Graphical abstract
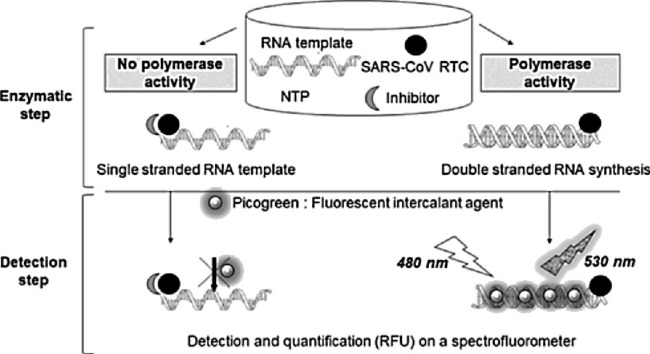
Principle of SARS-CoV RNA synthesis detection by a fluorescence-based High Throughput Screening assay.
During the enzymatic step, the RNA template was incubated with the SARS-CoV RTC, the nucleotides triphosphate and the compound to be tested (putative inhibitor). The polymerase activity released a double stranded RNA which was detected by adding a fluorescent intercalant agent (Picogreen®, ex: 480 nM, em: 530 nm). In presence of an inhibitor, the fluorescence signal decreased and is quantified.
NTP: Nucleotides Tri-Phosphate.
Keywords: Coronavirus, Small molecule, Screening, Replication complex
Highlights
-
•
A new non-radioactive SARS-CoV RNA polymerase assay was developed.
-
•
The robotized assay is suitable to identify RdRp inhibitors based on HTS.
-
•
A total of 1520 FDA-approved compounds were tested using the SARS-CoV-RTC assay.
Abstract
The Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) emergence in 2003 introduced the first serious human coronavirus pathogen to an unprepared world. To control emerging viruses, existing successful anti(retro)viral therapies can inspire antiviral strategies, as conserved viral enzymes (eg., viral proteases and RNA-dependent RNA polymerases) represent targets of choice. Since 2003, much effort has been expended in the characterization of the SARS-CoV replication/transcription machinery. Until recently, a pure and highly active preparation of SARS-CoV recombinant RNA synthesis machinery was not available, impeding target-based high throughput screening of drug candidates against this viral family. The current Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) pandemic revealed a new pathogen whose RNA synthesis machinery is highly (>96 % aa identity) homologous to SARS-CoV. This phylogenetic relatedness highlights the potential use of conserved replication enzymes to discover inhibitors against this significant pathogen, which in turn, contributes to scientific preparedness against emerging viruses. Here, we report the use of a purified and highly active SARS-CoV replication/transcription complex (RTC) to set-up a high-throughput screening of Coronavirus RNA synthesis inhibitors. The screening of a small (1520 compounds) chemical library of FDA-approved drugs demonstrates the robustness of our assay and will allow to speed-up drug discovery against the SARS-CoV-2.
1. Introduction
The current worldwide Coronavirus Disease 2019 (COVID-19) crisis highlights the urgent need of cost-effective drugs able to control the SARS-CoV-2 prophylactically or at the earliest stage of infection (Wu et al., 2020; Zhou et al., 2020). Towards this endeavor, one of the most potentially rapid ways is to screen chemical libraries of FDA-approved compounds in order to accelerate clinical evaluation (Cheng et al., 2019).
Target-based screening generally proposes to select ligands or inhibitors through the use of the purified target known to play a critical role for pathogen propagation. Viral RNA-dependent RNA polymerases (RdRp) represent an extremely attractive target for antiviral drugs due to their high level of sequence and structural conservation and vital role in viral replication (Venkataraman et al., 2018). This has been beautifully demonstrated by, amongst others, the design of Sofosbuvir, a nucleotide analogue occupying the foremost position in the fight against the hepatitis C virus (HCV) (Ji et al., 2019).
The situation is blatantly different for SARS-CoV, whose drug market has remained difficult to evaluate after its emergence in 2002 and subsequent vanishing in the middle of year 2003 (Pruijssers and Denison, 2019). Another reason for the lagging research of SARS-CoV is its sophisticated and multi-subunit RNA synthesis machinery (Snijder et al., 2003). It was only in 2014 that nsp7 and nsp8 were identified as co-factors of the RdRp core nsp12 and shown to confer full activity and processivity to nsp12 (Subissi et al., 2014).
These findings were corroborated by the Cryo-EM structure determination of the nsp7-8-12 SARS-CoV RNA synthesis complex in 2019, followed by that of SARS-CoV-2 in 2020 (Gao et al., 2020; Kirchdoerfer and Ward, 2019). The structure shows the nsp12 RdRp core bound to one molecule of nsp7 and two molecules of nsp8. Furthermore, the structure described by Hillen et al. suggest that nsp8 acts as a “sliding pole” for processivity of RNA synthesis (Hillen et al., 2020). Although it is not known if this structure corresponds to one of the biologically relevant form(s) of the SARS-CoV Replication Transcription Complex (RTC), this nsp7-8-12 complex represents so far the minimum RTC required for synthesis of the coronavirus ∼30,000 nt RNA genome.
