Abstract
Background
Patients with coronavirus disease 2019 (COVID-19) requiring mechanical ventilation have high mortality and resource utilisation. The ability to predict which patients may require mechanical ventilation allows increased acuity of care and targeted interventions to potentially mitigate deterioration.
Methods
We included hospitalised patients with COVID-19 in this single-centre retrospective observational study. Our primary outcome was mechanical ventilation or death within 24 h. As clinical decompensation is more recognisable, but less modifiable, as the prediction window shrinks, we also assessed 4, 8, and 48 h prediction windows. Model features included demographic information, laboratory results, comorbidities, medication administration, and vital signs. We created a Random Forest model, and assessed performance using 10-fold cross-validation. The model was compared with models derived from generalised estimating equations using discrimination.
Results
Ninety-three (23%) of 398 patients required mechanical ventilation or died within 14 days of admission. The Random Forest model predicted pending mechanical ventilation with good discrimination (C-statistic=0.858; 95% confidence interval, 0.841–0.874), which is comparable with the discrimination of the generalised estimating equation regression. Vitals sign data including SpO2/FiO2 ratio (Random Forest Feature Importance Z-score=8.56), ventilatory frequency (5.97), and heart rate (5.87) had the highest predictive utility. In our highest-risk cohort, the number of patients needed to identify a single new case was 3.2, and for our second quintile it was 5.0.
Conclusion
Machine learning techniques can be leveraged to improve the ability to predict which patients with COVID-19 are likely to require mechanical ventilation, identifying unrecognised bellwethers and providing insight into the constellation of accompanying signs of respiratory failure in COVID-19.
Keywords: COVID-19, critical care medicine, machine learning, mechanical ventilation, predictive models, respiratory insufficiency, respiratory failure
Editor's key points.
-
•
Being able to predict early when patients are likely to deteriorate with life-threating diseases such as COVID-19 could guide clinical management and improve patient outcomes.
-
•
Expert human gestalt and classic static prediction models can be useful, but do not take sufficient advantage of the numerous data elements, including time series data, in modern electronic health records.
-
•
This study evaluated machine learning approaches for predicting respiratory failure and death in patients with COVID-19.
-
•
In choosing the optimal machine learning techniques, it is important to consider both model performance and interpretability; the Random Forest model used in this study performed well and ranked features most strongly associated with the outcomes of interest.
Coronavirus disease 2019 (COVID-19) is the clinical disease caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).1 Although the virus can affect various organs and physiological functions, including the bowel, kidneys, heart, brain, and coagulation, its initial stereotypical clinical presentation is pulmonary with cough, dyspnoea, and hypoxaemia among the presenting features.2, 3, 4, 5, 6 Although respiratory symptoms can be mild, some patients progress to hypoxaemia, necessitating supplementary oxygen or even mechanical ventilation. Studies of invasive mechanical ventilation to treat COVID-19 respiratory failure have shown a mortality rate greater than 85%.7, 8, 9 Limited information is available about which patients admitted to the hospital not requiring mechanical ventilation will progress to mechanical ventilation and what clinical factors are associated with that progression.
Improved identification of patients likely to require mechanical ventilation will enable closer monitoring for signs of clinical deterioration and optimise allocation of resources such as ventilators and intensive care beds. Novel analytical techniques could also reveal previously unrecognised indicators of a worsening respiratory trajectory. This could guide treatment decisions (e.g. medications such as anticoagulants or corticosteroids, tighter haemodynamic regulation, or titration of supplemental oxygen) which may mitigate progression to respiratory failure.
Previous attempts to predict clinical deterioration of patients with COVID-19 have used traditional regression-based techniques,10, 11 failed to capitalise on the diversity of available data in the modern electronic health record,12 or been limited to a small, potentially non-generalisable population.13 Furthermore, heterogeneous outcomes such as critical illness or disease severity10,12 may mask the influence of a singular class of variables. A predictive algorithm leveraging machine learning techniques on the diverse data captured in the electronic health record to predict imminent mechanical ventilation in patients with COVID-19 may facilitate predictive accuracy. We hypothesise that an assessment metric, developed from a Random Forest decision algorithm, can predict which patients with COVID-19 will subsequently require mechanical ventilation.
