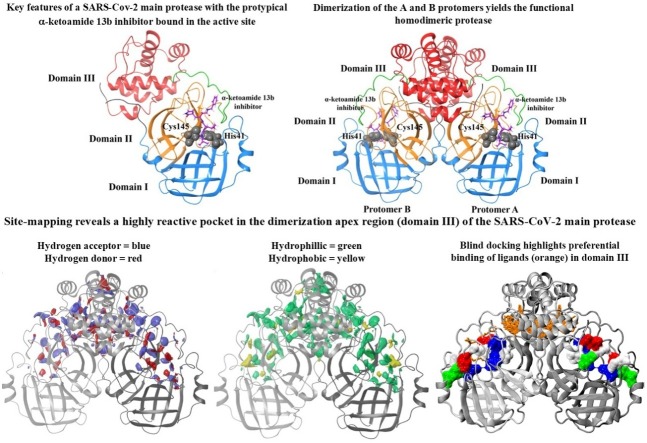
Graphical abstract
Keywords: Coronavirus, COVID-19, SARS-CoV-2, SARS-CoV-2 main protease, Site mapping, Blind docking
Abstract
The SARS-CoV-2 virus is causing COVID-19 resulting in an ongoing pandemic with serious health, social, and economic implications. Much research is focused in repurposing or identifying new small molecules which may interact with viral or host-cell molecular targets. An important SARS-CoV-2 target is the main protease (Mpro), and the peptidomimetic α-ketoamides represent prototypical experimental inhibitors. The protease is characterised by the dimerization of two monomers each which contains the catalytic dyad defined by Cys145 and His41 residues (active site). Dimerization yields the functional homodimer. Here, our aim was to investigate small molecules, including lopinavir and ritonavir, α-ketoamide 13b, and ebselen, for their ability to interact with the Mpro. The sirtuin 1 agonist SRT1720 was also used in our analyses. Blind docking to each monomer individually indicated preferential binding of the ligands in the active site. Site-mapping of the dimeric protease indicated a highly reactive pocket in the dimerization region at the domain III apex. Blind docking consistently indicated a strong preference of ligand binding in domain III, away from the active site. Molecular dynamics simulations indicated that ligands docked both to the active site and in the dimerization region at the apex, formed relatively stable interactions. Overall, our findings do not obviate the superior potency with respect to inhibition of protease activity of covalently-linked inhibitors such as α-ketoamide 13b in the Mpro active site. Nevertheless, along with those from others, our findings highlight the importance of further characterisation of the Mpro active site and any potential allosteric sites.
1. Introduction
On December 31st 2019, the World Health Organisation (WHO), was informed of cases of atypical pneumonia, originating in Wuhan City (Hubei Province), China (Hui et al., 2020). The outbreak was quickly identified to be caused by a novel coronavirus, provisionally designated as 2019-nCoV (Wang et al., 2020). Based on genetic relationships, including to severe acute respiratory syndrome (SARS-CoV-1) and Middle East respiratory syndrome (MERS-CoV), introduced to humans in 2002 and 2012, respectively, the novel coronavirus has been designated as SARS-CoV-2 (V. Coronaviridae Study Group of the International Committee on Taxonomy of, 2020). The SARS-CoV-2 virus is currently causing coronavirus disease 2019 (COVID-19). Although associated with a lower case fatality rate (CFR) than both SARS-CoV-1 (∼9.5 %) and MERS-CoV (∼34.4 %), SARS-CoV-2 (accurate CFR not yet established however, clearly lower than both SARS-CoV-1 and MERS-CoV), is highly transmissible and COVID-19 was declared a global pandemic by WHO on March 11th, 2020 (Rajgor et al., 2020; de Wit et al., 2016; Cucinotta and Vanelli, 2020).
The COVID-19 pandemic is ongoing and SARS-CoV-2 virus has now spread worldwide with confirmed cases in 188 countries and regions (Dong et al., 2020). To date, we are limited to hand hygiene, self-isolation of infected individuals, quarantine, global travel restrictions and lockdowns, and physical distancing to mitigate the spread of infection with significant health, social, and economic repercussions. Hospitalisation and respiratory care is required for more severe cases, and intensive care with mechanical ventilation necessary in critical cases leading to well-documented issues with resource allocation (Phua et al., 2020; Peters et al., 2020). The most important goal for disease prevention is the development of a safe and effective vaccine, and there is an intense global research effort in this direction with numerous preparations already progressing in human clinical trials (Callaway, 2020; Cohen, 2020). There is also intense interest in the investigation of potential prophylactic or therapeutic compounds. Drug repurposing approaches offer the opportunity to expedite the management of COVID-19 by utilising existing medications for which dosages and side effects, and in some cases therapeutic targets are known. A well-known example where this strategy has led to confusion is the suggested use of the antimalarial, hydroxychloroquine alone or in combination with the broad-spectrum antibiotic azithromycin. To date, clinical findings are limited and largely controversial, and anticipated to be further clarified with ongoing human clinical trials, although signs are not encouraging (Taccone et al., 2020; Funck-Brentano et al., 2020). Further, the antiviral remdesivir is progressing in clinical trials in people with COVID-19 (Antinori et al., 2020). Remdesivir, been previously shown to inhibit SARS-CoV-1 and MERS-CoV in vitro and in animal models, and was shown to be well tolerated in people with Ebola virus disease (Mulangu et al., 2019; Sheahan et al., 2017; Sheahan et al., 2020). Although remdesivir is currently the only drug to have received emergency use authorization for treatment of COVID-19, by the U.S. Food and Drug Administration, further human clinical trials are necessary (Grein et al., 2020).
In the context of drug repurposing or identification and evaluation of compounds the potential antiviral effects, a multitude of potential targets against SARS-CoV-2 have been characterized (Gordon et al., 2020). Most research efforts are focussed on the initial viral binding and fusion with host cells, and small molecules interfering with the SARS-CoV-2 surface spike protein interactions with the angiotensin-converting enzyme 2 and host cell protease, are being widely investigated (Hoffmann et al., 2020). Further, the viral RNA−dependent RNA polymerase (RdRp), which is required for replication of SARS-CoV-2 is a critical component, and a target for remdesivir (Yin et al., 2020). The recent release of a high resolution cryo-EM structure of the SARS-CoV-2 RdRp will expedite further investigation of potential inhibitors (Yin et al., 2020).
Another important target for SARS-CoV-2 is the main protease (Mpro, also known as the 3-chymotrypsin-like protease [3CLpro]). The ∼34 KDa (306 amino acid residues) Mpro is important for releasing functional polypeptides from translated RNA by processing viral polyproteins and therefore, has a critical role in the viral life cycle (Zhang et al., 2020a). The protease is active as a homodimer, comprised by dimerization of two protomers, designated as monomer A and monomer B, and the catalytic dyad on each protomer is defined by Cys145 and His (Kim et al., 2016) residues (Zhang et al., 2020a). Identification and development of potential inhibitors of the Mpro with antiviral effects against SARS-CoV-2 has been of particular research interest in an attempt to mitigate this current pandemic (Zhang et al., 2020a; Ton et al., 2020; Tsuji, 2020; Dai et al., 2020; Jin et al., 2020). In an interesting development, apart from targeting the active site, there has been suggestions of hindering the dimerization of the two protomers, either with small molecules or peptides, thereby, inhibiting the catalytic activity of the Mpro (Goyal and Goyal, 2020).
