Abstract
The novel severe acute respiratory syndrome coronavirus (SARS-CoV-2) has emerged to a pandemic and caused global public health crisis. Human angiotensin-converting enzyme 2(ACE2) was identified as the entry receptor for SARS-CoV-2. As a carboxypeptidase, ACE2 cleaves many biological substrates besides angiotensin II to control vasodilatation and vascular permeability. Given the nanomolar high affinity between ACE2 and SARS-CoV-2 spike protein, we investigated how this interaction would affect the enzymatic activity of ACE2. Surprisingly, SARS-CoV-2 trimeric spike protein increased ACE2 proteolytic activity ∼3-10 fold against model peptide substrates, such as caspase-1 substrate and Bradykinin-analog. The enhancement in ACE2 enzymatic function was mediated by the binding of SARS-CoV-2 spike RBD domain. These results highlighted the potential for SARS-CoV-2 infection to enhance ACE2 activity, which may be relevant to the cardiovascular symptoms associated with COVID-19.
Keywords: viral protein, carboxypeptidase, renin angiotensin system, fluorescence resonance energy transfer (FRET), pathogenesis, angiotensin converting enzyme 2, enzymatic activity, SARS-CoV-2 spike protein
The novel coronavirus, SARS-CoV-2 (1, 2, 3), has emerged as an unprecedented global pandemic resulting in over 35 million confirmed cases and more than 1 million deaths as of October 11, 2020 (WHO). The infection of SARS-CoV-2 causes fever, dry cough, severe respiratory illness and pneumonia, a disease recently named COVID-19 (4). Pathological studies have revealed all features of diffuse alveolar damage (DAD) with excessive fluid in the lungs of infected individuals (5). In addition, abnormal blood clots were observed in many hospitalized patients (6). However, the mechanistic understanding of the pathogenicity of SARS-CoV-2 and its complications is still lacking.
ACE2 was identified as the entry receptor for both SARS-CoV-2 2, and SARS-CoV (7, 8, 9). Structural studies revealed that both SARS-CoV-2 and SARS-CoV spike (S) glycoproteins bind ACE2 with higher affinity (10, 11, 12, 13). The overall structure of SARS-CoV-2 S resembles that of SARS-CoV S with the spike RBD domain contacting the extracellular region of ACE2. Physiologically, ACE2 is a zinc metalloprotease (carboxypeptidase), a homolog to dipeptidase angiotensin-converting enzyme (ACE) but with different substrate specificity (14). ACE cleaves the C-terminal of angiotensin I (Ang I) to produce the potent vasopressor octapeptide angiotensin II (Ang II), which is further cleaved at its C terminus by ACE2 to deactivate Ang II and produce Ang 1-7. Together, ACE and ACE2 regulate vasoconstriction and vasodilatation in the rennin-angiotensin system (RAS). In addition, ACE and ACE2 regulate kinin-kallikrein system to control vascular permeability and vasodilatation (15). ACE deactivates Bradykinin (BK) nonapeptide, the ligand for constitutively expressed bradykinin receptor B2. Bradykinin can be further processed by carboxypeptidase N or M to form des-Arg9-bradykinin (desBK), a potent ligand for bradykinin receptor B1(16). Beyond renin-angiotensin and kinin-kallikrein systems, ACE2 also cleaves other biological peptides such as Apelin-13 that activates apelin receptor to cause vasodilatation (17). Despite the importance of ACE2 in RAS, there is limited understanding to the impact of coronavirus infection to the physiological function of ACE2.
ACE2 is predominantly expressed on type II pneumocytes in lung (18). Clinical observations showed that COVID-19 patients often had dyspnea and accumulation of fluid in lung resembling local angioedema (7, 16, 19), suggesting a pathology driven by changes in vascular permeability and vasodilatation during SARS-CoV-2 infection. However, there was no direct assessment of ACE2 enzymatic activity during the coronavirus infection. Here we examined the effect of the binding of SARS-CoV-2 spike protein to the intrinsic enzymatic activity of ACE2 using two fluorogenic substrates, the caspase-1 substrate (Mca-YVADAPK-Dnp) (14), and a bradykinin analog (Mca-RPPGFSAFK-Dnp) (20). To our surprise, SARS-CoV-2 spike enhanced ACE2 proteolytic activity on both caspase-1 substrate and bradykinin-analog, and the enzymatic enhancement was mediated by the spike RBD domain binding. The ability of SARS-CoV-2 spike protein to alter ACE2 enzymatic activity may result in dysregulation of RAS and contribute to the pathogenesis of COVID-19.
Results
SARS-CoV-2 spike protein enhances ACE2 activity
SARS-CoV-2 is highly homologous to SARS-CoV and both use ACE2 as their entry receptor (2, 9). Further structural studies demonstrated that both SARS-CoV-2 and SARS-CoV used their spike protein RBD domain to interact with ACE2 in similar binding modes (11). However, SARS-CoV-2 spike protein exhibited higher binding affinity to ACE2 than that of SARS-CoV. Given the nanomolar affinity between ACE2 and SARS-CoV-2 S spike protein, we wonder if the binding of SARS-CoV-2 spike protein to ACE2 affected its function as a carboxypeptidase regulating both the rennin-angiotensin and kinin-kallikrein systems (15, 21).