Methods
Study design
For this retrospective observational study performed at our academic quaternary care centre, we obtained Institutional Review Board approval (University of Michigan, Ann Arbor, MI, USA; HUM00052066). As no patient care interventions were made through conducting the study, patient consent was waived. This manuscript follows multidisciplinary guidelines for reporting machine learning predictive models in biomedical research.14 Study outcomes, data collection, and statistical analyses were established a priori and presented at a multidisciplinary peer-review forum on May 20, 2020 before data access.15
Data collection
For all patients with COVID-19 admitted to the hospital, the electronic health record (Epic Systems, Verona, WI, USA) was queried for patient characteristics, baseline comorbidities, vital signs, laboratory values, medication administration record, and processes of care. The full list of features included in our model can be found in Supplementary Table S1. Medical comorbidities were categorised according to International Classification of Diseases-9/10 diagnoses present upon admission according to a previously described and validated classification system.16,17 Patients were excluded if they were receiving mechanical ventilation on arrival (via hospital transfer) or were intubated within 4 h of hospital admission. Data were grouped into 4 h windows and extended to the next window, if no new data were recorded. If supplementary O2 was expressed in L min−1, instead of FiO2, then L min−1 flow was converted to FiO2 by adding 0.038 for every L min−1 of supplemental oxygen.18 Hi-Flow nasal cannula and Venturi masks are recorded in the medical record as FiO2. Non-rebreather masks were considered to supply FiO2=0.70. The actual FiO2 for face masks and nasal cannula will vary from person to person depending on factors such as tidal volume and ventilatory frequency18; we used these conversion factors to be consistent across all patients. Data at a given time window, data from the immediately preceding time window, and the change between them (delta) were incorporated into our model. If preceding data were not available, data were imputed to population mean and the delta value was set to zero. Data for all patients were censored at 14 days after hospital admission.
Target output
Our target output (primary outcome) was mechanical ventilation or death within 24 h. As the clinical decompensation is likely more recognisable and less modifiable as the time window decreases, we also assessed and characterised the predictive utility of our model to predict mechanical ventilation or death within 4 and 8 h, and, for more notice, 48 h as secondary outcomes. Each outcome extended from whenever the prediction was being made to the end of the designated prediction window. Predictions were made every 4 h through the first 14 days of a patient's hospitalisation (or until the outcome was reached). For example at the 8 h prediction point, the primary outcome was intubation before the 32 h mark and 12, 16, and 56 h for the secondary outcomes. At the 24 h prediction point, the primary outcome was intubation before the 48 h mark, and the secondary outcomes 28, 32, and 72 h. The decision to intubate was left to the discretion of the clinical care team (typically fellowship-trained intensivists). There were no institutional criteria for intubation. Bi-level positive airway pressure was used as an escalation of respiratory management, but was not included as a primary outcome (i.e. invasive mechanical ventilation). The initial prediction window (i.e. 0 h) began with the first documented vital signs (which may have occurred upon presentation to the emergency department, before hospital admission).
Statistical analyses
Clinical data were summarised using means and standard deviations (sd) for normally distributed continuous covariates, medians and inter-quartile range for non-normally distributed continuous variables, and counts and percentages for categorical covariates. Statistical analysis was performed in SAS for Windows 9.4 (SAS Institute Inc., Cary, NC, USA).
Machine learning: model design
A Random Forest is a classification algorithm characterised by a set of many decision ‘trees’ uncorrelated to each other.19 A Random Forest was trained to predict when a patient would require mechanical ventilation (using randomForest V4.6-14 in R version 3.5.1; R Foundation for Statistical Computing, Vienna, Austria) using 500 trees and default parameters.20 For classifier training, 398 patients were monitored across 4-h time intervals resulting in 27 282 observations. The Random Forest used 73 predictive features grouped into demographic features, comorbidities, laboratory values, vital signs, and medications (Supplementary Table S1). The Comorbidities included in our static variables were derived from International Classification of Diseases (ICD)-9/10 diagnostic codes present upon admission (from previous hospitalisations, rather than the patient's current hospitalisation), the goal being to only include data that would be available to the clinical provider in real time at the point when the prediction is being made. Groupings for each class (such as renal failure and cardiac arrhythmias) are composite variables based upon these ICD-9/10 codes using previously validated Elixhauser Comorbidity Index.16 Missing laboratory values and vital signs were giving the average across all non-missing features. Missing medication values were given a value of zero. Delta values based on missing values were imputed to zero. The classifier was assessed for sensitivity, specificity, and balanced accuracy using 10-fold cross-validation. To ensure that performance was not overestimated, all time points from the same patient were restricted to the same fold.