The prototypical broad-spectrum protease inhibitors are the peptidomimetic α-ketoamides which have been investigated in numerous viruses, including betacoronaviruses (Hilgenfeld, 2014; Anand et al., 2003). Most recently, structural components of α-ketoamide analogues have been optimized for favourable pharmacokinetic properties, with a compound designated as α-ketoamide 13b emerging as a lead (Zhang et al., 2020a). In another pertinent study, over 10,000 compounds were screened for activity against Mpro and selected compounds for antiviral activity in cell-based assays (Jin et al., 2020). Six lead compounds were identified, and an interesting organoselenium compound, ebselen, was shown to be particularly effective (Jin et al., 2020). With respect to drug repurposing, the combination of lopinavir and ritonavir (Kaletra®), which represents a co-formulation of protease inhibitors approved by the US FDA for the treatment of human immunodeficiency (HIV) type-1 infection (Chandwani and Shuter, 2008). The aspartate protease inhibitors, were shown to have modest antiviral effects against SARS-CoV-1 and MERS-CoV (Chu et al., 2004, Spanakis et al., 2014). Although there is interest, in the potential of lopinavir and ritonavir – which have been investigated for their binding to the SARS-CoV-2 Mpro – for the treatment of COVID-19 findings from clinical trials to date are not encouraging (Muralidharan et al., 2020; Cao et al., 2020; Hung et al., 2020).
Here, our overall aim was to investigate the binding and stability of lopinavir and ritonavir, α-ketoamide 13b, and ebselen with the active site of the Mpro. Further, we extended our studies to explore interactions of the small molecules with the dimerization pocket at the apex of the Mpro. We also included the sirtuin 1 activator, SRT1720, which has been previously investigated for effects in metabolism and as a life extension compound in animal models, in our analyses (Minor et al., 2011; Liu et al., 2017). Our choice for this compound was motivated by the differing binding characteristics of SRT1720 to the active and dimerization sites of the Mpro compared to lopinavir and ritonavir, α-ketoamide 13b, and ebselen, enabling characterisation of a wider-range of compounds as described in this work.
2. Materials and methods
2.1. Docking to the active site of the SARS-CoV-2 Mpro
System preparation and docking calculations were performed using the Schrodinger Suite (Schrödinger, 2020a) molecular modelling package (version 2018-1) using default parameters unless otherwise specified. The published crystal structure of the SARS-CoV-2 Mpro (PDB ID: 6LU7) was utilized for docking to the monomeric protein (Jin et al., 2020). A homodimer complex of the SARS-CoV-2 Mpro was assembled using the PDBePISA (Proteins, Interfaces, Structures and Assemblies) server (Krissinel and Henrick, 2007). Protein structures were prepared using Protein Preparation Wizard (Sastry et al., 2013), where bond orders were assigned, and zero-order bonds to metals and disulphide bonds were created. Crystallographic water molecules were removed. Hydrogen bonds were assigned and optimised, followed by restrained energy minimization. Ligand structures were obtained from the PubChem database (Kim et al., 2016), with the exception of α-ketoamide 13b which was obtained from PDB 6Y2F (Zhang et al., 2020a). Ligand structures were processed using LigPrep (Sastry et al., 2013), to obtain structures with optimised geometry.
Receptor grids with dimensions of 20 × 20 × 20 Å were generated around the active site of the protease, centroid to residues Gly23, Thr24, Gly143, His163, Thr190, and Ala191. Docking was carried out using the Quantum Mechanics-Polarized Ligand Docking (QPLD) workflow (Cho et al., 2005) of Schrodinger. Initial docking utilized the extra precision (XP) scoring function of Glide (Friesner et al., 2006). Partial charges on ligand atoms were then calculated using quantum mechanical methods using the ‘accurate’ setting in Jaguar (Bochevarov et al., 2013). Ligands were re-docked using the calculated charges with XP docking mode of Glide, and final pose selection was based on GlideScore.
2.2. Mapping potential ligand binding sites on the SARS-CoV-2 Mpro using schrödinger’s SiteMap tool and PrankWeb
Potential binding sites on the SARS-CoV-2 Mpro were explored and characterized using the default settings in Schrödinger’s SiteMap (Halgren, 2009; Halgren, 2007) tool and the PrankWeb binding site prediction tool (Jendele et al., 2019; Krivák and Hoksza, 2018). Proteins were prepared as described above, with calculations performed on the protease monomer (PDB ID: 6LU7) and dimer, which was assembled using the PDBePISA (Krissinel and Henrick, 2007).
2.3. Blind docking using AutoDock Vina
Blind docking was performed using AutoDock Vina (Trott and Olson, 2010). Protein and ligand structures were processed to generate PDBQT files, where rotatable torsions for ligands were activated and proteins assumed to be rigid. The receptor grid encompassed the entire protein, and docking was performed at an exhaustiveness level of 2000.
2.4. Molecular dynamics simulations
Classical molecular dynamics (MD) simulations were carried out using GROMACS 2018.2 software (Berendsen et al., 1995; Abraham et al., 2015) with the CHARMM27 force field (Bjelkmar et al., 2010; Vanommeslaeghe et al., 2010). Docked ligands served as starting structures, and their topology was generated using SwissParam (Zoete et al., 2011). Protein-ligand complexes were solvated using TIP3P water (Jorgensen et al., 1983) in a dodecahedral box, with a minimum distance of 2.0 nm between any protein atom to the box edge. The solvated system was neutralized with sodium ions. Energy minimisation was performed using the steepest-descent gradient method for a maximum of 50,000 steps. The solvated systems were restrained using an isothermal-isochloric (NVT) ensemble followed by an isothermal-isobaric ensemble (NPT) for 100 ps. Temperature was maintained at 310 K with a modified Berendsen thermostat (Berendsen et al., 1984), and pressure at 1.0 bar with the Parrinello-Rahman barostat (Parrinello and Rahman, 1980). Bond lengths were constrained using the LINCS algorithm (Hess et al., 1997), and long-range electrostatic forces were calculated using the particle-mesh Ewald scheme (PME) (Darden et al., 1993) (grid spacing 0.16 nm). Cutoff ratios of 1.2 nm for Coulomb and van der Waals potentials were used for the calculation of short-range nonbonded interactions. Simulations were carried out for 100 ns with a time-step of 2 fs in triplicate, with random generation of velocities according to a Maxwell distribution.
Visual Molecular Dynamics 1.9.3 was used for visualisation and analysis (Humphrey et al., 1996; Roberts et al., 2006). Molecular Mechanics-Poisson Boltzmann Surface Area (MM-PBSA) was employed for the quantification of free energy calculations (Baker et al., 2001) using the g_mmpbsa tool (Kumari et al., 2014). Energy contributions from electrostatic, van der Waals, and polar solvation terms were calculated using the adaptive Poisson-Boltzmann Solver (APBS) (Robert et al., 2012). Grid spacing was set to 0.05 nm, and values of 80 and 2 were used for solvent dielectric and solute dielectric constants, respectively. The non-polar energy contribution was approximated using solvent-accessible surface area (SASA), with the probe radius set to 0.14 nm. Calculations were performed on 1000 ps segments of the stabilised trajectory (Hou et al., 2011). Entropic energy terms were excluded from the calculations.