The SARS-CoV and SARS-CoV-2 viruses are highly homologous, with a 79 % overall amino acid sequence identity along the genome (Wang et al., 2020). Remarkably, the SARS-CoV nsp7, nsp8 and nsp12 amino acid sequences are 98.8 %, 97.4 %, and 96 % identical to that of SARS-CoV-2, and changes are conservative. The genome is translated into two large polyproteins originating from Orf1a and Orf1b, the latter being expressed from a ribosomal frameshift occurring at the end of Orf1a. Orf1b codes for a set of five conserved replication proteins, among which nsp12 carries the polymerase activity when supplemented with Orf1a products nsp7 and nsp8. In the core polymerase domain, amino acid differences between SARS-CoV-1 and 2 map to the surface of the protein, not onto any of the conserved motif A to G, indicating that SARS-CoV and SARS-CoV-2 RNA synthesis properties should be nearly identical (Shannon et al., 2020).
In this paper, we use the nsp7, nsp8, and nsp12 proteins of the SARS-CoV to assemble a highly active RNA synthesis complex. We optimize reaction conditions to set-up a non-radioactive nucleotide polymerization assay with a signal-to-noise ratio appropriate for a robotized high-throughput screening (HTS) assay. We validate this HTS assay through a screening campaign using the Prestwick Chemical Library® of 1520 FDA-EMA approved compounds, and report detailed inhibition profiles of two series of hits.
2. Materials and methods
2.1. Products and reagents
Homopolymeric adenine (Poly (A)) RNA template was obtained from GE Healthcare. Hinokiflavone (ref 1017), Apigenine (ref 1102S) and Amentoflavone (ref 1057S) were purchased from Extrasynthese and from SigmaAldrich for Quercetin (ref Q4951). Compounds were resuspended in 100 % DMSO at 20 mM and stored at −20 °C. 3’dUTP (ref N-3005) was purchased from Trilink. Quant-it Picogreen® dsDNA assay kit (ref P11496) was obtained from Thermofisher scientific.
2.2. Cloning, expression and proteins purification
The fusion protein nsp7-nsp8 was generated by inserting a GSGSGS linker sequence between the nsp7 and nsp8 coding sequences, and was named nsp7L8 (Subissi et al., 2014) (Fig. A1). The nsp7L8 and nsp12 proteins were produced and purified independently. Competent cells Escherichia coli C2523 or BL21 (New England Biolabs) were transformed with the recombinant plasmid pQE30-nsp7L8 or pj404-nsp12 respectively. The expressed constructs contained a poly-histidine tag (6His for nsp7L8 and 8His for nsp12) and a Tobacco Etch Virus (TEV) cleavage site at the carboxy-terminal. The transformation was plated on Luria Broth (LB) agar with 100 μg/ml Ampicillin and 17 μg/ml Chloramphenicol. One isolated clone from the transformation was used to start a preculture (LB medium, 100 μg/ml Ampicillin, 17 μg/ml Chloramphenicol) overnight at 37 °C. Afterward, the preculture was used to start the culture with LB medium and then grown at 37 °C until the optical density at 600 nm reached 0.6 to 0.8. Expression was then induced by using Isopropyl β-d-1-thiogalactopyranoside (IPTG) at 100 μM (nsp7L8) or 250 μM (nsp12) final concentration and incubated overnight at 17 °C or 23 °C for nsp7L8 and nsp12 respectively. Nsp12 cultures were supplemented with ethanol 2 % final concentration. The cells were harvested by centrifugation and then frozen and stored at −20 °C until they were used. Proteins were purified in a two-step chromatography. Buffer compositions were the same for both nsp7L8 and nsp12, except adding glycerol 10 % final concentration in the case of nsp12. Briefly, cells were resuspended by using lysis buffer (Tris 50 mM pH 8.0, NaCl 300 mM, Imidazole 10 mM, β-Mercapto-ethanol 5 mM). Lysozyme (0.25 mg/ml final), Benzamidine 0.2 mM, MgSO4 20 mM and DNase (10 μg/ml final) were added to the suspended lysate following by incubation at 4 °C for 30 min. Powder NaCl was added directly into the lysate to reach final concentration of 1 M and incubation at 4 °C for 40 more minutes. Lysates were then sonicated 3 times and centrifuged for 30 min at 4 °C. The supernatant was purified by metal affinity chromatography using HisPur Cobalt resin (Thermo scientific) equilibrated with buffer A (Tris 50 mM pH 8.0, NaCl 300 mM, Imidazole 10 mM, β-Mercapto-ethanol 5 mM). Washing step was performed by using 5 column volumes (CV) of buffer A with 1 M NaCl, then 5 CV of buffer A. Protein was eluted by using buffer A containing 500 mM of Imidazole instead of 10 mM. Eluted protein was concentrated using Amicon Ultra falcon to 1 mg/ml–2 mg/ml. The protein was finally purified by gel filtration chromatography HiLoad S200 26/60 using GF buffer (Hepes 25 mM pH 8.0, NaCl 300 mM, MgCl2 5 mM, TCEP 5 mM). The protein was concentrated to obtain a final concentration more than 20 μM for enzymatic activity tests. Sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) was used to follow the different steps of the purification and check the size and purity of final protein.
Fig. A1.
Complete amino acid sequence of nsp7L8.
The fusion protein nsp7-nsp8 was generated by inserting a GSGSGS linker sequence, underlined, between the nsp7 and nsp8 coding sequences, and is named nsp7L8.