The Random Forest Feature Importance Z-score19 was used to rank all candidate features. As data from the immediately preceding time window, and the change between them (delta) were also included, this is larger than the feature list presented in Supplementary Table S2. Briefly, the Random Forest Feature Importance Z-score calculates the number of correct votes on the out-of-bag cases for a particular model feature compared with a randomly permuted set of values from that same feature. [In Breiman's original implementation of the random forest algorithm, each tree is trained on about two-thirds of the total training data.19 As the forest is built, each tree can thus be tested (similar to leave one out cross-validation) on the samples not used in building that tree. This is the out of bag error estimate – an internal error estimate of a random forest as it is being constructed.]
During the initial development, we considered several machine learning approaches but ultimately selected a Random Forest. Although a deep neural network would in theory provide the highest performance for real-time classification, fewer than 400 patients would not be a sufficient number of training examples to properly train the model. In addition, Random Forests are more capable of handling categorical features compared with support vector machines (SVMs). Random Forests are more interpretable and transparent than deep learning or SVMs. To facilitate interpretability of our model, predictive features were ranked according to Z-score. In addition, the highest predictive score for each patient was graphed with visualisation of the primary outcome after the time that score occurred.
Generalised linear modelling
The Random Forest model was then compared with generalised estimating equations (GEE) models at each of the four prediction windows. GEE was selected to account for the longitudinal structure of the data. To create this model, we first used least absolute shrinkage and selection operator (LASSO) using the proc hpgenselect procedure in SAS to select variables for inclusion at each prediction window as previously described.17 LASSO regression also provided the reported c-statistics, as GEE does not provide these. In brief, this method estimates the parameters of a generalised linear regression model by using maximum likelihood techniques with exchangeable correlation structure and logit link. The hpgenselect procedure is a high-performance procedure that provides model fitting and model building for generalised linear models. It fits models for standard distributions in the exponential family, such as the binomial distributions.
Results
Patient characteristics
A total of 398 patients met our inclusion criteria, with 90 patients requiring mechanical ventilation (23%) and three patients dying without mechanical ventilation (0.8%). The dataset included patients admitted from March 1, 2020 to May 5, 2020. After compiling dynamic model features into 4 h increments, we assessed our primary outcome at 27 282 observations, with 431 positive observations. For our secondary outcomes, we had 93, 171, and 715 positive observations at 4, 8, and 48 h, respectively. Patients meeting our composite outcome tended to be older (mean [sd]: 65 [14] vs 59 [17], P=0.001), male (70% vs 48%, P<0.001), and had higher incidence of: renal failure (58% vs 28%, P<0.001), diabetes (58% vs 37%, P<0.001), and cardiac arrhythmias (69% vs 47%, P<0.001). Furthermore, patients meeting the composite outcome had higher serum creatinine (mean, 2.2 vs 1.4; P=0.019) and ventilatory frequency (23 [6] vs 20 [4], P<0.001) and lower SpO2 (94% [3%] vs 96% [3%], P<0.001) and SpO2/FiO2 ratio (271 [114] vs 367 [107], P<0.001) upon presentation than those not meeting the outcome. Patients requiring subsequent ventilation were administered tocilizumab (15% vs 7%, P=0.021) and norepinephrine (10% vs 2%, P=0.002) more frequently than those not progressing to ventilation or death. Additional details on our patient population can be found in Table 1.
Table 1.
Characteristics of patients requiring intubation or dying within 24 h. Laboratory studies and vital signs are presenting or initial values. Note that not all patients have full laboratory results or vital signs within the first 4 h of admission. The medications counts/percentages listed are based upon administration at any point from admission until data collection was censored at either primary outcome or 14 days after admission. COPD, chronic obstructive pulmonary disease; FiO2, fraction of inspired oxygen; sd, standard deviation; SpO2, blood oxygen saturation level .