3. Results and discussion
3.1. Docking to the SARS-CoV-2 Mpro active site highlights high binding affinity for structurally diverse small molecules
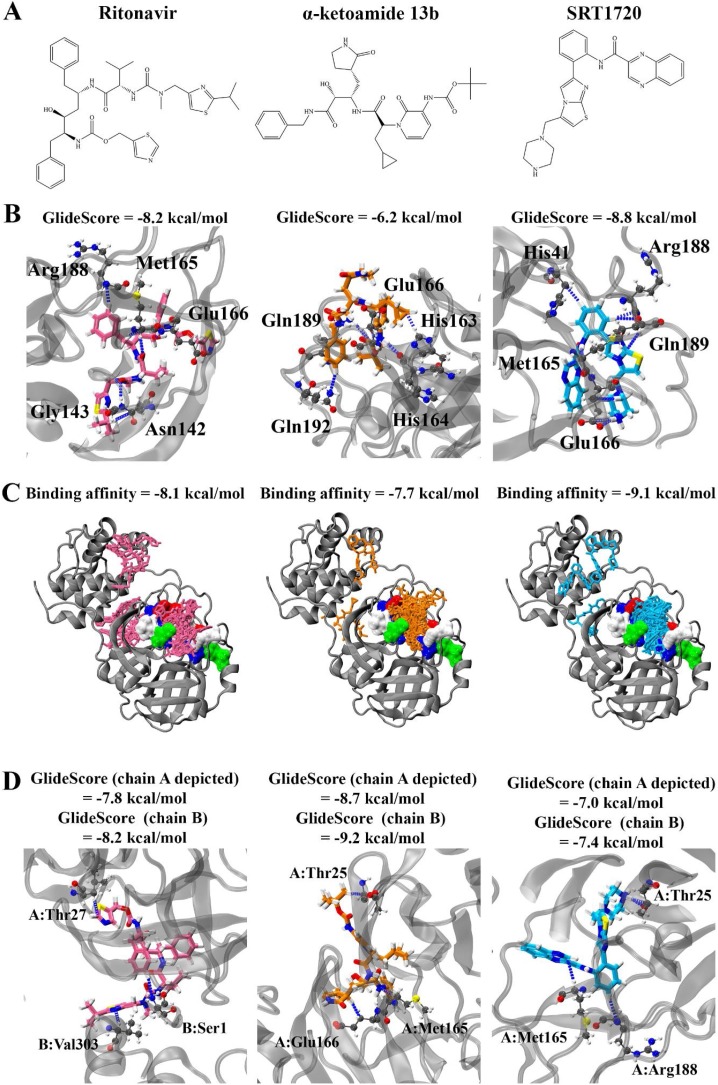
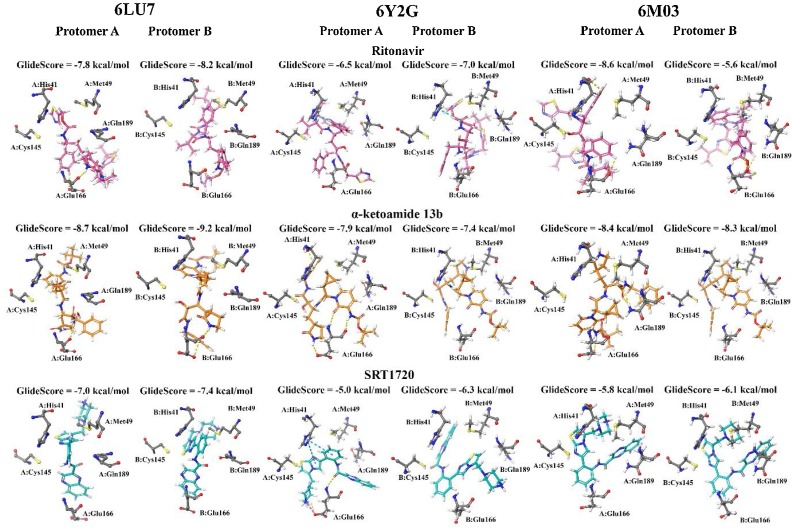
Initially, one protomer of the SARS-CoV-2 Mpro (PDB: 6LU7) was available for in silico analysis of potential ligands. Therefore, our initial work and most of the studies employing high-throughput methodologies have focused on binding of small molecules to the active site of the SARS-CoV-2 Mpro. We employed this methodology utilising Glide in this study and our findings indicated that ritonavir, α-ketoamide 13b, and SRT1720 (Fig. 1 A) bound to the active site with GlideScores of -8.2, -6.2, and -8.8. kcal/mol respectively (Fig. 1B). The compounds formed hydrogen bonds with key residues of the protease active site, including Met165, Glu166, and Gln189 (Zhang et al., 2020a). Blind docking was performed using AutoDock Vina to the monomer to identify the likelihood of the compounds to binding to the active site of the protease. Ritonavir, α-ketoamide 13b, and SRT1720 were shown to have a preference for binding to the active site (Fig. 1C). Top ranking poses produced binding affinities of -8.1, -7.7, and -9.1 kcal/mol respectively in the active site, with 10, 18, and 17 out of 20 poses found to bind in the active site in the monomer. Blind docking to the monomer was also performed with the drugs lopinavir and ebselen, with a lesser number of poses found to bind in the active site (1 and 7 poses out of 20, respectively) (Figure S1, Figure S2). Docking to the active site of the dimeric protein using Glide similarly indicated strong binding affinities for the main compounds in this study: -7.8 and -8.2 kcal/mol for ritonavir, -8.7 and -9.2 kcal/mol for α-ketoamide 13b, and -7.0 and -7.4 kcal/mol for protomers A and B with SRT1720, respectively (Fig. 1D).
Fig. 1.
Molecular docking of ritonavir, α-ketoamide 13b, and SRT1720 to the active site of SARS-CoV-2 main protease. Compound structures are shown in A). B) depicts the compounds docked to the active site of the monomer using Glide, with hydrogen bond forming residues highlighted. Blind docking was performed using Autodock Vina in C), with binding affinities of the top compound and number of poses in the active site (brown surface representation) shown. D) Docking to the active site of the dimer using Glide.
Overall, our findings indicate that the structurally diverse compounds had comparable binding affinities to the SARS-CoV-2 active site. Indeed, taking into account our findings from this study and much larger screens, with other high through-put studies, it is evident that many structurally diverse compounds are being identified, in many cases with stronger binding affinities than the prototypical α-ketoamide 13b inhibitor (Ton et al., 2020; Tsuji, 2020). A striking example, is a recent study in which screening of over 1.3 billion compounds, identified 585 unique scaffolds not associated with known protease inhibitors, and offered the top 1000 potential inhibitors of the SARS-CoV-2 Mpro (Ton et al., 2020). Taken together these findings highlight the need for finer discrimination of potential targets against the SARS-CoV-2 Mpro.
Site-mapping reveals a highly reactive binding pocket at the dimerization apex (domain III) attracts ligands away from the active site of the SARS-CoV-2 Mpro
To further explore potential sites associated with the SARS-CoV-2 Mpro, we performed site-mapping using commercial SiteMap software in the Schrodinger suite (Halgren, 2009; Halgren, 2007; Schrödinger, 2020b). SiteMap examines the entire protein to locate potential binding sites, usually in cases where the location of binding sites are not yet known, or to identify allosteric sites. Predictive binding tools have been used in conjunction with molecular docking to elucidate potential allosteric sites in various systems (Nayal and Honig, 2006; McCarthy et al., 2015; Kim et al., 2012). As well as a hydrophilic and hydrophobic contour map of the predicted binding site, SiteMap also generates a SiteScore which characterises the binding site in terms of its size, exposure to solvent, tightness of interaction between site points and the receptor, balance between the site hydrophobicity and hydrophilicity, and the degree of hydrogen bond donation and acceptance (Halgren, 2009; Halgren, 2007). A SiteScore greater than 1 is suggestive of a promising binding site, with a score of 0.80 previously shown to distinguish between drug-binding and non-binding sites (Schrödinger, 2020b); Nayal and Honig, 2006).