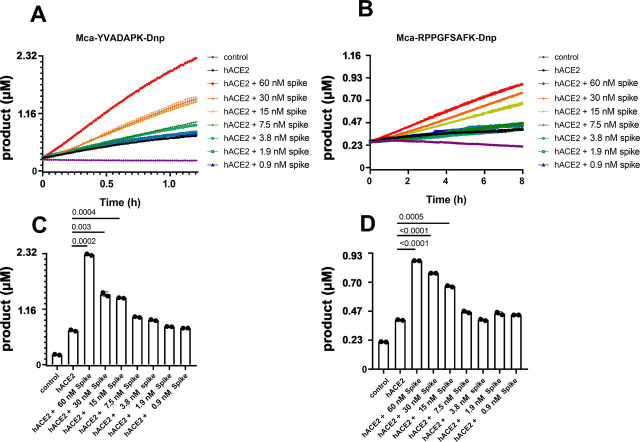
As previously reported, ACE2 efficiently hydrolyzes peptides with Pro-X(1-3 residues)-Pro-hydrophobic sequences between proline and the hydrophobic residue as exemplified by Ang II (DRVYIHP↓F) and des-Arg9-BK (RPPGFSP↓F) (14). ACE2 also cleaves peptides with a basic residue at P1′ position such as Dynorphin A (YGGFLRRIRPKL↓K) and Neurotensin 1-8(pE-LYENKP↓R). We measured ACE2 proteolytic activity using fluorogenic capase-1 substrate Mca-YVADAPK(Dnp) (14), and bradykinin analog Mca-RPPGFSAFK(Dnp)-OH(20). Indeed, ACE2 efficiently hydrolyzed Mca-YVADAPK(Dnp) but cleaved bradykinin analog Mca-RPPGFSAFK(Dnp)-OH less (Fig. 1 A and B). Within 2 h, ACE2 cleaved ∼0.5 μm of Mca-YVADAPK (Dnp), and ∼ 0.05 μm of Mca-RPPGFSAFK(Dnp)-OH. Surprisingly, addition of SARS-CoV-2 spike protein at 14 µg/ml (∼30nM) concentration to the enzymatic assay resulted in ∼3 fold increase in ACE2 cleavage of both the caspase-1 substrate and the BK-analog (Fig. 1 A and B). Consistent with reported NaCl concentration-dependent ACE2 activity (14), SARS-CoV-2 spike protein further enhanced ACE2 cleavage of both peptides at 0.3M and 1M NaCl (Fig. S1), indicating the enhancement was resulted by the interaction between SARS-CoV-2 spike protein and ACE2. In addition, the spike protein-mediated enhancement of ACE2 activity depended on the concentration of the spike. Dilution of SARS-CoV-2 spike protein from 60 nm to 0.9 nm gradually mitigated the enhancement to ACE2 activity with a transition between 7.5 nm and 15 nm of SARS-CoV-2 spike protein (Fig. 1 C and D). This dose dependent enhancement of ACE2 activity was consistent with a 24 nm binding affinity between ACE2 and SARS-CoV-2 spike protein (Fig. S2B). Previous studies showed that SARS-CoV infection may lead to internalization of ACE2 receptor and thus decrease cell surface level of ACE2 (7). However, immunostaining of SARS-CoV-2 infected ACE2 transgenic lung showed cell surface colocalization of the viral spike and ACE2 receptor 3 days post infection (8, 18), supporting the presence of a stable spike-ACE2 complex on the surface of infected cells.
Figure 1.
SARS-CoV-2 spike protein enhanced ACE2 cleavage of fluorogenic caspase-1 substrate and bradykinin analog in a concentration dependent manner.A and B, Kinetic reading of the hydrolysis of Mca-YVADAPK-Dnp(20 μm) and Mca-RPPGFSAFK-Dnp (20 μm) in the presence of SARS-CoV-2 spike protein at indicated final concentrations in the enzymatic assays. The maximum reading is limited to 100000 RFU on Synergy H1 plate reader. RFU was converted to product concentrations with 1 μm equal to 42942 RFU based on the calibration standard. C and D, Comparison of the cleavage of Mca-YVADAPK-Dnp and Mca-RPPGFSAFK-Dnp at 1.5h and 8 h, respectively. Individual p-values for pairwise students' t test were displayed between each pair. The mean and standard deviation of each reaction were shown as columns with each RFU reading repetition displayed as a black dot.
SARS-CoV-2 RBD enhanced ACE2 activity better than SARS-CoV RBD
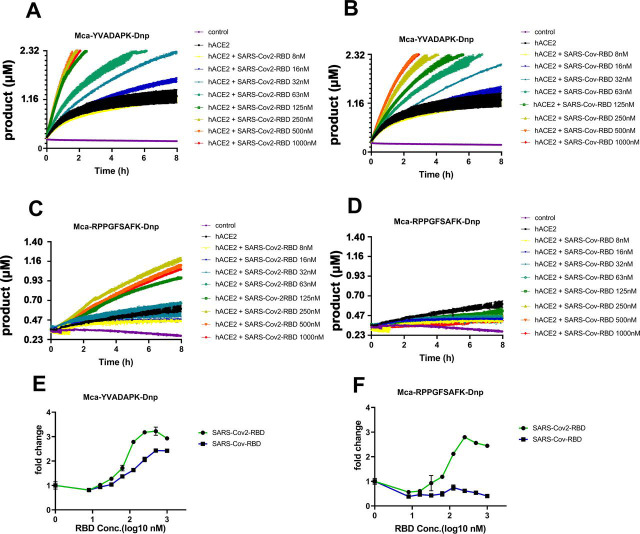
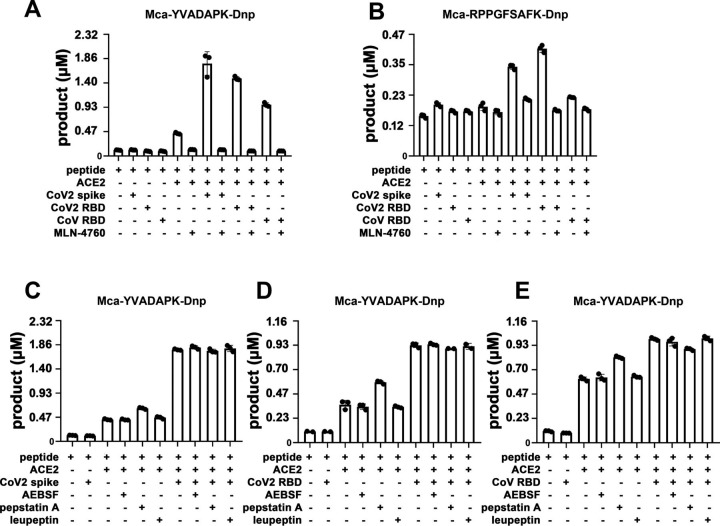
SARS-CoV-2 and SARS-CoV spike protein are highly homologous in their prefusion trimeric architectures and both bind ACE2 with one receptor binding domain (RBD). We then determined whether RBD domain was sufficient to boost ACE2 activity. Indeed, both SARS-CoV-2 RBD and SARS-CoV RBD enhanced ACE2 cleavage of caspase-1 substrate (Fig. 2 A and B), demonstrating that RBD alone was sufficient to enhance ACE2 activity. However, SARS-CoV-2 spike protein exhibited ∼5-10-fold higher ACE2 binding affinity than SARS-CoV (10, 12). The enhancement of ACE2 activity by SARS-CoV-2 RBD showed a concentration dependent saturation with half maximal en-hancement at ∼70nM, whereas the enhancement by SARS-CoV RBD