2.3. Setting up of the RTC activity experimental conditions based on a fluorescent readout
2.3.1. Determination of the Poly(A) template concentration
Picogreen kinetic assay was based on polymerase activity of SARS nsp12 in complex with nsp7L8, which catalyzed the reaction using a poly (A) template and uridine triphosphate (UTP). The reaction (160 μl) was carried out at 30 °C in 50 mM Hepes pH 8, 10 mM DTT, 10 mM KCl, 2 mM MgCl2, 2 mM MnCl2, 500 μM UTP and 150 nM nsp12 as final concentrations. The final Poly(A) concentration varied from 5 nM to 500 nM. To reconstitute an active replicase, nsp7L8 was used in a 10-fold molar concentration excess compared to nsp12 as described. Nsp12 and nsp7L8 were incubated for 5 min in the presence of Poly(A) before starting the reaction with UTP addition.
30 μl of Ethylenediaminetetraacetic acid (EDTA) 100 mM were added into each well of a 96-well black flat bottom plate (Greiner Bio-One ref 675076). At each time interval (5;7.5;10;15;20;30 and 45 min), 20 μl of reaction was added into wells on the plate with EDTA to stop the reaction. The plate was then incubated 5 min in the dark with 1/800 Picogreen® in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.5). The plate was read on Tecan Safire II using software Magellan 6 (excited light at 480 nm, emitted light at 530 nm, optimal gain).
The assay was done in triplicate. Velocity values of each condition were determined using the Prism software by calculating the slope of the linear phase and then plotted against the Poly(A) template concentration to determine the Km (Poly(A)) and Vmax values by using Michaelis-Menten fitting.
2.3.2. Optimizing UTP concentration
UTP variation assay was performed in the same conditions as described above, with the defined Poly(A) optimum concentration of 350 nM, except for the final UTP concentration, which varied from 50 μM to 1 mM. At each time interval (5;7.5;10;15;20; 30 and 45 min), 20 μl of reaction was added into wells on the plate with 100 mM EDTA to stop the reaction. The assay was tested three times., Velocity values of each condition were determined using the Prism software by calculating the slope of the linear phase and then plotted against the UTP concentration to determine the apparent Km app (UTP) value by using Michaelis-Menten fitting or Hill fitting.
2.3.3. Optimizing enzyme concentration
Enzyme variation assays were performed in the same conditions as described above (Poly(A) 350 nM, UTP 500 μM) except for the final enzyme concentration, which varied from 20 nM to 250 nM. Background and optimal gain values (given by TecanSafire 2) of each assay were determined and analyzed, using Prism software, to obtain the final best condition.
2.3.4. Time course of the SARS-COV RTC assay
To assess the optimized conditions of the polymerase activity of the SARS nsp12-nsp7L8 complex, a reaction time course was performed using 350 nM poly (A) template, 500 μM UTP and 150 nM nsp12 enzyme, testing nsp12 alone, nsp7L8 alone or the nsp12/nsp7L8 complex. Nsp7L8 was in a 10-fold molar concentration excess compared to nsp12 concentration. Nsp12 and nsp7L8 were incubated at room temperature for 5 min in the presence of Poly (A) before starting the reaction by the addition of UTP. As a comparison, the assay was performed with the DV-2 polymerase (100 nM Poly (U) template, 300 μM ATP, 10 nM Enzyme as already described (Benmansour et al., 2016). Experiments, performed in triplicate, were analyzed using the Prism 5 software.
2.4. Robotization of the RTC assay
The assay was performed in 96-well Nunc plates. The chemical library is from Prestwick Chemical. The 1520 compounds were distributed in 19 plates, with 80 compounds per plate and the first and 12th columns with 5 % DMSO. Each of the 80 compounds were added to the reaction mix using a BioMek I5 workstation (Beckman) to a final concentration of 20 μM in 5 % DMSO. Reactions were conducted in 40 μL final volume. The enzyme mix containing both nsp12, nsp7L8 and the Poly (A) template was incubated 5 min at room temperature to form the active complex. For each assay, 20 μL of this enzyme mix was distributed in wells using a Biomek 4000 (Beckman), containing 2 μL of the compounds. Reactions were initiated by addition of 18 μL of the nucleotide mix (500 μM UTP) and incubated at 30 °C for 20 min. Assays were stopped by the addition of 20 μL of EDTA (100 mM). Reaction mixes were transferred to a Greiner plate using a Biomek I5 Automate (Beckman). Picogreen fluorescent reagent was diluted to 1/800 in TE buffer according to manufacturer’s data, and 60 μL of reagent was distributed into each well of the Greiner plate. The plate was incubated for 5 min in the dark at room temperature, and the fluorescence signal was read at 480 nm (excitation) and 530 nm (emission) using a TecanSafire2.
Positive and negative controls consisted respectively of a reaction mix with 5 % DMSO final concentration or EDTA 100 mM or Hinokiflavone 10 μM instead of compounds.
For each compound, the percentage of inhibition was calculated as follows: Inhibition % = 100 (raw_data_of_compound − av(pos)/(av(neg) − av(pos)).
Compounds leading to a 30 % inhibition or more at 20 μM were selected to further investigations.