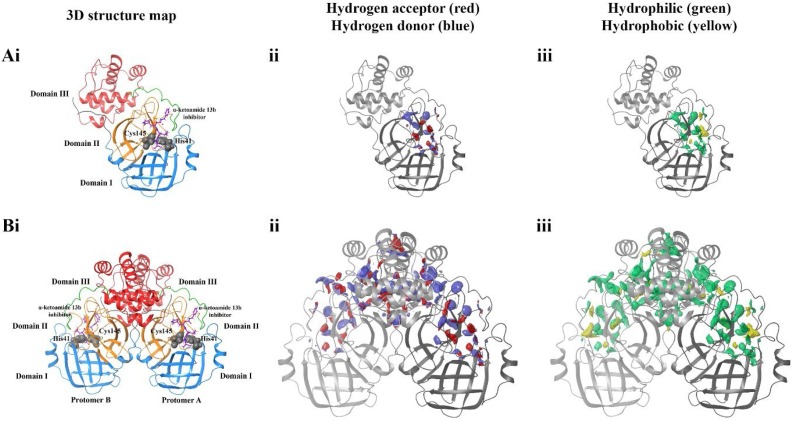
Here, we analysed utilised SiteMap to calculate potential binding sites on both monomer and dimeric forms of the SARS-CoV-2 Mpro (Fig. 2 ). The predicted binding site encompasses the known substrate binding site on the monomeric protease (Fig. 2A), with hydrophobic and hydrophilic, and hydrogen acceptor and donor sites surrounding the docked α-ketoamide 13b inhibitor and catalytic residues His41 and Cys145 with a SiteScore of 1.0. When SiteMap analysis was performed on the dimeric protease, the predicted binding site had a similarly high SiteScore of 1.0. Likewise, druggability scores were also high, with Dscores of 1.1 for the monomer, and 1.0 for the dimer (Halgren, 2009). Surprisingly, in addition the regions surrounding the docked inhibitor, clusters of hydrophilic and hydrophobic areas were observed at the dimerization apex, defined as domain III. It is noted that the predicted binding site of the dimeric protein is vastly larger compared to the monomer, with volumes of 1977 and 275 Å (V. Coronaviridae Study Group of the International Committee on Taxonomy of, 2020) for the dimer and monomer, respectively. Overall, the mapping of predicted sites highlights a highly reactive pocket at the dimerization apex (domain III) of the SARS-CoV-2 Mpro.
Fig. 2.
SiteMap analysis of the SARS-CoV-2 main protease in its A) monomeric and B) dimeric form. Domains of the protease are coloured in i), and catalytic residues His41 and Cys145 of the active site highlighted in grey. ii) depicts hydrogen acceptor (red) and hydrogen donor (blue) regions, while iii) shows hydrophilic (green) and hydrophobic (yellow) regions of predicted ligand binding sites.
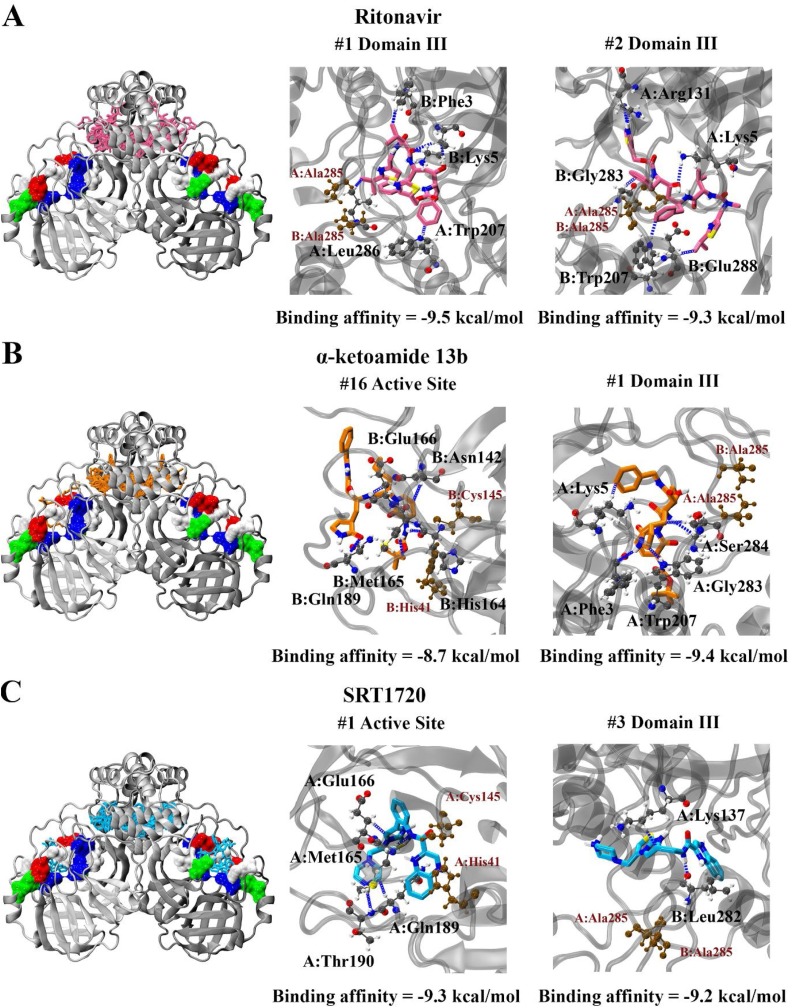
Given these findings we performed blind docking on the protease dimer to identify the likelihood of the compounds to bind to the active site. For all compounds, it was found that they were less likely to target the catalytic site compared to blind docking results with the monomer. In all cases, the compounds clustered near the apex of the protease in domain III (Fig. 3 ). Out of twenty poses produced, none of the poses were in the active site for ritonavir, with binding affinities ranging from -9.5 to -8.9 kcal/mol binding entirely within domain III. Only one pose was in the active site for α-ketoamide 13b. This pose was ranked 16th, and produced a binding affinity of -8.7 kcal/mol, compared to the highest ranked pose at in domain III with a predicted affinity of -9.4 kcal/mol. For SRT1720, four out of twenty poses were positioned in the active sites of protomer A and B, with two of these being the most highly ranked producing a binding affinity of -9.3 kcal/mol. The other two poses were ranked 19th and 20th, with binding affinities of -8.8 kcal/mol in the active sites of protomer A and B respectively.
Fig. 3.
Blind docking of A) ritonavir, B) α-ketoamide 13b, and C) SRT1720 to the SARS-CoV-2 Mpro protease dimer. The binding affinities are stated, along with interaction diagrams depicting hydrogen bond formation. Proximity of the ligand to the active site is indicated by catalytic dyad residues His41 and Cys145, while proximity to the domain III dimerization apex is indicated by residue Ala285.