| Variable | Level | All data (n=398) |
Control group (n=305) |
Ventilation or death (n=93) |
P-value |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | % | Mean | sd | N | % | Mean | sd | N | % | Mean | sd | χ2 | t-test | ||
| Age (yr) | 398 | 100.0 | 60 | 17 | 305 | 100.0 | 59 | 17 | 93 | 100.00 | 65 | 14 | 0.001 | ||
| BMI (kg m−2) | 398 | 100.0 | 31.5 | 8.5 | 305 | 100 | 31.2 | 8.6 | 93 | 100 | 32.6 | 8.3 | 0.171 | ||
| Height (cm) | 398 | 100.0 | 170.0 | 11.4 | 305 | 100.0 | 169.5 | 11.5 | 93 | 100.0 | 171.7 | 10.9 | 0.105 | ||
| Weight (kg) | 398 | 100.0 | 91.1 | 26.5 | 305 | 100.0 | 89.7 | 27.1 | 93 | 100.0 | 95.6 | 24.3 | 0.063 | ||
| Sex | Female | 187 | 47.0 | 159 | 52.1 | 28 | 30.1 | <0.001 | |||||||
| Male | 211 | 53.0 | 146 | 47.9 | 65 | 69.9 | |||||||||
| Race | African American | 139 | 34.9 | 99 | 32.5 | 40 | 43.0 | 0.433 | |||||||
| American Indian | 1 | 0.3 | 1 | 0.3 | 0 | 0.0 | |||||||||
| Asian | 13 | 3.3 | 11 | 3.6 | 2 | 2.2 | |||||||||
| Caucasian | 208 | 52.3 | 166 | 54.4 | 42 | 45.2 | |||||||||
| Other | 16 | 4.0 | 13 | 4.3 | 3 | 3.2 | |||||||||
| Unknown | 21 | 5.3 | 15 | 4.9 | 6 | 6.5 | |||||||||
| Elixhauser comorbidities | Alcohol abuse | 21 | 5.3 | 17 | 5.6 | 4 | 4.3 | 0.631 | |||||||
| Blood loss anaemia | 52 | 13.1 | 36 | 11.8 | 16 | 17.2 | 0.176 | ||||||||
| Cardiac arrhythmias | 207 | 52.0 | 143 | 46.9 | 64 | 68.8 | <0.001 | ||||||||
| COPD | 140 | 35.2 | 104 | 34.1 | 36 | 38.7 | 0.415 | ||||||||
| Coagulopathy | 99 | 24.9 | 74 | 24.3 | 25 | 26.9 | 0.609 | ||||||||
| Congestive heart failure | 97 | 24.4 | 67 | 22.0 | 30 | 32.3 | 0.043 | ||||||||
| Anaemia (iron deficiency) | 77 | 19.3 | 60 | 19.7 | 17 | 18.3 | 0.766 | ||||||||
| Depression | 132 | 33.2 | 103 | 33.8 | 29 | 31.2 | 0.643 | ||||||||
| Complicated diabetes mellitus | 94 | 23.6 | 62 | 20.3 | 32 | 34.4 | 0.005 | ||||||||
| Uncomplicated diabetes mellitus | 166 | 41.7 | 112 | 36.7 | 54 | 58.1 | <0.001 | ||||||||
| Drug abuse | 28 | 7.0 | 25 | 8.2 | 3 | 3.2 | 0.101 | ||||||||
| Fluid and electrolyte disorders | 224 | 56.3 | 151 | 49.5 | 73 | 78.5 | <0.001 | ||||||||
| Complicated hypertension | 121 | 30.4 | 80 | 26.2 | 41 | 44.1 | 0.001 | ||||||||
| Uncomplicated hypertension | 266 | 66.8 | 191 | 62.6 | 75 | 80.6 | 0.001 | ||||||||
| Hypothyroidism | 67 | 16.8 | 49 | 16.1 | 18 | 19.4 | 0.458 | ||||||||
| Liver disease | 66 | 16.6 | 48 | 15.7 | 18 | 19.4 | 0.412 | ||||||||