was almost linearly correlated with the RBD concentration with half maximal enhancement at ∼170nM (Fig. 2 E). Interestingly, only SARS-CoV-2 RBD but not SARS-CoV RBD enhanced ACE2 cleavage of the BK-analog (Fig. 2 C, D, and F). SARS-CoV-2 RBD bound to ACE2 at nanomolar affinities (∼30–50 nM), ∼5–10 fold better than that of SARS-CoV RBD (∼180–400 nM) (12). The different capabilities of SARS-CoV-2 and SARS-CoV RBD proteins to enhance ACE2 enzymatic activity suggested that the stronger interaction between RBD and ACE2 is necessary for its cleavage of nonoptimal substrates such as bradykinin analog. Further, the viral spike protein-mediated cleavage enhancement is specific to carboxypeptidase activity of ACE2 rather than potential contaminating protease present in the spike preparation as the enzymatic enhancement was abolished in the presence of a potent ACE2 specific inhibitor MLN-4760 (10 μm) (Fig. 3 A and B). In contrast, none of the category protease inhibitors specific for serine protease (AEBSF), asparagine protease (pepstatin A) and cysteine protease (leupeptin) inhibited ACE2 cleavage at 40 μm concentrations nor its enhancement by the spike RBD (Fig. 3 C–E). Thus, the spike RBD specifically enhanced ACE2 carboxypeptidase activity.
Figure 2.
SARS-CoV-2 RBD but not SARS-CoV RBD enabled ACE2 to cleave bradykinin analog.A and B, hydrolysis of Mca-YVADAPK-Dnp(20 μm) in the presence of SARS-CoV-2 RBD and SARS-CoV RBD proteins over time at indicated concentrations. C and D, hydrolysis of Mca-RPPGFSAFK-Dnp (20 μm) in the presence of SARS-CoV-2 RBD and SARS-CoV RBD proteins at indicated concentrations. Data are shown as relative fluorescence units (RFU) where the maximum reading is 100000 RFU on Synergy H1 plate reader. E, Fold change of RFU during Mca-YVADAPK-Dnp cleavage in the presence of SARS-CoV-2 RBD or SARS-CoV RBD at the time point of 1.5h because of instrument overflow. F, Fold change of RFU during Mca-RPPGFSAFK-Dnp cleavage in the presence of SARS-CoV-2 RBD or SARS-CoV RBD at time point of 8h. All RFU readings at different concentrations of RBD proteins were normalized to that of ACE2 cleavage without RBD proteins correspondingly.
Figure 3.
A and B, ACE2 specific inhibitor MLN-4760 (10 μm) completely blocked ACE2 activity and abolished the enhancement mediated by SARS-CoV-2 spike and RBD protein binding.C–E, various protease inhibitors at 40 μm (AEBSF, pepstatin A, leupeptin) had no effects on ACE2 activity.
Effect of SARS-CoV-2 spike to the kinetic rates of ACE2 cleavage
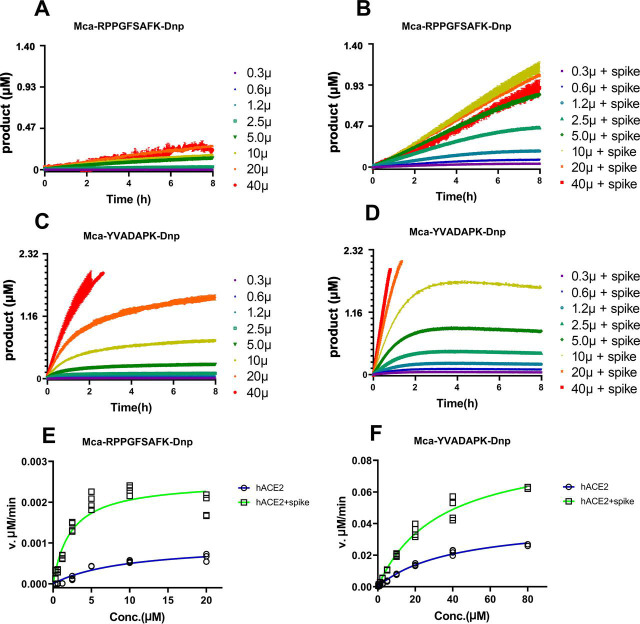
To further characterize how efficiently ACE2 cleaves fluorogenic caspase-1 substrate and bradykinin analog in the presence of SARS-CoV-2 spike protein, we determined the kinetic rate constants of ACE2 cleavage under physiological pH7.5 with 150 mm NaCl. The measurements were carried out with 20 ng ACE2 in the presence or absence of 14 µg/ml (∼30nM) SARS-CoV-2 spike protein, and the hydrolysis was limited to 15% product formed as initial velocity conditions. Michaelis-Menten plots showed that SARS-CoV-2 spike protein resulted in a ∼4-fold reduction in binding constant Km from 10.2 μm to 2.4 μm for ACE2 hydrolysis of the BK-analog (Fig. 4 and Table 1 ). Likewise, SARS-CoV-2 spike protein reduced the Km for ACE2 hydrolysis of Mca-YVADAPK(Dnp) from 46.6 μm to 28.2 μm. This results in a ∼3- and 10-fold increase to k cat/Km for ACE2 hydrolysis of the caspase-1 substrate and BK-analog, respectively. It is worth mention that k cat/Km value for ACE2 hydrolysis of bradykinin analog Mca-RPPGFSAFK(Dnp)-OH in the presence of SARS-CoV-2 spike protein was 7.55 × 103 M−1s−1, similar to that of ACE2 hydrolysis of Angiotensin I decapeptide (DRVYIHPFH↓L), a biological substrate for ACE214. Taken together, SARS-CoV-2 increased the substrate binding and catalytic rate of ACE2.