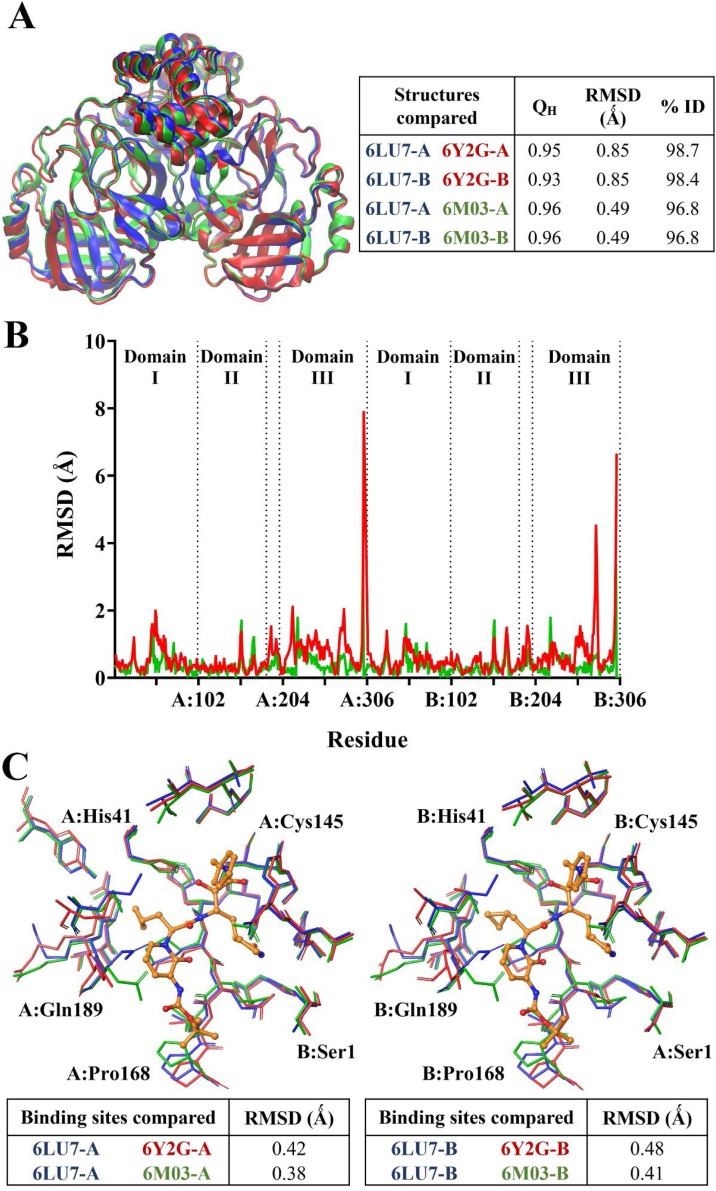
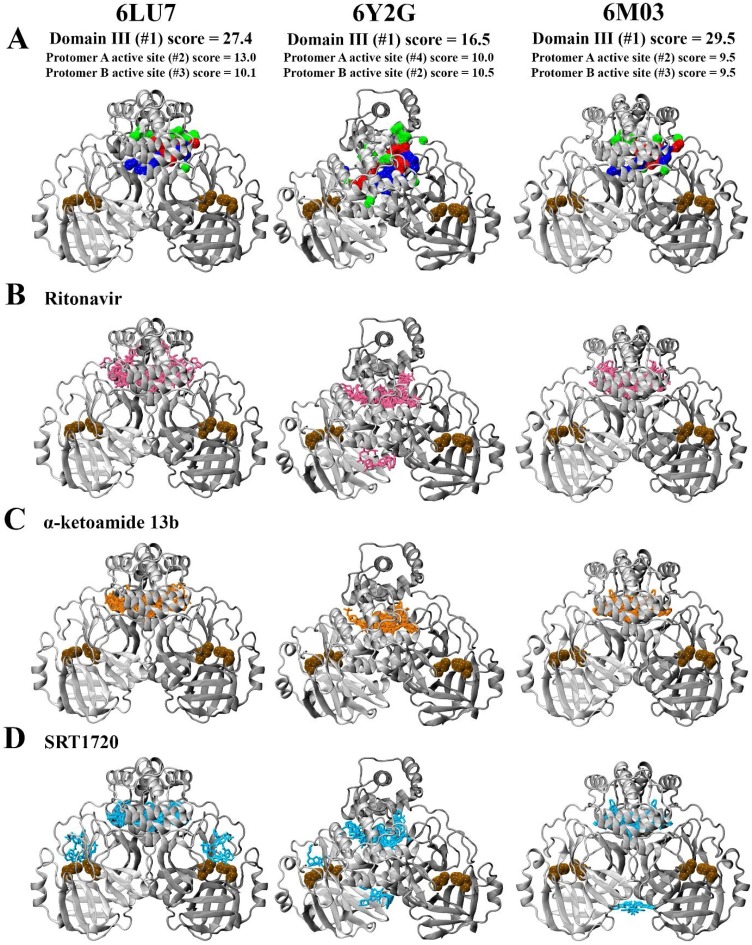
We extended this data to other available PDB structures of the Mpro including α-ketoamide 13b-bound 6Y2G (Zhang et al., 2020a) and apo 6M03 (Zhang et al., 2020b). The findings highlight a high similarity between 6LU7, 6Y2G, and 6M03 with similarity (QH) scores close to 1, and RMSD values of 0.85 and 0.49 Ǻ between the C-α atoms of 6LU7 superimposed with 6Y2G and 6M03, respectively (Fig. 4 A). Percentage identity between the structures was greater than 96 % (Roberts et al., 2006). When decomposed on a per-residue basis (Fig. 4B), RMSD between structures remained lower than 2 Ǻ in general, with the exception of C-terminal residues of each protomer. 6LU7 was fairly consistently similar to the apo 6M03 structure. While the α-ketoamide 13b-bound 6Y2G was also similar to 6LU7, slight fluctuations in RMSD were observed in loop residues of 6Y2G in protomer A at Leu50 (1.98 Ǻ) in domain I, and Asp216 (2.09 Ǻ) and Gly278 (2.02 Ǻ) in domain III. Larger RMSDs were observed in domain III of protomer B at residues Asn277 (4.51 Ǻ) and Gly278 (3.45 Ǻ). It should be noted that these domain III residues are located on an outer loop at the apex of the protein. Alignment of binding site residues indicated a low RMSD between C-α atoms, with values of less than 0.50 Ǻ across the three structures in both protomers (Fig. 4C). Further, docking to the active site of the different structures of SARS-CoV-2 Mpro indicated that ligands were consistently binding in proximity to key active site residues, including the catalytic dyad residues His41 and Cys145 (Fig. 5 ). A covalent bond is formed between α-ketoamide 13b and Cys145 in the original crystal structure (Zhang et al., 2020a), however we have focussed on non-covalent interactions to identify other potential binding sites on the SARS-CoV-2 Mpro. Using an additional ligand binding site prediction tool (PrankWeb) (Jendele et al., 2019), the findings indicate the highly reactive pocket in the region at the domain III apex (Fig. 6 ). This was consistent for all three structures of the Mpro. As noted above in Fig. 3, preferential binding of the ligands in this domain III pocket was observed for 6LU7, 6Y2G, and 6M03 (Fig. 6).
Fig. 4.
Comparison between structures of SARS-CoV-2 Mpro (PDB ID: 6LU7, 6Y2G, and 6M03). Protein structures are aligned, with qualitative similarity scores indicated A). The RMSD between the structures are separated on a per-residue basis in B). The binding sites of Mpro were also compared between the structures in C), with the binding site was defined by residues within 5 Ǻ of α-ketoamide 13b (orange) in the native crystal structure in each protomer of PDB ID 6Y2G. RMSD values shown are for the C-α atoms of the residues.
Fig. 5.
Comparison between docking to the active site of SARS-CoV-2 Mpro structures (PDB ID: 6LU7, 6Y2G, and 6M03) in protomers A and B. Hydrogen bonds are indicated by yellow, pi-pi stacking in cyan, and salt bridge formation shown by magenta lines.
Fig. 6.