The Z’ factor was calculated using the following equation: Z’ = 1 – [3(SD of max) + 3(min SD)] /[(mean max signal) - (mean min signal)], where SD is the standard deviation.
2.5. IC50 determination based on the fluorescent Picogreen® assay
The compound’s concentration leading to a 50 % inhibition of polymerase mediated RNA synthesis was determined in IC50 buffer (50 mM HEPES pH 8.0, 10 mM KCl, 2 mM MnCl2, 2 mM MgCl2, 10 mM DTT) containing 350 nM of Poly(A) template, 150 nM of nsp12 in complex with 1.5 μM nsp7L8 using 7 different concentrations of compound. Five ranges of inhibitor concentration were available (0,01–5 μM / 0,1–50 μM / 0,5–50 μM / 1–100 μM / 5–400 μM). According to the inhibitory potency of the compound tested, a range was selected to determine the IC50. Reactions were conducted in 40 μl on a 96-well Nunc plate. All experiments were robotized by using a BioMek 4000 automate (Beckman). 2 μl of each diluted compound in 100 % DMSO were added in wells with the selected concentration (5 % DMSO final concentration). For each assay, the enzyme mix was distributed in wells after a 5 min incubation at room temperature to form the active complex. Reactions were started by the addition of the UTP mix and were incubated at 30 °C for 20 min. Reaction assays were stopped by the addition of 20 μl EDTA 100 mM. Positive and negative controls consisted respectively of a reaction mix with 5 % DMSO final concentration or EDTA 100 mM instead of compounds. Reaction mixes were then transferred to Greiner plate using a Biomek I5 automate (Beckman). Picogreen® fluorescent reagent was diluted to 1/800° in TE buffer according to the data manufacturer and 60 μl of reagent were distributed into each well of the Greiner plate. The plate was incubated 5 min in the dark at room temperature and the fluorescence signal was then read at 480 nm (excitation) and 530 nm (emission) using a TecanSafire2.
IC50 was determined using the equation: % of active enzyme = 100/(1+(I)2/IC50), where I is the concentration of inhibitor and 100 % of activity is the fluorescence intensity without inhibitor. IC50 results were determined from average of triplicates using curve-fitting Prism software.
3. Results
3.1. Determination of optimized experimental conditions of the SARS-CoV RTC fluorescent assay
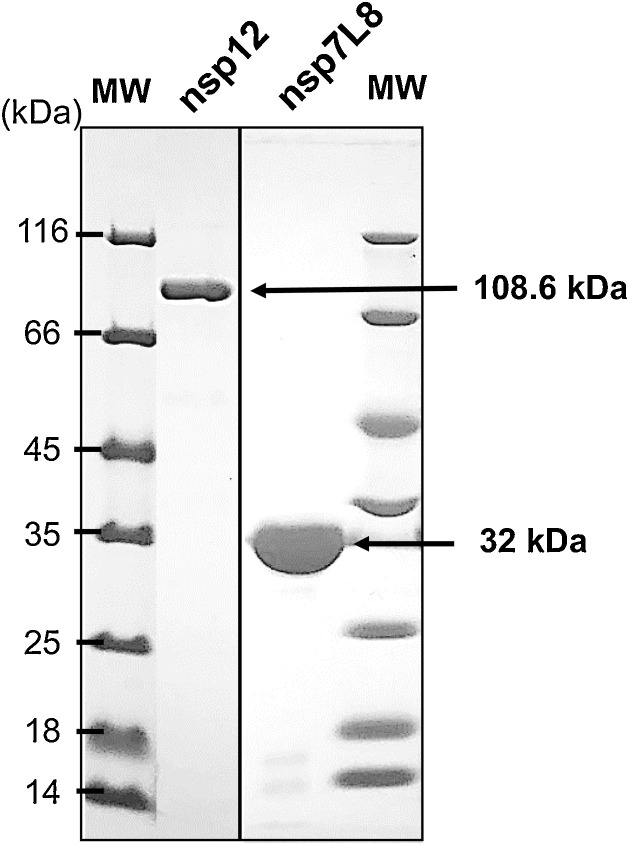
The nsp12 RdRp activity is dependent on the presence of the nsp7 and nsp8 proteins (Subissi et al., 2014). Since the exact and biologically relevant stoichiometry of the complex is unknown, we made use of a 1:10 ratio of nsp12:nsp7L8 which provides appropriately high levels of RdRp activity. A HTS assay was developed with a non-radioactive readout. We used conditions similar to the HTS Dengue Virus (DV) polymerase assay previously developed on our platform (Benmansour et al., 2016). Briefly, the homo-polymeric NTP template is incubated with nsp12:nsp7L8, and the polymerase activity is detected through a fluorescent dye which intercalates upon the synthesis of double stranded RNA (dsRNA). Nsp12 protein and nsp7L8 complex were expressed and purified separately (Fig. A2).
Fig. A2.
Characterization of the purified recombinant SARS nsp12 and nsp7L8 proteins.
5 μg of SARS nsp12 and 5 μg of nsp7L8 were loaded on a 10 % and 12 % SDS-PAGE gel respectively, stained by Coomassie blue. (MW, molecular size markers).