Figure 4.
Measurement of kinetic constants for hydrolysis of Mca-RPPGFSAFK-Dnp and Mca-YVADAPK-Dnp in the absence or presence of SARS-CoV-2 spike protein (14 µg/ml).A and B, ACE2 hydrolysis of Mca-RPPGFSAFK-Dnp at different concentrations in the absence (A) and presence (B) of SARS-CoV-2 spike protein. C and D, ACE2 hydrolysis of Mca-YVADAPK-Dnp at different concentrations in the absence (C) and presence (D) of SARS-CoV-2 spike protein. Background RFU readings were subtracted. E and F, Michaelis plots for ACE2 hydrolysis of Mca-RPPGFSAFK-Dnp and Mca-YVADAPK-Dnp in the absence (blue) or presence (green)of SARS-CoV-2 spike protein. The initial velocity conditions were limited to 30 min for Mca-YVADAPK-Dnp and 60 min for Mca-RPPGFSAFK-Dnp because of different cleavage rate. All determinations were repeated with duplicates.
Table 1.
Kinetics constants for hydrolysis of fluorogenic peptides Mca-YVADAPK-Dnp and Mca-RPPGFSAFK-Dnp by ACE2 in the presence of SARS-CoV-2 spike protein (30 nM)
| Substrate | SARS-CoV-2 Spike (30nm) | NaCl (mm) | Km (µM) | Kcat (s−1) | Kcat/Km (M−1s−1) | IC50 (µM) for Competitive substrates | ||
|---|---|---|---|---|---|---|---|---|
| BK | desBK | Ang II | ||||||
| YVADAPK | - | 150 | 46.6 ± 10.3 | 0.33 ± 0.05 | 7.08 × 103 | n.d | n.d | 47 ± 9 |
| + | 150 | 28.2 ± 0.6 | 0.58 ± 0.07 | 2.06 × 104 | 680 ± 87 | 1428 ± 655 | 6.4 ± 0.9 | |
| - | 300 | 24.8 ± 9.9 | 0.058 ± 0.01 | 1.66 × 104 | ||||
| + | 300 | 21.1 ± 6.8 | 0.11 ± 0.014 | 3.71 × 104 | ||||
| RPPGFSAFK | - | 150 | 10.2 ± 2.7 | 0.007 ± 0.002 | 7.06 × 102 | n.d | n.d | 129 ± 52 |
| + | 150 | 2.41 ± 0.85 | 0.018 ± 0.002 | 7.55 × 103 | n.d | n.d | 23 ± 5.0 | |
| - | 300 | 10.0 ± 3.0 | 0.01 ± 0.005 | 1.02 × 103 | ||||
| + | 300 | 1.3 ± 0.55 | 0.015 ± 0.004 | 1.14 × 104 | ||||
Notes: blank cell (not measured); n.d (not detectable).
Competitive ACE2 cleavage in the presence of ang II, desBK and BK
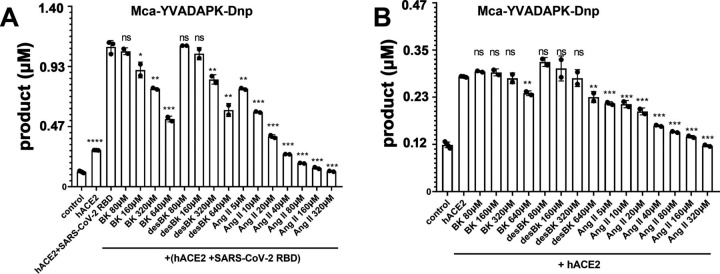
Although SARS-CoV-2 spike protein enhanced ACE2 cleavage of caspase-1 substrate and BK-analog, it was not clear if the spike binding enhanced ACE2 cleavage of its physiological substrates desBK and Ang II. Because of the difficulty to conjugate FRET fluorophores to the physiological ligands without affecting their enzymatic cleavage, we used an unlabeled substrate competition assay to assess the effect of the spike RBD binding on ACE2 cleavage of caspase-1 substrate in the presence of nonfluorogenic BK, desBK or Ang II peptides. In the absence of SARS-CoV-2 RBD binding, Ang II but not desBK inhibited the cleavage of caspase-1 substrate (Mca-YVADAPK(Dnp)-OH) with an inhibition constant IC50 of 47 μm, consistent with observed better ACE2 binding to Ang II (Km ∼2.0 μm) than to desBK (Km ∼290 μm) (Fig. 5 ) (14). In the presence of SARS-CoV-2 RBD, however, desBK inhibited the ACE2 cleavage with a IC50 of 1428 μm (Fig. 5 A), suggesting that SARS-CoV-2 enhanced ACE2 binding to desBK. Surprisingly, despite no detectable ACE2 cleavage of BK under physiological condition (14), BK inhibited ACE2 cleavage of caspase-1 substrate in the presence of the spike RBD with a IC50 of 680 μm (Fig. 5). As for ACE2 cleavage of the BK-analog, Ang II competed with a IC50 constant of 129 μm and 23 μm in the absence and presence of the SARS-CoV-2 RBD, respectively (Fig. S3), similar to that of caspase-1 substrate cleavage. However, both BK and desBK failed to inhibit ACE2 cleavage of BK-analog. As shown before, BK-analog bound to endothelin converting enzyme-1 (ECE-1) much better than BK because of its C-terminal modification (20). It is likely that BK-analog also bound better to ACE2 than BK.
Figure 5.
Competition of Bradykinin(BK), des-Arg9-BK, and Ang II peptide with fluorogenic Mca-YVADAPK-Dnp (10 μm) for ACE2 hydrolysis.A, Competitive substrates inhibition in the presence of SARS-CoV-2 RBD protein at time 1hof fluorogenic Mca-YVADAPK-Dnp cleavage. The pairwise p-value statistics were calculated between (hACE2 + SARS-CoV-2 RBD) and its addition of corresponding concentrations of competitive peptides. B, Competitive substrates inhibition in the absence of SARS-CoV-2 RBD protein at time 1 h of fluorogenic Mca-YVADAPK-Dnp cleavage. The pairwise p-value statistics were calculated between hACE2 and corresponding concentrations of competitive peptides.