Analysis of binding sites in structures of the SARS-CoV-2 Mpro (PDB ID: 6LU7, 6Y2G, and 6M03) using the PrankWeb Server A). The most likely binding pocket indicated by pocket score is depicted, highlighted in surface representation with non-polar residues in white, basic residues in blue, acidic residues in red, and polar residues in green. Blind docking of B) ritonavir, C) α-ketoamide 13b, and D) SRT1720 to the protease dimer is shown. Catalytic dyad residues His41 and Cys145 are indicated in ochre to denote the location of the active site.
Overall, our observations are in line with accumulating evidence which demonstrates the difficulty associated with targeting the SARS-CoV-2 Mpro with small molecule inhibitors (Bzowka et al., 2020). Indeed, the region between domains II and III, which is essential for dimerization is emerging as an important allosteric site (Bzowka et al., 2020). Further, the findings from direct inhibition of the SARS-CoV-2 Mpro by small molecules, and antiviral assays need to be considered (Zhang et al., 2020a; Dai et al., 2020; Jin et al., 2020). The SARS-CoV-2 Mpro and in vitro antiviral assays indicate a ligand concentration-dependence and biological effects with pronounced protease inhibition activity displayed by covalently-linked inhibitors such as α-ketoamide 13b (Zhang et al., 2020a; Dai et al., 2020; Jin et al., 2020). It will be interesting to see how small molecules interact with the active site and the potential sites that are distinct from this region, including the domain III interface. These observations can only be precisely reconciled with further in silico work, and biochemical and detailed ligand binding studies.
3.2. Molecular dynamics simulations highlight the stability of ligand complexes in the active site and in domain III of the SARS-CoV-2 Mpro
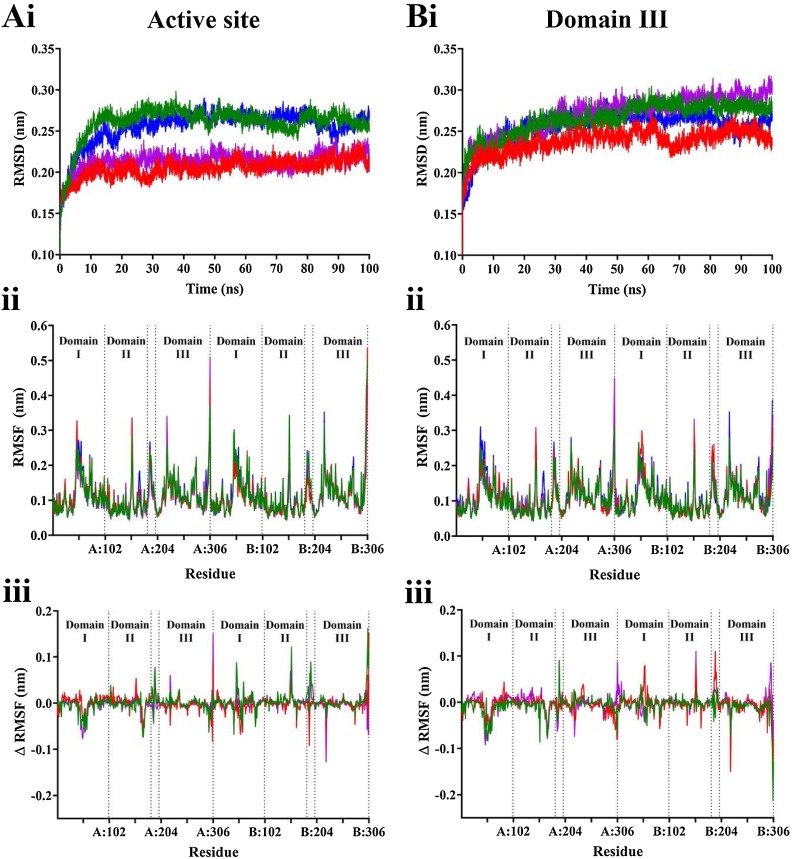
Classical molecular dynamics (MD) simulations were performed to assess the stability of the SARS-CoV-2 Mpro in complex with two ligands bound to active sites on each protomer of the dimer, and in complex with a single ligand bound to domain III. From root mean square deviation (RMSD) analysis, the protein structure reaches equilibrium after 40 ns when ligands are bound to the active site, and after 70 ns when ligands are bound to domain III (Fig. 7 A and B). Subsequent analysis was performed on the stabilised trajectory. As well as stabilising at a faster rate, the proteases complexed with the ligands in the active sites demonstrated lower average RMSD values: 0.21 compared to 0.26 nm for ritonavir, and 0.22 and 0.29 nm for α-ketoamide 13b, and to a lesser extent for SRT1720 with values of 0.26 nm and 0.28 nm for active site-bound and domain III-bound complexes, respectively. This suggests that binding to the active site may have a modest stabilising effect compared with binding to domain III. However, root mean square fluctuation (RMSF) analysis does not indicate that the differences can be attributed to particular regions, with similar trends in protein fluctuation across all ligands and their binding location (Fig. 7C–E). Visual analysis of the trajectories indicate that the docked ligands mainly remain stable in both the active sites and domain III of the protease (Movie S1-7).
Fig. 7.
Stability of SARS-CoV-2 Mpro complex with ligands bound to the A) active site, and B) domain III apex. Values for the apo protein are shown in blue, ritonavir in purple, α-ketoamide 13b in red, and SRT1720 in green. i) Average root mean square deviation (RMSD) was calculated for the protein with respect to backbone for triplicate 100 ns simulations. ii) Following stabilisation, average root mean square fluctuation (RMSF) for the whole protein was calculated. iii) RMSF difference was calculated by subtracting values from the apo form from the ligand bound forms of the protein.