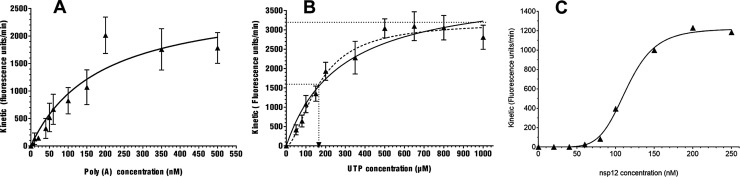
We first evaluated the effect of the poly(A) concentration. Maximal SARS-CoV RTC activity was obtained with a Poly(A) concentration of 350 nM, allowing a stable and reproducible signal (Fig. 1 A). To define the optimal UTP concentration, the velocity of the RTC was assessed at different UTP concentrations. We observed a non Michaelis-Menten curve with a significant lag-phase (Fig. 1B, full line), and therefore it was not possible to calculate a reliable Km concentration for the robotized assay using standard kinetic modeling. Using a Hill equation fit, we observed an allosteric cooperation with a Hill coefficient of 1.72 (Fig. 1B, dotted line). The UTP concentration to be used was defined at 500 μM, as it corresponds to the beginning of the saturation phase. Finally, to reach 80 % of the maximum activity, the nsp12 concentration was set to 150 nM (Fig. 1C).
Fig. 1.
Setting up of the SARS-CoV-RTC activity experimental conditions based on a fluorescent readout.
(A)Poly(A) template variation with SARS nsp12 in complex with nsp7L8.
Velocity values of SARS nsp12 (150 nM) in complex with nsp7L8 (1.5 μM) was measured for various concentrations in Poly (A) template (5; 10; 20; 40; 60; 80; 100; 150; 200; 350; 500 nM). Using the Prism software, velocity values of each condition were determined by calculating the slope of the linear phase of the kinetic and then plotted against the Poly(A) template concentration by using Michaelis-Menten fitting. Data were the results of three independent experiments.
(B) Nucleotide variation with SARS nsp12 in complex with nsp7L8.
Velocity values of SARS nsp12 (150 nM) in complex with nsp7L8 (1.5 μM) was measured for various concentrations in UTP (50;80;100;150;200;350;500 ;650;800 and 1000 nM). Using the Prism software, velocity values of each condition were determined by calculating the slope of the linear phase of the kinetic and then plotted against the UTP concentration by using Michaelis-Menten equation (full line) or Hill equation (dot line) to determine the apparent Km (UTP) of the SARS-CoV-RTC. Data were the results of three independent experiments.
(C) Variation in SARS nsp12 in complex with nsp7L8.
Velocity values obtained during a time course with 350 nM Poly (A) and 500 μM UTP were plotted against different concentrations in SARS nsp12 in complex with nsp7L8. The concentration ratio nsp12:nsp7L8 was conserved at 1:10. Curve was fitted according the Hill equation.
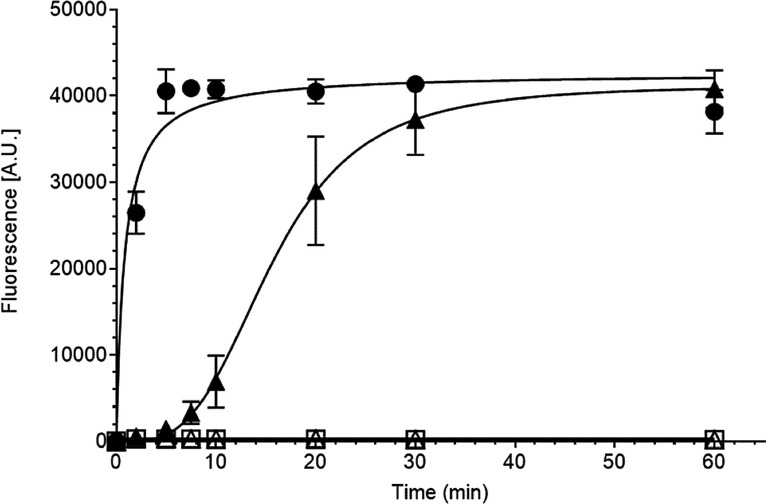
Using the conditions established above (350 nM poly(A) template, 500 μM UTP with 150 nM nsp12 and 1.5 μM nsp7L8) a time course up to 60 min was performed (Fig. 2 ). In contrast to the DV2 polymerase, the SARS-CoV RTC complex exhibits a significant lag phase (Fig. 2). The observed lag phase is anticipated to be due to the RTC complex formation, nsp12:nsp7L8 (Shannon et al., 2020). To obtain sufficient levels of activity, a reaction time of 20 min. was determined to be suitable for inhibitor screening. As expected, nsp12 alone, or nsp7L8 alone, in the same conditions, does not exhibit any RNA synthesis activity (Fig. 2).
Fig. 2.
SARS nsp12 in complex with nsp7L8 polymerase activity on a Picogreen kinetic assay.
The polymerase activity of 100 nM DV2-NS5 pol (●), 150 nM nsp12 in complex with 1.5 μM nsp7L8 (▲), 150 nM nsp12 alone (Δ) or 1.5 μM nsp7L8 alone (□) were measured in a time course (0;2;5;7.5;10;20;30 and 60 min). The produced doubled strand RNA was detected by adding an intercalant reagent (Picogreen®) and by measuring the fluorescence emission at 530 nM. Each assay was performed three times (mean value ± SD).