Discussion
The new coronavirus SARS-CoV-2 infects human cells via the binding of its spike (S) glycoprotein to the human angiotensin-converting enzyme-2 (ACE2). Here we examined the influence of the high affinity binding of SARS-CoV-2 spike protein on the enzymatic activity of ACE2. Surprisingly, SARS-CoV-2 spike protein significantly enhanced the enzymatic activity of ACE2 to cleave caspase-1 (YVADAPK) peptide in a concentration and RBD dependent manner. It is likely that the spike protein binding to ACE2 would lead to more efficient cleavage of similar substrates such as Ang II (DRVYIHP↓F) and Apelin-13(QRPRLSHKGPMP↓F). Similarly, the binding of SARS-CoV-2 spike enhanced ACE2 cleavage of bradykinin analog, and bradykinin (RPPGFSPFR) and des-Arg9-bradykinin competed similarly for ACE2 cleavage of the caspase-1 substrate.
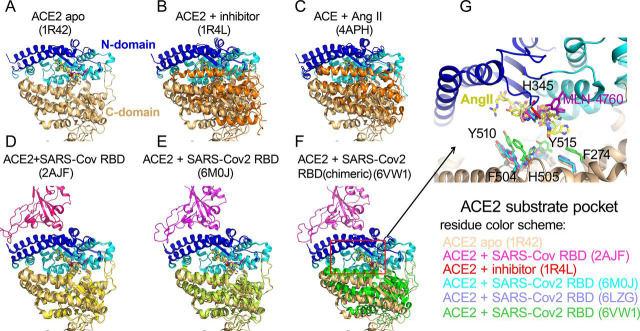
To further gain insight into how the binding of the viral spike protein affected the catalytic activity of ACE2, we compared structures of ACE2 in the presence and absence of the viral spike protein. The native structure of ACE2 extracellular proteolytic domain comprised two subdomains, the N-terminal and C-terminal subdomain with a wide cleft in between for substrate binding and catalysis (22) (Fig. 6 A). The structure of ACE2 with a potent inhibitor MLN-4760 bound at the active site showed a clear ligand-induced hinge bending movement between the N- and C-terminal domains (Fig. 6 B). This closed conformation of ACE2 proteolytic domain on inhibitor binding was consistent with that of ACE bound to its hydrolyzed product Ang II (23), suggesting a common mode to bind substrate and reaction intermediates among zinc metalloproteases (Fig. 6 C). The comparison between native and inhibitor bound ACE2 structures revealed a critical substrate binding pocket including residues F274, H345, F504, H505, Y510, and Y515. SARS-CoV-2 and SARS-CoV spike protein both use the receptor binding domain (RBD) to recognize ACE2 N-terminal domain (11, 12, 13, 24, 25). The structural superposition between various RBD bound ACE2 and native ACE2 structures revealed a close agreement in the N-domain of ACE2 conformation with r.m.s.d less than 0.5 Å (Fig. 6 D and E). However, the SARS-CoV-2 RBD but not SARS-CoV RBD binding caused the hinge movement between the N- and C-terminal domains reminiscent of a closing clam. We then assessed such relative movement by measuring the angle formed by Asn136 on the rim of native ACE2 C-terminal domain, zinc atom at the catalytic center, and Asn136 of superimposed ACE2 bound by RBD of SARS-CoV-2 or SARS-CoV. Compared with SARS-CoV RBD binding (0.3°), this angle upon SARS-CoV-2 RBD binding was increased to 5°. Particularly, the binding of a chimeric SARS-CoV-2 RBD to ACE2 (12), resulted in a ∼12°movement of ACE2 C-terminal domain toward the N-terminal domain (Fig. 6 F). Furthermore, this hinge angle was 29° resulting in a closed conformation of ACE2 in the presence of inhibitor MLN-4760, suggesting that SARS-CoV-2 spike binding initiated a conformational change to energetically facilitate ACE2 catalysis. The comparison of substrate binding pocket on ACE2 showed that residues F274, H345, F504, H505, Y510, and Y515 in SARS-CoV-2-ACE2 complexes moved closer to aligned Ang II or MLN-4760 inhibitor, consistent with the RBD binding affected the substrate binding (Fig. 6 G). This is consistent with a decreasing in Km values of ACE2 hydrolysis in the presence of SARS-CoV-2 spike binding. In addition, in SARS-CoV-2 RBD bound ACE2, H345 residue that was proposed to stabilize the hybridized nitrogen upon catalysis adopted the same conformation as that bound by inhibitor. This active conformation of H345 was not observed in native or SARS-CoV RBD bound ACE2 structure, suggesting that the tighter binding of SARS-CoV-2 RBD could also induce a conformation change to position ACE2 substrate pocket residues for efficient catalysis.
Figure 6.
Conformational changes of ACE2 upon SARS-CoV-2 binding.A, apo structure of ACE2 (PDB ID: 1R42).The N-terminal subdomain was colored in cyan with the secondary structures of SARS-CoV-2 spike binding sites highlighted in blue. The C-terminal domain of apo ACE2 was colored in wheat. All subsequent structural superpositions were based the alignment of ACE2 residues 20-84 that formed the first 2 helix for RBD domain interaction and displayed at the same orientation. The ACE2 inhibitor MLN-4760 (purple) and Angiotensin II (yellow) were positioned in the substrate pocket based on the structural alignment. B, ACE2 structure with inhibitor MLN-4760 binding (PDB ID: 1R4L). The C-terminal domain was highlighted in orange. C, hACE in complex with Ang II(PDB ID: 4APH). The C-terminal domain was highlighted in orange. D, ACE2 structure in complex with SARS-CoV RBD(PDB ID: 2AJF). The C-terminal domain was highlighted in yellow. E, ACE2 structure in complex with SARS-CoV-2 RBD (PDB ID: 6M0J). The C-terminal domain was highlighted in lime green. F, ACE2 structure in complex with a chimeric SARS-CoV-2 RBD domain (PDB ID: 6VW1). The C-terminal domain was highlighted in green. G, Enlarged view of ACE2 substrate binding pocket. One additional ACE2-SARS-CoV-2 RBD complex (PDB ID: 6LZG) was included. The residue color scheme was listed in the bottom panel.