| Metastatic cancer | 66 | 16.6 | 50 | 16.4 | 16 | 17.2 | 0.854 | ||||||||
| Obesity | 158 | 39.7 | 114 | 37.4 | 44 | 47.3 | 0.086 | ||||||||
| Neurological disorders | 103 | 25.9 | 74 | 24.3 | 29 | 31.2 | 0.182 | ||||||||
| Peripheral vascular disorders | 78 | 19.6 | 62 | 20.3 | 16 | 17.2 | 0.507 | ||||||||
| Pulmonary/circulation disorder | 80 | 20.1 | 54 | 17.7 | 26 | 28.0 | 0.031 | ||||||||
| Renal failure | 139 | 34.9 | 85 | 27.9 | 54 | 58.1 | <0.001 | ||||||||
| Solid tumour without metastasis | 74 | 18.6 | 61 | 20.0 | 13 | 14.0 | 0.191 | ||||||||
| Valvular diseases of the heart | 46 | 11.6 | 37 | 12.1 | 9 | 9.7 | 0.517 | ||||||||
| Weight loss | 97 | 24.4 | 73 | 23.9 | 24 | 25.8 | 0.713 | ||||||||
| Laboratory studies | Alanine transaminase (ALT) | 349 | 87.7 | 60.0 | 181.4 | 268 | 67.3 | 51.4 | 136.9 | 81 | 87.1 | 88.4 | 282.1 | 0.258 | |
| (Initial/Presenting) | Aspartate transaminase (AST) | 349 | 87.7 | 67.8 | 131.7 | 268 | 67.3 | 57.7 | 94.9 | 81 | 87.1 | 101.2 | 209.6 | 0.073 | |
| Brain natriuretic peptide | 127 | 31.9 | 300.7 | 808.8 | 93 | 23.4 | 296.1 | 843.4 | 34 | 36.6 | 313.2 | 717.2 | 0.916 | ||
| Serum creatinine (Cr) | 378 | 95.0 | 1.6 | 1.9 | 292 | 73.4 | 1.4 | 1.4 | 86 | 92.5 | 2.2 | 2.9 | 0.019 | ||
| C-reactive protein | 264 | 66.3 | 11.8 | 9.5 | 194 | 48.7 | 11.4 | 9.2 | 70 | 75.3 | 13.2 | 10.3 | 0.174 | ||
| D-dimer | 242 | 60.8 | 3.6 | 7.2 | 176 | 44.2 | 4.0 | 7.8 | 66 | 71.0 | 2.6 | 5.1 | 0.123 | ||
| Glucose | 376 | 94.5 | 143.5 | 76.8 | 288 | 72.4 | 140.0 | 75.6 | 88 | 94.6 | 154.8 | 79.7 | 0.115 | ||
| High-sensitivity troponin | 225 | 56.5 | 62.6 | 205.5 | 170 | 42.7 | 57.5 | 221.0 | 55 | 59.1 | 78.4 | 148.2 | 0.514 | ||
| Total bilirubin | 341 | 85.7 | 0.7 | 1.1 | 261 | 65.6 | 0.7 | 1.2 | 80 | 86.0 | 0.7 | 0.5 | 0.940 | ||
| White blood cell | 374 | 94.0 | 8.6 | 4.8 | 290 | 72.9 | 8.6 | 4.5 | 84 | 90.3 | 8.7 | 5.6 | 0.865 | ||
| Procalcitonin | 256 | 64.3 | 2.2 | 10.3 | 189 | 47.5 | 2.5 | 11.8 | 67 | 72.0 | 1.4 | 3.9 | 0.472 | ||
| Vital signs | Ventilatory frequency (bpm) | 367 | 92.2 | 21 | 5 | 284 | 71.4 | 20 | 4 | 83 | 89.2 | 23 | 6 | <0.001 | |
| (Initial/Presenting) | Systolic blood pressure (mm Hg) | 396 | 99.5 | 134 | 22 | 303 | 76.1 | 135 | 23 | 93 | 100.0 | 131 | 21 | 0.194 | |
| Diastolic blood pressure (mm Hg) | 396 | 99.5 | 73 | 12 | 303 | 76.1 | 74 | 12 | 93 | 100.0 | 72 | 11 | 0.129 | ||
| Heart rate (beats min−1) | 370 | 93.0 | 87 | 17 | 287 | 72.1 | 87 | 17 | 83 | 89.2 | 88 | 18 | 0.452 | ||