3.2. IC50 determination of reference inhibitors of the SARS-CoV RTC
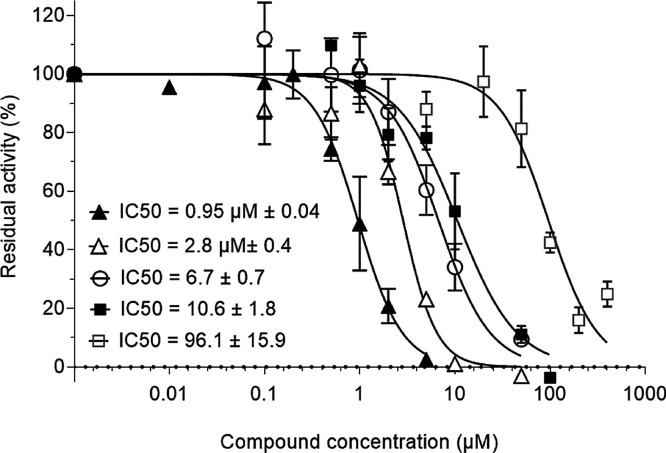
To develop the HTS assay of the SARS-CoV RTC, inhibitors of its activity had to be selected. In absence of specific inhibitors of the SARS-CoV RTC we first tested a nucleotide analogue (NA), 3’dUTP, along with a large spectrum of non-specific RNA synthesis inhibitors: Hinokiflavone, Amentoflavone, Quercetin and Apigenin (Coulerie et al., 2012). IC50s values range from 0.95 μM for Hinokiflavone to 96 μM for Apigenin (Fig. 3 ), demonstrating the discriminating capacity of the HTS assay. The 3’dUTP IC50 is measured at 6.7 μM under our experimental conditions.
Fig. 3.
Quantification of SARS-CoV-RTC enzymatic activity inhibitors.
Increasing concentrations of Hinokiflavone (▲), Amentoflavone (Δ), 3’dUTP (○), Quercetin (◼) and Apigenin (□) were incubated with 150 nM nsp12, 1.5μ M nsp7Lnsp8, 500 μM UTP, 350 nM Poly (A) at 30 °C during 20 min. The Inhibitory concentrations 50 (IC50s) were then calculated using graphPad Prism equation (Experiments were done twice in triplicate; Mean value ± SD).
3.3. High-throughput screening
The total of 1520 compounds from the Prestwick Chemical Library® (PCL) were tested using the SARS-CoV-RTC assay. The PCL® is a unique collection of small molecules, mostly approved drugs (FDA, EMA and other agencies) selected by a team of medicinal chemists and pharmacists for their high chemical and pharmacological diversity as well as for their known bioavailability and safety in humans.
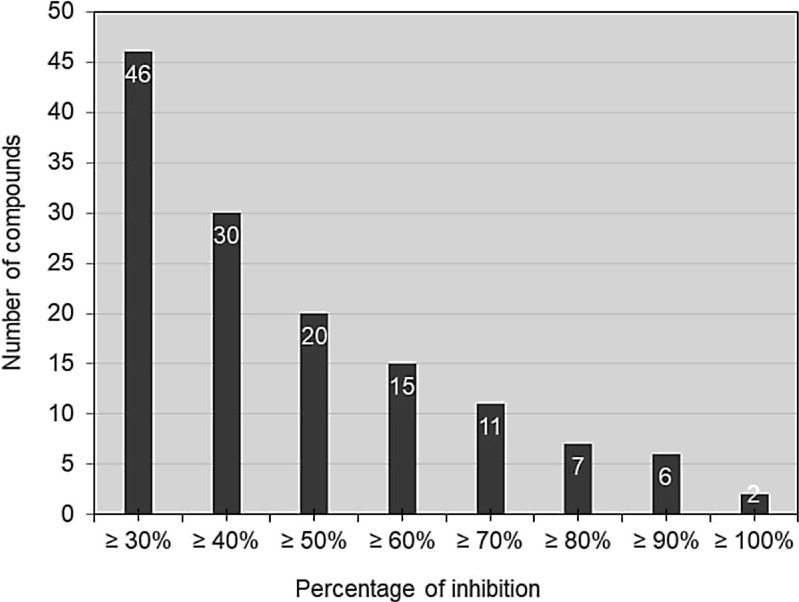
The Z’ value was calculated based on the ten control wells on each microplate, resulting in an overall Z’-score of 0,8 +/−0,06 for all the chemical library used in the screen. The screening was performed using 20 μM of compounds in 5 % DMSO in a single assay. Based on a cut-off of 30 % inhibition, the calculated hit rate was 3 % and the repartition of identified compounds according their percentage of inhibition was indicated in Fig. 4 . Based on these results, two main families of molecules were identified, anthracyclines and tetracyclines, which were further investigated (Table 1, Table 2 ).
Fig. 4.
Number of compounds from PCL on the screening of SARS-CoV RTC, according to their inhibitory potential.
Based on their efficiency on the SARS-CoV RTC assay, the number of compounds with more of 30 %; 40 %; 50 %; 60 %; 70 %; 80 %; 90 % and 100 % inhibition were evaluated. The exact value was indicated in white above each bar graph. (Only compounds with more 30 % inhibition of the polymerase activity in the assay were represented. Frequent Hitter and fluorescent compounds were excluded). The Z’ value was calculated based on the ten control wells on each microplate, resulting in an overall Z’ score of 0,8 ± 0,06.