With increasing number of confirmed cases worldwide, the major manifestation of SARS-CoV-2 mediated COVID-19 disease and its complications generated critical concerns because of the lack of highly effective antiviral drugs or vaccines. Compared with SARS-CoV and MERS, SARS-CoV-2 is more contagious and the clinical symptoms of COVID-19 arevariable (4). In addition to the accumulation of fluid in the lung, a considerable fraction of critically ill COVID-19 patients developed deep venous thrombosis or pulmonary embolism (6). These clinical observations shared a common prognostic feature that pointed to the cardiovascular system. Vasodilation, vasoconstriction, and permeability are the essential components of the cardiovascular system, which are regulated by the cross-talk of many hormones and biological active peptides. Among them, renin-angiotensin system and kinin-kallikrein system have major effects on cardiovascular function (15, 21). Importantly, ACE2 catalyzes the conversion of these regulatory peptides in these systems to maintain the homeostatic state. Among the biological active substrates, ACE2 cleaves Ang II (DRVYIHP↓F), Apelin-13(QRPRLSHKGPMP↓F), and dynorphin A 1-13(YGGFLRRIRPKL↓K) with high proteolytic efficiency (Km <10 μm and k cat/Km >1 × 106M−1s−1). ACE2 also has substantial activity against des-Arg9-bradykinin (RPPGFSP↓F), and neurotensin 1-8 (ELYENKP↓R). Our results suggest that SARS-CoV-2 may alter ACE2 cleavage of these substrates in infected individuals. During SARS-CoV-2 infection, the enhancement of ACE2 activity could produce more of their cleaved products, Ang 1-9 and Ang 1-7. Beside the opposing effects of Ang II and Ang 1-7 on vasoconstriction and vasodilatation, they regulated the expression of ACE2. Several in vivo and in vitro studies documented that Ang II could reduce ACE2 expression through activation of MAPK pathways, whereas Ang 1-7 increased the receptor expression (26, 27). Indeed, ACE2 expression was increased in the lungs of severe COVID-19 patients with comorbidities, compared with control individuals (18, 28). This is consistent with the binding of the viral spike protein resulting in enhanced ACE2 cleavage of Ang II.
Apelin 1-13(QRPRLSHKGPMP↓F) is an endogenous ligand for angiotensin II protein J receptor (APJR), a homolog of the angiotensin receptor AT1(29). Apelin 1-13 contributes to nitric oxide dependent arterial dilatation to counterbalance Ang II effects. The cleavage of its carboxyl-terminal phenylalanine weakens Apelin 1-13 interaction with APJR (30), resulting in 2- to 5-fold decrease of its potency (17). The enhancement of ACE2 activity would accelerate the metabolism of Apelin 1-13, further dampening its beneficial effects to cardiac functions. The proteolysis of bradykinin may be relevant to COVID-19 as ACE2 hydrolyzes des-Arg9-bradykinin (15). The viral spike enhanced ACE2 activity may affect rate of des-Arg9-bradykinin degradation (31). The ACE2 cleavage of bradykinin analog in the presence of SARS-CoV-2 RBD suggested the increased ACE2 activity upon SARS-CoV-2 spike binding may allow efficient cleavage of otherwise suboptimal sequences and thus alter their pharmacological effects in vivo. In addition to vascular symptoms, COVID-19 patients present with neurological sequalae including body aches and loss of taste and smell. The neuropeptides Dynorphin A 1-13(YGGFLRRIRPKL↓K) and β–Casomorphin(YPFVEP↓I) are good ACE2 substrate, which are endogenous ligands for opioid receptors and have antinociceptive effects (32). Interestingly, Dynorphin A 1-13 was also implicated as an appetite stimulant (33). The removal of the C-terminal lysine residue of Dynorphin A 1-13 resulted in 90% loss of its potency (34). It is interesting to speculate that altered ACE2 hydrolysis of these neuropeptides would affect the homeostatic functions of these opioid receptors as well.
The lack of widely available and highly effective antiviral drugs or vaccines makes the understanding SARS-CoV-2 infection critical to the development of COVID-19 treatments. Our study demonstrated the direct influence of SARS-CoV-2 spike protein on the enzymatic activity of ACE2 and warranted further investigation into the biological roles of ACE2 in individuals infected with SARS-CoV-2. Given the central role of ACE2 in the homeostatic regulation of cardiovascular system, our data suggest that enhanced ACE2 activity may explain some of the vascular complications associated with COVID-19 disease. Future studies of animal models and COVID-19 patients will be required to determine the physiological effect of this viral spike enhanced ACE2 activity.
Materials and methods
Reagents
Recombinant proteins of human ACE2 extracellular enzymatic domain with >90% purity were purchased from R&D systems, USA (933-ZN) and Sinobiological, Inc, USA (10108-H08H). Recombinant proteins of SARS-CoV RBD (40150-V08B2) and SARS-CoV-2 RBD proteins (40592-V08H) were obtained from Sinobiological, Inc. Fluorogenic peptide substrates were obtained from R&D systems, where Mca-YVADAPK(Dnp)-OH (ES007) was used as an angiotensin II like substrate and Mca-RPPGFSAFK(Dnp)-OH(ES005) was a fluorogenic peptide derivative of Bradykinin according to manufacturer's instruction. Nonfluorogenic bradykinin(BK, RPPGFSPFR), des-Arg9-bradykinin (desBK, RPPGFSPF) and angiotensin II (Ang II, DRVYIHPF) peptides were purchased from Genscript, Inc, USA. ACE2 specific inhibitor MLN-4760 and various protease inhibitors AEBSF, pepstatin A and leupeptin were obtained from Sigma-Aldrich, Inc. Fluorogenic peptide Mca-Pro-Leu-OH (Bachem, USA, M-1975) was used as calibration standard to obtain the molarity (pmol) to relative fluorescence unit (RFU) conversion factor on Synergy H1 fluorescent plate reader.
