the box. MD simulations were carried out for 100ns in triplicates using standard protocols published earlier [48]. Binding energy calculations were carried out over the last 50ns of trajectories using protocols published earlier [48]. Simulation trajectories were visualized using VMD [49] and figures were generated using Pymol [50].
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. SK and CSV are founder directors of Sinopsee Therapeutics and Aplomex; neither company has any conflict with current work.
Acknowledgement
We thank Mahalakshmi Harish for careful reading of the manuscript and input. We thank all authors who would have contributed to the field but could not be cited here. We thank Venkatraman G Mangasuli for initiating PV into this project. SK and CSV would like to thank Agency for Science Technology and Research (A*STAR), Singapore and National Super Computing Center (NSCC), Singapore for the support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2021.100907.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Reperant L.A., Kuiken T., Osterhaus A.D. Adaptive pathways of zoonotic influenza viruses: from exposure to establishment in humans. Vaccine. 2012;30(30):4419–4434. doi: 10.1016/j.vaccine.2012.04.049. [DOI] [PubMed] [Google Scholar]
- 2.Mostafa A. Zoonotic potential of influenza A viruses: a comprehensive overview. Viruses. 2018;10(9) doi: 10.3390/v10090497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ye Z.W. Zoonotic origins of human coronaviruses. Int. J. Biol. Sci. 2020;16(10):1686–1697. doi: 10.7150/ijbs.45472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey E.S. A mini review of the zoonotic threat potential of influenza viruses, coronaviruses, adenoviruses, and enteroviruses. Front. Public Health. 2018;6:104. doi: 10.3389/fpubh.2018.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li X. Evolutionary history, potential intermediate animal host, and cross-species analyses of SARS-CoV-2. J. Med. Virol. 2020 doi: 10.1002/jmv.25731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heald-Sargent T., Gallagher T. Ready, set, fuse! the coronavirus spike protein and acquisition of fusion competence. Viruses. 2012;4(4):557–580. doi: 10.3390/v4040557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casasnovas J.M. Virus-receptor interactions and receptor-mediated virus entry into host cells. Subcell. Biochem. 2013;68:441–466. doi: 10.1007/978-94-007-6552-8_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belouzard S. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4(6):1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bottcher-Friebertshauser E., Klenk H.D., Garten W. Activation of influenza viruses by proteases from host cells and bacteria in the human airway epithelium. Pathog. Dis. 2013;69(2):87–100. doi: 10.1111/2049-632X.12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bottcher E. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J. Virol. 2006;80(19):9896–9898. doi: 10.1128/JVI.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menachery V.D. Trypsin treatment unlocks barrier for zoonotic bat coronavirus infection. J. Virol. 2020;94(5) doi: 10.1128/JVI.01774-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braun E., Sauter D. Furin-mediated protein processing in infectious diseases and cancer. Clin. Trans. Immunol. 2019;8(8) doi: 10.1002/cti2.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Y. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41(9):1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park J.E. Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc. Natl. Acad. Sci. U. S. A. 2016;113(43):12262–12267. doi: 10.1073/pnas.1608147113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffmann M., Kleine-Weber H., Pohlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020 doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann M. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280 e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Licitra B.N. Mutation in spike protein cleavage site and pathogenesis of feline coronavirus. Emerg. Infect. Dis. 2013;19(7):1066–1073. doi: 10.3201/eid1907.121094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas G. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell Biol. 2002;3(10):753–766. doi: 10.1038/nrm934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamada Y., Liu D.X. Proteolytic activation of the spike protein at a novel RRRR/S motif is implicated in furin-dependent entry, syncytium formation, and infectivity of coronavirus infectious bronchitis virus in cultured cells. J. Virol. 2009;83(17):8744–8758. doi: 10.1128/JVI.00613-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andersen K.G. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Q. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020 doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walls A.C. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292 e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Z. Temperature Dependence of the SARS-CoV-2 affinity to human ACE2 determines COVID-19 progression and clinical outcome. Comput. Struct. Biotechnol. J. 2020 doi: 10.1016/j.csbj.