Table 1.
Percentage of inhibition and Inhibitory Concentration 50 of compounds related to the anthracyclin chemical family.
| Compound | % Inhibition at 20 μM | IC50 (μM) |
|---|---|---|
| Prestw-385 | 101.12 % | 0.34 ± 0.06 |
| Prestw-1752 | 41.13 % | 15.2 ± 3.4 |
| Prestw-1224 | 50.08 % | 20.4 ± 4.4 |
| Prestw-438 | 52.12 % | 26.2 ± 7.2 |
| Prestw-487 | 64.84 % | 44.5 ± 6.5 |
| Prestw-1399 | 23.72 % | N.D. |
| Prestw-1167 | 17.63 % | N.D. |
| Prestw-1703 | 0 % | N.D. |
The compounds are listed according to their inhibitory effect (N.D.: Not Determined).
Table 2.
Percentage of inhibition and Inhibitory Concentration 50 of compounds related to the tetracyclin chemical family.
| Compound | % Inhibition at 20 μM | IC50 (μM) |
|---|---|---|
| Prestw-456 | 40.94 % | 3.2 ± 0.3 |
| Prestw-1799 | 46.92 % | 5.7 ± 1.4 |
| Prestw-1000 | 74.77 % | 22 ± 1.3 |
| Prestw-145 | 65.99 % | 22.8 ± 1.9 |
| Prestw-140 | 43.79 % | 77.1 ± 12.7* |
| Prestw-1399 | 23.72 % | N.D. |
| Prestw-964 | 8.82 % | N.D. |
| Prestw-315 | 5.75 % | N.D. |
| Prestw-307 | 1.42 % | N.D. |
| Prestw-753 | 0 % | N.D. |
The compounds are listed according to their inhibitory effect.
(* approximate IC50): (N.D.: Not Determined).
3.4. IC50 determination
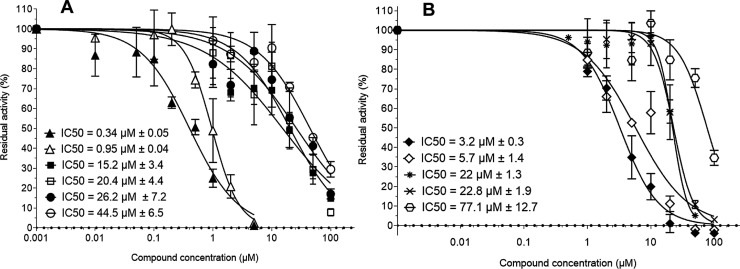
Eight anthracyclins were identified with inhibitory potential from zero to 100 % at 20 μM (Table 1). Regarding the five determined IC50s, these correlated perfectly with the calculated percentage of inhibition at 20 μM of the HTS assays. Four of the five anthracyclins tested were in the same range of IC50s (from 15 to 44 μM). The remaining one, Prestw-385, exhibited a sub-micromolar IC50 (0.34 μM ± 0.05) which was in the same range as Hinokiflavone, the inhibitor control (Fig. 5 A).
Fig. 5.
Quantification of SARS-CoV-RTC enzymatic activity inhibitors belonging to the anthracycline and tetracyclin chemical families.
(A) Anthracyclin chemical family. Increasing concentrations of Prestw-385 (▲), Prestw-1752 (◼), Prestw-1224 (□), Prestw-438 (●) and Prestw-487 (○) were used to determine the IC50s of each compound. Hinokiflavone (Δ) was indicated as a control.
(B) Tetracycline chemical family. Increasing concentrations of Prestw-456 (♦), Prestw-1799 (◊), Prestw-1000 (*), Prestw-145 (X) and Prestw-140 () were used to determine the IC50s of each compound.
Compounds were incubated with 150 nM nsp12, 1.5 μM nsp7Lnsp8, 500 μM UTP, 350 nM Poly (A) at 30 °C during 20 min. The IC50s were then calculated using graphPad Prism equation (Experiment were done twice in triplicate; Mean value ± SD). IC50: concentration for 50 % inhibition. The IC50 of Prestw-140 was approximate.
Ten tetracyclins were identified in the PCL, with inhibitory potential up to 74 % at 20 μM (Table 2). Again, the calculated IC50 values for these compounds correlated perfectly with the percentage of inhibition at 20 μM with the HTS assay. Three tetracyclins show IC50s of 20 μM or higher (Prestw-1000; Prestw-145; Prestw-140) while Prestw-456 and Prestw-1799 exhibit lower IC50s of 3.2 and 5.7 μM respectively, comparable to the 3’dUTP IC50 (6.7 μM) (Fig. 5B).
4. Discussion
In this study we have established a robust HTS assay with a fluorescent readout for the SARS-CoV RTC. The overall hit rate for the PCL screen was ∼3 %, with a cut-off 30 % of inhibition. Obviously, at this stage the identified hits are active only on the SARS-CoV RTC and have by no means been deemed suitable for medication.