SARS-CoV-2 trimeric spike protein expression and purification
The vector to express the prefusion S ectodomain, a gene encoding residues 1-1208 of SARS-CoV-2(GenBank: MN908947) was kindly provided by Jason S. McLellan (the University of Texas at Austin) and the protein was expressed as described previously (10). In brief, FreeStyle293 F cells (Thermo Fisher, USA) were transiently transfected with this expression vector using polyethylenimine (PEI). Protein was purified from filtered cell supernatants using StrepTactin XT resin (Germany, IBA, Inc) and subsequently subjected to further purification by size exclusion chromatography using a Superose 6 10/300 column (GE Healthcare, USA) with the buffer of 75 mm Tris (pH7.5), 0.15M NaCl. Before enzymatic assays, SARS-CoV-2 spike and commercial RBD recombinant proteins were dialyzed extensively against 75 mm Tris(pH7.5) and 0.15M NaCl.
Surface plasmon resonance solution binding experiments
Surface plasmon resonance measurements were performed using a BIAcore 3000 instrument and analyzed with BIAevaluation 4.1 software (Biacore AB). Human ACE2 was immobilized on carboxylated dextran CM5 chips (Biacore AB) to 200–1000 response units (RU) using a primary amine-coupling in 10 mm sodium acetate (pH 4.0). The analytes consisted of serial dilutions of SARS-CoV-2 spike protein between 1000 nm and 31 nm in a buffer containing 50 mm Tris (pH7.4) and 0.15M NaCl. The dissociation constants were obtained by kinetic curve fitting with 1:1 binding model using BIAevaluation 4.1 (BIAcore Inc.).
Enzymatic assays of ACE2 hydrolysis of biological peptides
Reactions were performed in black microtiter plates at ambient temperature (26 °C) according to manufacturer's instruction. To each well, 50 µl of 0.2 or 0.4 µg/ml ACE2 in assay buffer containing 75 mm Tris(pH7.5) plus NaCl at 0.15M, 0.3M or 1.0M were added, respectively. Then 10 µl dialysis buffer or SARS-CoV-2 S spike/RBD domain proteins at various final concentrations ranging from 70 µg/ml to 0.4 µg/ml were added to wells and incubated for 20 min after mixing by shaking. The reactions were initiated by adding 50 µl of fluorogenic peptides at 40 μm or with 2-fold serial dilutions ranging from 160 μm to 0.6 μm to determine the kinetic constants for ACE2 hydrolysis. The relative fluorescence units (RFU) were read at excitation and emission wavelengths of 320 nm and 405 nm (top read), respectively in kinetic mode at 1-min intervals for 8 h.
To calculate specific activity of ACE2, the substrate blank adjusted relative fluorescent units were converted to molarities according to the conversion factor on Synergy H1 plate reader that was calibrated by standard Mca-pro-Leu-OH (Fig. S2A). To obtain the kinetic constants, the hydrolysis was limited to ≤15% product formed as the initial velocity conditions. Initial velocities were plotted versus substrate concentration and fit to the Michaelis-Menten equation v=V max[S]/(Km +[S]) using GraphPad Prism software. Turnover numbers (k cat) were calculated from the equation k cat=V max/[E], using a calculated ACE2 molecular mass of 85,314 Da and assuming the enzyme sample to be essentially pure and fully active.
Substrate competition assays using nonfluorogenic BK, desBK and ang II peptides
To compare the relative proteolytic activity of ACE2 with different substrates, various concentrations of nonfluorogenic BK, desBK or Ang II peptides were added to the reaction mixture with fluorogenic peptides as competitive substrates. As mentioned above, 50 µl of ACE2 at 0.4 µg/ml was preincubated with buffers or SARS-CoV-2 RBD protein at a final concentration of 125 nm for 20 min. Subsequently, 50 µl of fluorogenic peptides at 20 μm supplemented with a serial dilution of nonfluorogenic BK, desBK or Ang II peptides ranging 640 μm to 80 μm were added to initiate enzymatic cleavage. In addition, 10 μm of ACE2 specific inhibitor MLN-4760 (Sigma-Aldrich) was used as positive control to block ACE2 activity in the absence or presence of SARS-CoV-2 spike(15 nm), SARS-CoV-2 RBD(125 nm) or SARS-CoV RBD (125 nm) proteins. Various protease inhibitors at a final concentration of 40 μm such as AEBSF, pepstatin A and leupeptin (Sigma-Aldrich) were also used to verify the specific activity of ACE2 enzyme. Fluorescence changes were measured at 1 min intervals for 1 h at 26 degree as the initial velocity conditions. The dose dependent inhibition was fitted with a competitive inhibition model to obtain IC50.
Statistical analysis
Student's t test was used to determine the statistical significance in each pair wise comparison. Each experiment was repeated at least with duplicates. p < 0.05 was considered statistically significant (* p < 0.05; ** p < 0.01; ***p < 0.001; **** p < 0.0001).
Data Availability Statement
All data described in the manuscript are contained within the manuscript and can be shared upon request.
Acknowledgments
We thank Dr. Malcolm Sim for proofreading the manuscript.
Division of Intramural Research, National Institute of Allergy and Infectious Diseases (DIR, NIAID) (AI000697) to Jinghua Lu, and Peter D. Sun
Edited by Ruma Banerjee
Footnotes
Author contributions—JL conceptualization; JL and PDS data curation; JL formal analysis; JL investigation; JL and PDS writing-original draft; PDS supervision; PDS funding acquisition
Funding and additional information—The funding of this work is provided by the Intramural Research Program (IRP) of National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents f this article.
- DAD
- diffuse alveolar damage
- ACE
- angiotensin-converting enzyme
- RAS
- rennin-angiotensin system
- BK
- Bradykinin
- RBD
- receptor binding domain
- ECE
- endothelin converting enzyme.