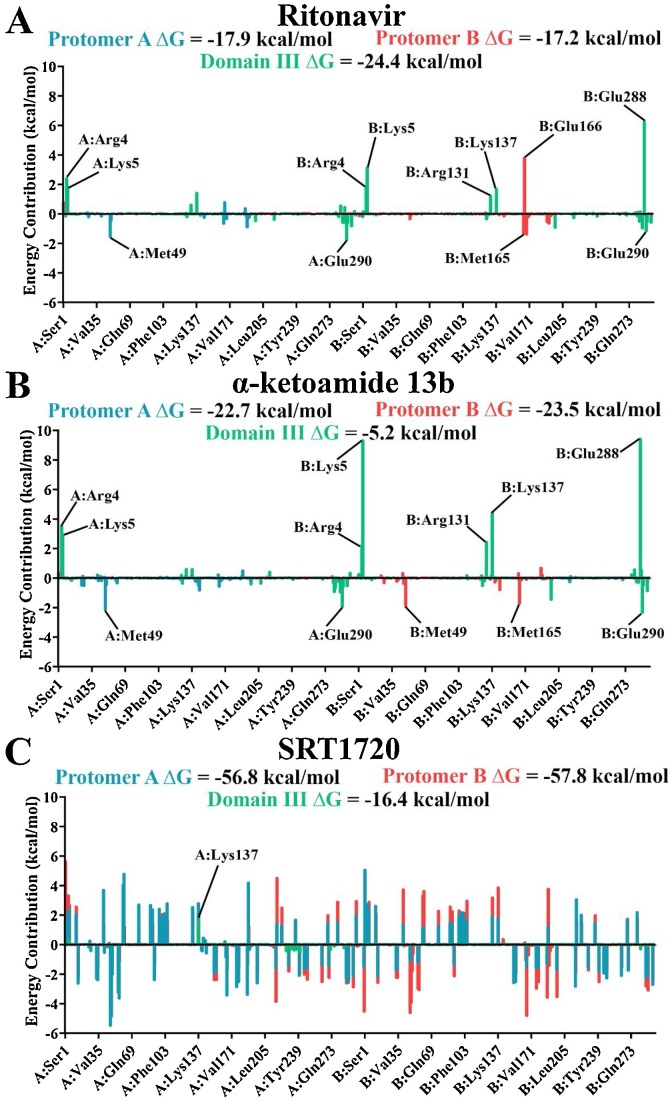
The complexes were further analysed using molecular mechanics-Poisson Boltzmann surface area (MM-PBSA) to calculate binding free energy. α-ketoamide 13b and SRT1720 bound with a stronger ΔG to the active site of the protease (α-ketoamide 13b: -22.7 and -23.5 kcal/mol to protomers A and B, compared to -5.2 kcal/mol to domain III; SRT1720: -56.8 and -57.8 kcal/mol to protomers A and B, compared to -16.4 kcal/mol to domain III) (Fig. 8 ), with SRT1720 demonstrating the strongest binding energy out of the ligands. Van der Waals forces were the main driving factor for ligand binding, along with contributions from electrostatic and solvent accessible surface area (SASA) energy (Table S1). When binding energy was decomposed into a per-residue basis, α-ketoamide 13b demonstrated clear trends with active site residues, such as Met49 and Met165, favourably contributing to binding in their respective protomers (Fig. 8B). Similarly binding of α-ketoamide 13b to domain III of the protease demonstrated slightly favourable contributions from Glu290. However, strongly unfavourable energy contributions from apex residues Arg4 and Lys5 on both protomers, and from Arg131, Lys137, and Glu288 on protomer B resulted in a weaker ΔG of α-ketoamide 13b. SRT1720 displayed large fluctuations across the protease when it was bound to the active site regardless of protomer (Fig. 8C). Overall, favourable energy contributions were more prominent, yielding strong binding free energies of SRT1720 to the active site. When bound to domain III, energy contribution peaks were less apparent, with the strongest contribution being unfavourable from Lys137 in protomer A, resulting in a weak binding energy.
Fig. 8.
Average energy contributions from MM-PBSA analysis were decomposed into a per-residue basis for binding of SARS-CoV-2 Mpro with A) ritonavir, B) α-ketoamide 13b, and C) SRT1720 to the active site in protomer A (blue), active site of protomer B (red), and domain III apex (green). Energy contributions were calculated in triplicate on 1000 ps segments of stabilised trajectories.
On the other hand, ritonavir demonstrated a stronger binding energy with domain III compared to the active site (−24.4 kcal/mol to domain III, −17.9 and −17.2 kcal/mol to protomers A and B, respectively). Decomposition of energy contribution from residues follows a similar pattern to that of α-ketoamide 13b (Fig. 8), with favourable energy contributions from active site residues Met49 and Met165, and unfavourable energy contributions from Glu166 in protomer B (Zhang et al., 2020a; Liang et al., 2020). Similar trends are also observed for binding to domain III, albeit with less pronounced unfavourable contributions from Lys5 and Glu288 of protomer B, contributing to a relatively stronger overall binding energy (Fig. 8A). This suggests that although ritonavir may bind strongly to the active site through molecular docking (Fig. 1), blind docking (Fig. 3, Fig. 5) and MM-PBSA analysis demonstrate a preference for binding to domain III instead, highlighting the importance of using multiple methods to verify potential lead compounds.
Overall, site-mapping highlights a highly reactive binding pocket at the dimerization apex (domain III) of the SARS-CoV-2 Mpro, and the attraction of small molecules to this site is consistent with the blind docking analyses. Regarding the preferential binding of small molecules in domain III, it will be interesting to determine if those ligands 1) impose allosteric effects, and 2) may potentially act to destabilize dimerization of the homodimeric SARS-Cov-2 Mpro.
Declaration of Competing Interest
The authors report no declarations of interest.
CRediT authorship contribution statement
Julia Liang: Data curation, Formal analysis, Writing - original draft, Writing - review & editing. Chris Karagiannis: Data curation, Formal analysis, Writing - review & editing. Eleni Pitsillou: Data curation, Formal analysis, Writing - review & editing. Kevion K. Darmawan: Methodology, Writing - review & editing. Ken Ng: Conceptualization, Supervision, Writing - review & editing. Andrew Hung: Conceptualization, Supervision, Writing - review & editing. Tom C. Karagiannis: Conceptualization, Supervision, Writing - review & editing.
Acknowledgements
We would like to acknowledge intellectual and financial support by McCord Research (Iowa, USA). JL and KKD are supported by an Australian Government Research Training Program Scholarship. We are indebted to Alfonso Perez Escudero and the team at Crowdfightcovid for enabling access to supercomputing facilities and to Matthew Gasperetti and the team at Hypernet Labs (Galileo), for enabling cloud computing for this project. We thank the National Computing Infrastructure (NCI), and in particular the Pawsey Supercomputing Centre (which provided specific time for COVID-19 related work on the Topaz GPU cluster), in Australia (funded by the Australian Government). We are grateful for the expert support provided by Marco De La Pierre (supercomputing application specialist). Further, we thank the Spartan High Performance Computing service (University of Melbourne), and the Partnership for Advanced Computing in Europe (PRACE) for awarding the access to Piz Daint, hosted at the Swiss National Supercomputing Centre (CSCS), Switzerland. All data is available in the main text or supplementary materials.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.compbiolchem.2020.107372.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Hui D.S., et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health - the latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V. Coronaviridae Study Group of the International Committee on Taxonomy of The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajgor D.D., Lee M.H., Archuleta S., Bagdasarian N., Quek S.C. The many estimates of the COVID-19 case fatality rate. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30244-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucinotta D., Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020;91:157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phua J., et al. Intensive care management of coronavirus disease 2019 (COVID-19): challenges and recommendations. Lancet Respir. Med. 2020;8:506–517. doi: 10.1016/S2213-2600(20)30161-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A.W., Chawla K.S., Turnbull Z.A. Transforming ORs into ICUs. N. Engl. J. Med. 2020;382:e52. doi: 10.1056/NEJMc2010853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E. The race for coronavirus vaccines: a graphical guide. Nature. 2020;580:576–577. doi: 10.1038/d41586-020-01221-y. [DOI] [PubMed] [Google Scholar]
- Cohen J. Vaccine designers take first shots at COVID-19. Science. 2020;368:14–16. doi: 10.1126/science.368.6486.14. [DOI] [PubMed] [Google Scholar]
- Taccone F.S., Gorham J., Vincent J.L. Hydroxychloroquine in the management of critically ill patients with COVID-19: the need for an evidence base. Lancet Respir. Med. 2020 doi: 10.1016/S2213-2600(20)30172-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funck-Brentano C., Salem J.E., Nguyen L.S., Drici M.D., Roden D.M. Response to the editorial “COVID-19 in patients with cardiovascular diseases”: Covid-19 treatment with hydroxychloroquine or chloroquine and azithromycin: A potential risk of Torsades de Pointes. Arch. Cardiovasc. Dis. 2020;113:367–368. doi: 10.1016/j.acvd.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antinori S., et al. Compassionate remdesivir treatment of severe Covid-19 pneumonia in intensive care unit (ICU) and Non-ICU patients: clinical outcome and differences in post_treatment hospitalisation status. Pharmacol. Res. 2020;104899 doi: 10.1016/j.phrs.2020.104899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulangu S., et al. A randomized, controlled trial of ebola virus disease therapeutics. N. Engl. J. Med. 2019;381:2293–2303. doi: 10.1056/NEJMoa1910993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P., et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P., et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020;11:222. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grein J., et al. Compassionate use of remdesivir for patients with severe Covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020 doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W., et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020 doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton A.T., Gentile F., Hsing M., Ban F., Cherkasov A. Rapid identification of potential inhibitors of SARS-CoV-2 main protease by deep docking of 1.3 billion compounds. Mol. Inform. 2020 doi: 10.1002/minf.202000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji M. Potential anti-SARS-CoV-2 drug candidates identified through virtual screening of the ChEMBL database for compounds that target the main coronavirus protease. FEBS Open Bio. 2020 doi: 10.1002/2211-5463.12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W., et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020 doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., et al. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020 doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Goyal B., Goyal D. Targeting the dimerization of main protease of coronaviruses: a potential broad-spectrum therapeutic strategy. ACS Comb. Sci. 2020 doi: 10.1021/acscombsci.0c00058. [DOI] [PubMed] [Google Scholar]
- Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- Chandwani A., Shuter J. Lopinavir/ritonavir in the treatment of HIV-1 infection: a review. Ther. Clin. Risk Manag. 2008;4:1023–1033. doi: 10.2147/tcrm.s3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C.M., et al. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004;59:252–256. doi: 10.1136/thorax.2003.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanakis N., et al. Virological and serological analysis of a recent Middle East respiratory syndrome coronavirus infection case on a triple combination antiviral regimen. Int. J. Antimicrob. Agents. 2014;44:528–532. doi: 10.1016/j.ijantimicag.2014.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan N., Sakthivel R., Velmurugan D., Gromiha M.M. Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. J. Biomol. Struct. Dyn. 2020:1–6. doi: 10.1080/07391102.2020.1752802. [DOI] [PubMed] [Google Scholar]
- Cao B., et al. A trial of Lopinavir-Ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung I.F., et al. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020 doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor R.K., et al. SRT1720 improves survival and healthspan of obese mice. Sci. Rep. 2011;1:70. doi: 10.1038/srep00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., et al. SRT1720 promotes survival of aged human mesenchymal stem cells via FAIM: a pharmacological strategy to improve stem cell-based therapy for rat myocardial infarction. Cell Death Dis. 2017;8:e2731. doi: 10.1038/cddis.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger. (LLC, New York 2018), vol.11.5.
- Krissinel E., Henrick K. Protein interfaces, surfaces and assemblies service PISA at European Bioinformatics Institute. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Sastry G.M., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- Kim S., et al. PubChem substance and compound databases. Nucleic Acids Res. 2016;44:D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho A.E., Guallar V., Berne B.J., Friesner R. Importance of accurate charges in molecular docking: quantum mechanical/molecular mechanical (QM/MM) approach. J. Comput. Chem. 2005;26:915–931. doi: 10.1002/jcc.20222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner R.A., et al. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- Bochevarov A.D., et al. Jaguar: a high‐performance quantum chemistry software program with strengths in life and materials sciences. Int. J. Quantum Chem. 2013;113:2110–2142. [Google Scholar]
- Halgren T.A. Identifying and characterizing binding sites and assessing druggability. J. Chem. Inf. Model. 2009;49:377–389. doi: 10.1021/ci800324m. [DOI] [PubMed] [Google Scholar]
- Halgren T. New method for fast and accurate binding-site identification and analysis. Chem. Biol. Drug Des. 2007;69:146–148. doi: 10.1111/j.1747-0285.2007.00483.x. [DOI] [PubMed] [Google Scholar]
- Jendele L., Krivak R., Skoda P., Novotny M., Hoksza D. PrankWeb: a web server for ligand binding site prediction and visualization. Nucleic Acids Res. 2019;47:W345–W349. doi: 10.1093/nar/gkz424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivák R., Hoksza D. P2Rank: machine learning based tool for rapid and accurate prediction of ligand binding sites from protein structure. J. Cheminform. 2018;10:39. doi: 10.1186/s13321-018-0285-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen H.J.C., van der Spoel D., van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995;91:43–56. [Google Scholar]
- Abraham M.J., et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2. 2015:19–25. [Google Scholar]
- Bjelkmar P., Larsson P., Cuendet M.A., Hess B., Lindahl E. Implementation of the CHARMM force field in GROMACS: analysis of protein stability effects from correction maps, virtual interaction sites, and water models. J. Chem. Theory Comput. 2010;6:459–466. doi: 10.1021/ct900549r. [DOI] [PubMed] [Google Scholar]
- Vanommeslaeghe K., et al. CHARMM General Force Field (CGenFF): A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoete V., Cuendet M.A., Grosdidier A., Michielin O. SwissParam: a fast force field generation tool for small organic molecules. J. Comput. Chem. 2011;32:2359–2368. doi: 10.1002/jcc.21816. [DOI] [PubMed] [Google Scholar]
- Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- Berendsen H.J.C., Postma J.P.M., Gunsteren W.Fv., DiNola A., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- Parrinello M., Rahman A. Crystal structure and pair potentials: a molecular-dynamics study. Phys. Rev. Lett. 1980;45:1196–1199. [Google Scholar]
- Hess B., Bekker H., Berendsen H.J., Fraaije J.G. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- Darden T., York D., Pedersen L. Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14(33–38):27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Roberts E., Eargle J., Wright D., Luthey-Schulten Z. MultiSeq: unifying sequence and structure data for evolutionary analysis. BMC Bioinformatics. 2006;7:382. doi: 10.1186/1471-2105-7-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari R., Kumar R., Lynn A. g_mmpbsa—a GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014;54:1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- Robert K., Nathan A.B., McCammon J.A. iAPBS: a programming interface to the adaptive Poisson–boltzmann solver. Comput. Sci. Discov. 2012;5 doi: 10.1088/1749-4699/5/1/015005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou T., Wang J., Li Y., Wang W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J. Chem. Inf. Model. 2011;51:69–82. doi: 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger. (LLC, New York 2018), vol. 11.5.
- Nayal M., Honig B. On the nature of cavities on protein surfaces: application to the identification of drug-binding sites. Proteins Struct. Funct. Bioinform. 2006;63:892–906. doi: 10.1002/prot.20897. [DOI] [PubMed] [Google Scholar]
- McCarthy M., Prakash P., Gorfe A.A. Computational allosteric ligand binding site identification on Ras proteins. Acta Biochim. Biophys. Sin. 2015;48:3–10. doi: 10.1093/abbs/gmv100. [DOI] [PubMed] [Google Scholar]
- Kim M., Lee K.W., Cho A.E. Elucidation of allosteric inhibition mechanism of 2-Cys human peroxiredoxin by molecular modeling. J. Chem. Inf. Model. 2012;52:3278–3283. doi: 10.1021/ci3004495. [DOI] [PubMed] [Google Scholar]
- Zhang B., Zhao Y., Jin Z., Liu X., Yang H., Rao Z. 2020. The Crystal Structure of COVID-19 Main Protease in Apo Form. [DOI] [Google Scholar]
- Bzowka M., et al. Structural and evolutionary analysis indicate that the SARS-CoV-2 mpro is a challenging target for small-molecule inhibitor design. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21093099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J., et al. Interaction of the prototypical α-ketoamide inhibitor with the SARS-CoV-2 main protease active site in silico: molecular dynamic simulations highlight the stability of the ligand-protein complex. Comput. Biol. Chem. 2020;107292 doi: 10.1016/j.compbiolchem.2020.107292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.