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou H. A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr. Biol. 2020;30(11):2196–2203 e3. doi: 10.1016/j.cub.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu P. Are pangolins the intermediate host of the 2019 novel coronavirus (SARS-CoV-2)? PLoS Pathog. 2020;16(5) doi: 10.1371/journal.ppat.1008421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wahba L. An extensive meta-metagenomic search identifies SARS-CoV-2-homologous sequences in pangolin lung viromes. mSphere. 2020;5(3) doi: 10.1128/mSphere.00160-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lau S.K.P. Possible bat origin of severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020;26(7) doi: 10.3201/eid2607.200092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi J. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020 doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shiryaev S.A. High-resolution analysis and functional mapping of cleavage sites and substrate proteins of furin in the human proteome. PloS One. 2013;8(1) doi: 10.1371/journal.pone.0054290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simmons G. Different host cell proteases activate the SARS-coronavirus spike-protein for cell-cell and virus-cell fusion. Virology. 2011;413(2):265–274. doi: 10.1016/j.virol.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venkatraman P. A sequence and structure based method to predict putative substrates, functions and regulatory networks of endo proteases. PloS One. 2009;4(5) doi: 10.1371/journal.pone.0005700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wadhawan V. From prediction to experimental validation: desmoglein 2 is a functionally relevant substrate of matriptase in epithelial cells and their reciprocal relationship is important for cell adhesion. Biochem. J. 2012;447(1):61–70. doi: 10.1042/BJ20111432. [DOI] [PubMed] [Google Scholar]
- 34.Dhamne H., Chande A.G., Mukhopadhyaya R. Lentiviral vector platform for improved erythropoietin expression concomitant with shRNA mediated host cell elastase down regulation. Plasmid. 2014;71:1–7. doi: 10.1016/j.plasmid.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 35.Rolas L. NADPH oxidase depletion in neutrophils from patients with cirrhosis and restoration via toll-like receptor 7/8 activation. Gut. 2018;67(8):1505–1516. doi: 10.1136/gutjnl-2016-313443. [DOI] [PubMed] [Google Scholar]
- 36.Dye C., Temperton N., Siddell S.G. Type I feline coronavirus spike glycoprotein fails to recognize aminopeptidase N as a functional receptor on feline cell lines. J. Gen. Virol. 2007;88(Pt 6):1753–1760. doi: 10.1099/vir.0.82666-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis C.S. Genotyping coronaviruses associated with feline infectious peritonitis. J. Gen. Virol. 2015;96(Pt 6):1358–1368. doi: 10.1099/vir.0.000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tse L.V. A novel activation mechanism of avian influenza virus H9N2 by furin. J. Virol. 2014;88(3):1673–1683. doi: 10.1128/JVI.02648-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baron J. Matriptase, HAT, and TMPRSS2 activate the hemagglutinin of H9N2 influenza A viruses. J. Virol. 2013;87(3):1811–1820. doi: 10.1128/JVI.02320-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beaulieu A. Matriptase proteolytically activates influenza virus and promotes multicycle replication in the human airway epithelium. J. Virol. 2013;87(8):4237–4251. doi: 10.1128/JVI.03005-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng Y.W. Furin inhibitors block SARS-CoV-2 spike protein cleavage to suppress virus production and cytopathic effects. Cell Rep. 2020;33(2):108254. doi: 10.1016/j.celrep.2020.108254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pickett B.E. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 2012;40:D593–D598. doi: 10.1093/nar/gkr859. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Madeira F. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47(W1):W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huerta-Cepas J., Serra F., Bork P. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 2016;33(6):1635–1638. doi: 10.1093/molbev/msw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maier J.A. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theor. Comput. 2015;11(8):3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Case D.A.e.a. University of California; San Francisco: 2018. AMBER 18. [Google Scholar]
- 47.Jorgensen W.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79(2):926–935. [Google Scholar]
- 48.Kannan S. Inhibiting S100B(ββ) for activating wild-type p53: design of stapled peptides. ACS Omega. 2019;4(3):5335–5344. [Google Scholar]
- 49.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 50.DeLano W.L. De Lano Scientific; San Carlos CA, USA: 2002. The PyMOL Molecular Graphics System. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.