Supplementary Material
References
- 1.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. 32015508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. 32015507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. 31978945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L., Tai Y., Bai C., Gao T., Song J., Xia P. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wichmann D. Autopsy findings and venous thromboembolism in patients with COVID-19. Ann. Intern. Med. 2020;173:268–277. doi: 10.7326/L20-1206. [DOI] [PubMed] [Google Scholar]
- 7.Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., Bao L., Zhang B., Liu G., Wang Z., Chappell M. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005;11:875–879. doi: 10.1038/nm1267. 16007097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bao L. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Natjure. 2020 doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 9.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. 14647384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. 32075877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. 32225176. [DOI] [PubMed] [Google Scholar]
- 12.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. 32225175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.-Y., Wang Q., Zhou H., Yan J., Qi J. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. e899. 32275855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vickers C., Hales P., Kaushik V., Dick L., Gavin J., Tang J., Godbout K., Parsons T., Baronas E., Hsieh F., Acton S., Patane M., Nichols A., Tummino P. Hydrolysis of biological peptides by human angiotensin-converting enzyme-related carboxypeptidase. J. Biol. Chem. 2002;277:14838–14843. doi: 10.1074/jbc.M200581200. 11815627. [DOI] [PubMed] [Google Scholar]
- 15.Kaplan A.P., Joseph K. Pathogenic mechanisms of bradykinin mediated diseases: dysregulation of an innate inflammatory pathway. Adv. Immunol. 2014;121:41–89. doi: 10.1016/B978-0-12-800100-4.00002-7. 24388213. [DOI] [PubMed] [Google Scholar]
- 16.van de Veerdonk F.L., Netea M.G., van Deuren M., van der Meer J.W., de Mast Q., Brüggemann R.J., van der Hoeven H. Kallikrein-kinin blockade in patients with COVID-19 to prevent acute respiratory distress syndrome. Elife. 2020;9 doi: 10.7554/eLife.57555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang P. Apelin-13(1-12) Is a Biologically Active ACE2 Metabolite of the Endogenous Cardiovascular Peptide [Pyr(1)] Apelin-13. Front. Neurosci. 2017;11:92. doi: 10.3389/fnins.2017.00092. [Pyr(1)] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hou Y.J., Okuda K., Edwards C.E., Martinez D.R., Asakura T., Dinnon K.H., Kato T., Lee R.E., Yount B.L., Mascenik T.M., Chen G., Olivier K.N., Ghio A., Tse L.V., Leist S.R. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell. 2020;182:429–446.e14. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imai Y., Kuba K., Rao S., Huan Y., Guo F., Guan B., Yang P., Sarao R., Wada T., Leong-Poi H., Crackower M.A., Fukamizu A., Hui C.-C., Hein L., Uhlig S. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. 16001071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson G.D., Ahn K. Development of an internally quenched fluorescent substrate selective for endothelin-converting enzyme-1. Anal. Biochem. 2000;286:112–118. doi: 10.1006/abio.2000.4772. 11038281. [DOI] [PubMed] [Google Scholar]
- 21.Burrell L.M., Johnston C.I., Tikellis C., Cooper M.E. ACE2, a new regulator of the renin-angiotensin system. Trends Endocrinol. Metab. 2004;15:166–169. doi: 10.1016/j.tem.2004.03.001. 15109615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Towler P. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J. Biol. Chem. 2004;279:17996–18007. doi: 10.1074/jbc.M311191200. 14754895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masuyer G., Schwager S.L., Sturrock E.D., Isaac R.E., Acharya K.R. Molecular recognition and regulation of human angiotensin-I converting enzyme (ACE) activity by natural inhibitory peptides. Sci. Rep. 2012;2:717. doi: 10.1038/srep00717. 23056909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. 16166518. [DOI] [PubMed] [Google Scholar]
- 25.Song W., Gui M., Wang X., Xiang Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007236. 30102747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varagic J., Ahmad S., Nagata S., Ferrario C.M. ACE2: angiotensin II/angiotensin-(1-7) balance in cardiac and renal injury. Curr. Hypertens. Rep. 2014;16:420. doi: 10.1007/s11906-014-0420-5. 24510672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gallagher P.E., Ferrario C.M., Tallant E.A. MAP kinase/phosphatase pathway mediates the regulation of ACE2 by angiotensin peptides. Am. J. Physiol. Cell Physiol. 2008;295:C1169–C1174. doi: 10.1152/ajpcell.00145.2008. 18768926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pinto B.G. ACE2 Expression is Increased in the Lungs of Patients with Comorbidities Associated with Severe COVID-19. medRxiv. 2020 doi: 10.1093/infdis/jiaa332. 03.21.20040261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang W., McKinnie S.M.K., Farhan M., Paul M., McDonald T., McLean B., Llorens-Cortes C., Hazra S., Murray A.G., Vederas J.C., Oudit G.Y. Angiotensin-converting enzyme 2 metabolizes and partially inactivates Pyr-Apelin-13 and Apelin-17: physiological effects in the cardiovascular system. Hypertension. 2016;68:365–377. doi: 10.1161/HYPERTENSIONAHA.115.06892. 27217402. [DOI] [PubMed] [Google Scholar]
- 30.Ma Y., Yue Y., Ma Y., Zhang Q., Zhou Q., Song Y., Shen Y., Li X., Ma X., Li C., Hanson M.A., Han G.W., Sickmier E.A., Swaminath G., Zhao S. Structural basis for apelin control of the human apelin receptor. Structure. 2017;25:858–866. doi: 10.1016/j.str.2017.04.008. e854. 28528775. [DOI] [PubMed] [Google Scholar]
- 31.Sheikh I.A., Kaplan A.P. The mechanism of degradation of bradykinin (lysyl-bradykinin) in human serum. Adv Exp Med Biol. 1989;247:331–336. doi: 10.1007/978-1-4615-9543-4_49. [DOI] [PubMed] [Google Scholar]
- 32.Caudle R.M., Mannes A.J. Dynorphin: friend or foe? Pain. 2000;87:235–239. doi: 10.1016/s0304-3959(00)00360-2. 10963903. [DOI] [PubMed] [Google Scholar]
- 33.Lambert P.D., Wilding J.P., Al-Dokhayel A.A., Bohuon C., Comoy E., Gilbey S.G., Bloom S.R. A role for neuropeptide-Y, dynorphin, and noradrenaline in the central control of food intake after food deprivation. Endocrinology. 1993;133:29–32. doi: 10.1210/endo.133.1.8100519. [DOI] [PubMed] [Google Scholar]
- 34.Chavkin C., Goldstein A. Specific receptor for the opioid peptide dynorphin: structure–activity relationships. Proc Natl Acad Sci U S A. 1981;78:6543–6547. doi: 10.1073/pnas.78.10.6543. 6118865. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data described in the manuscript are contained within the manuscript and can be shared upon request.