Regarding the selected two families of molecules, the anthracyclins and the tetracyclins, the number of identified molecules did not allow Structure Activity Relationship (SAR) studies. Rather, it is important to outline the accuracy of the IC50s of the primary screening, as well as the reproducibility of the IC50 s regarding the calculated standard errors. With the exception of the anthracyclins, which have been described as potential intercalating molecules, it is not known at this stage if these compounds are indeed intercalating, denaturing, or low specificity agents having a mechanism of action suitable for advanced drug design.
In summary, this work, to the best of our knowledge, is the first description of a robust HTS assay based on a SARS-CoV RTC. It provides a new strategy for the rapid identification of potential anti-SARS inhibitors. The next steps already under development include increasing the output by scaling up the assay to 384 and potentially 1536 wells plates. Furthermore, the screening of large chemical libraries derived from innovative approaches as virtual screenings or Protein Protein Interaction inhibitors (iPPI) will broaden the scope of potential antivirals and constitute a key step to speed up these objectives. Identification of candidate anti-coronavirus drugs, based on a diversity of chemical libraries, should expedite drug discovery and design, and constitute an invaluable help to confirm the target of hits identified during cell-based or phenotypic screens.
CRediT authorship contribution statement
Cecilia Eydoux: Conceptualization, Data curation, Formal analysis, Writing - original draft, Project administration, Methodology. Veronique Fattorini: Methodology, Writing - review & editing. Ashleigh Shannon: Methodology, Writing - review & editing. Thi-Tuyet-Nhung Le: Methodology. Bruno Didier: Data curation. Bruno Canard: Conceptualization, Funding acquisition, Project administration, Writing - original draft. Jean-Claude Guillemot: Conceptualization, Formal analysis, Funding acquisition, Writing - original draft, Project administration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the Fondation pour la Recherche Médicale (Aide aux équipes), the SCORE project H2020 SC1-PHE-Coronavirus-2020 (grant#101003627), Inserm through the REACTing initiative (REsearch and ACTion targeting emerging infectious diseases), and the ANR-Flash-COVID (ANR-20-COVI-0047-01, TAMAC), supported by the Fondation de France. We thank ML Jung, E. Decroly for helpful comments.
Appendix A
.
References
- Benmansour F., Eydoux C., Querat G., de Lamballerie X., Canard B., Alvarez K., Guillemot J.-C., Barral K. Novel 2-phenyl-5-[(E)-2-(thiophen-2-yl)ethenyl]-1,3,4-oxadiazole and 3-phenyl-5-[(E)-2-(thiophen-2-yl)ethenyl]-1,2,4-oxadiazole derivatives as dengue virus inhibitors targeting NS5 polymerase. Eur. J. Med. Chem. 2016;109:146–156. doi: 10.1016/j.ejmech.2015.12.046. [DOI] [PubMed] [Google Scholar]
- Cheng Y.-S., Williamson P.R., Zheng W. Improving therapy of severe infections through drug repurposing of synergistic combinations. Anti-Infect. New Technol. Neurosci. -CNS Dis. Microbiome. 2019;48:92–98. doi: 10.1016/j.coph.2019.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulerie P., Eydoux C., Hnawia E., Stuhl L., Maciuk A., Lebouvier N., Canard B., Figadère B., Guillemot J.-C., Nour M. Biflavonoids of Dacrydium balansae with potent inhibitory activity on dengue 2 NS5 polymerase. Planta Med. 2012:672–677. doi: 10.1055/s-0031-1298355. [DOI] [PubMed] [Google Scholar]
- Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat L.W., Wang Q., Lou Z., Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020 doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020 doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- Ji F., Yeo Y.H., Wei M.T., Ogawa E., Enomoto M., Lee D.H., Iio E., Lubel J., Wang W., Wei B., Ide T., Preda C.M., Conti F., Minami T., Bielen R., Sezaki H., Barone M., Kolly P., Chu P., Virlogeux V., Eurich D., Henry L., Bass M.B., Kanai T., Dang S., Li Z., Dufour J.-F., Zoulim F., Andreone P., Cheung R.C., Tanaka Y., Furusyo N., Toyoda H., Tamori A., Nguyen M.H. Sustained virologic response to direct-acting antiviral therapy in patients with chronic hepatitis C and hepatocellular carcinoma: a systematic review and meta-analysis. J. Hepatol. 2019;71:473–485. doi: 10.1016/j.jhep.2019.04.017. [DOI] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruijssers A.J., Denison M.R. Nucleoside analogues for the treatment of coronavirus infections. Curr. Opin. Virol. 2019;35:57–62. doi: 10.1016/j.coviro.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon A., Le N.T.-T., Selisko B., Eydoux C., Alvarez K., Guillemot J.-C., Decroly E., Peersen O., Ferron F., Canard B. Remdesivir and SARS-CoV-2: Structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites. Antiviral Res. 2020;178 doi: 10.1016/j.antiviral.2020.104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L.M., Guan Y., Rozanov M., Spaan W.J.M., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/s0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E., Snijder E.J., Canard B., Imbert I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman S., Prasad B.V.L.S., Selvarajan R. RNA dependent RNA polymerases: insights from structure, function and evolution. Viruses. 2018;10:76. doi: 10.3390/v10020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Liu Z., Chen Z., Huang X., Xu M., He T., Zhang Z. The establishment of reference sequence for SARS-CoV-2 and variation analysis. J. Med. Virol. 2020;92:667–674. doi: 10.1002/jmv.25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E.C., Zhang Y.-Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]