| Temperature (°C) | 355 | 89.2 | 37.1 | 0.6 | 280 | 70.4 | 37.1 | 0.6 | 75 | 80.6 | 37.2 | 0.6 | 0.021 | ||
| SpO2 (%) | 366 | 92.0 | 96 | 3 | 283 | 71.1 | 96 | 3 | 83 | 89.2 | 94 | 3 | <0.001 | ||
| SpO2/FiO2 | 366 | 92.0 | 345 | 116 | 283 | 71.1 | 367 | 107 | 83 | 89.2 | 271 | 114 | <0.001 | ||
| Medications | Hydrocortisone | 9 | 2.3 | 8 | 2.6 | 1 | 1.1 | 0.379 | |||||||
| Heparin (s.c.) | 87 | 21.9 | 58 | 19.0 | 29 | 31.2 | 0.013 | ||||||||
| Heparin (i.v.) | 53 | 13.3 | 41 | 13.4 | 12 | 12.9 | 0.893 | ||||||||
| Enoxaparin | 16 | 4.0 | 11 | 3.6 | 5 | 5.4 | 0.447 | ||||||||
| Tocilizumab | 36 | 9.0 | 22 | 7.2 | 14 | 15.1 | 0.021 | ||||||||
| Remdesivir | 22 | 5.5 | 19 | 6.2 | 3 | 3.2 | 0.267 | ||||||||
| Norepinephrine | 16 | 4.0 | 7 | 2.3 | 9 | 9.7 | 0.002 | ||||||||
| Hydroxychloroquine | 92 | 23.1 | 68 | 22.3 | 24 | 25.8 | 0.482 | ||||||||
Machine learning
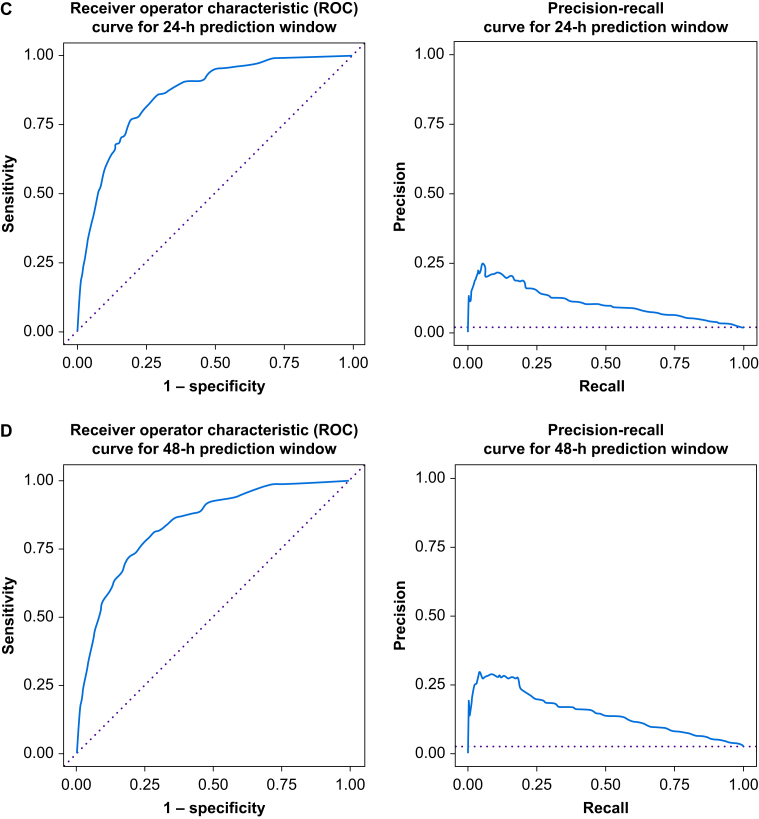
The Random Forest algorithm found several variables associated with receipt of mechanical ventilation or death. The variables with the best predictive ability were: (1) current SpO2/FiO2 (Z-score=8.55), (2) previous SpO2/FiO2 (Z=6.25), (3) current ventilatory frequency (Z=5.97), (4) current heart rate (Z=5.87), (5) previous heart rate (Z=5.83), (6) current diastolic blood pressure (Z=5.76), and (7) current blood glucose (Z=5.76) (Supplementary Table S2). Our algorithm is able to predict subsequent ventilation or death with very good discrimination (c-statistic for the 4 h time window=0.885, 95% confidence interval [CI], 0.858–0.924; 8 h window=0.881, 95% CI 0.856–0.906; 24 h window=0.858, 95% CI 0.841–0.874; and 48 h window=0.839, 95% CI 0.825–0.854). The areas under the precision recall curve were 0.038, 0.060, 0.106, and 0.147 at 4, 8, 24, and 48 h prediction windows, respectively. Receiver operator characteristic curves and precision–recall curves for each of our prediction windows are shown in Figure 1. Notably at Youden's point, the sensitivity for the 24 h prediction window was 0.77 and the specificity was 0.80 (compared with sensitivity of 0.84 and specificity of 0.80 for the 4 h prediction window).
Fig 1.
Receiver operator characteristic curves and precision recall curves for each prediction window. (a) Four hour prediction window. (b) Eight hour prediction window. (c) Twenty-four hour prediction window. (d) Forty-eight hour prediction window.
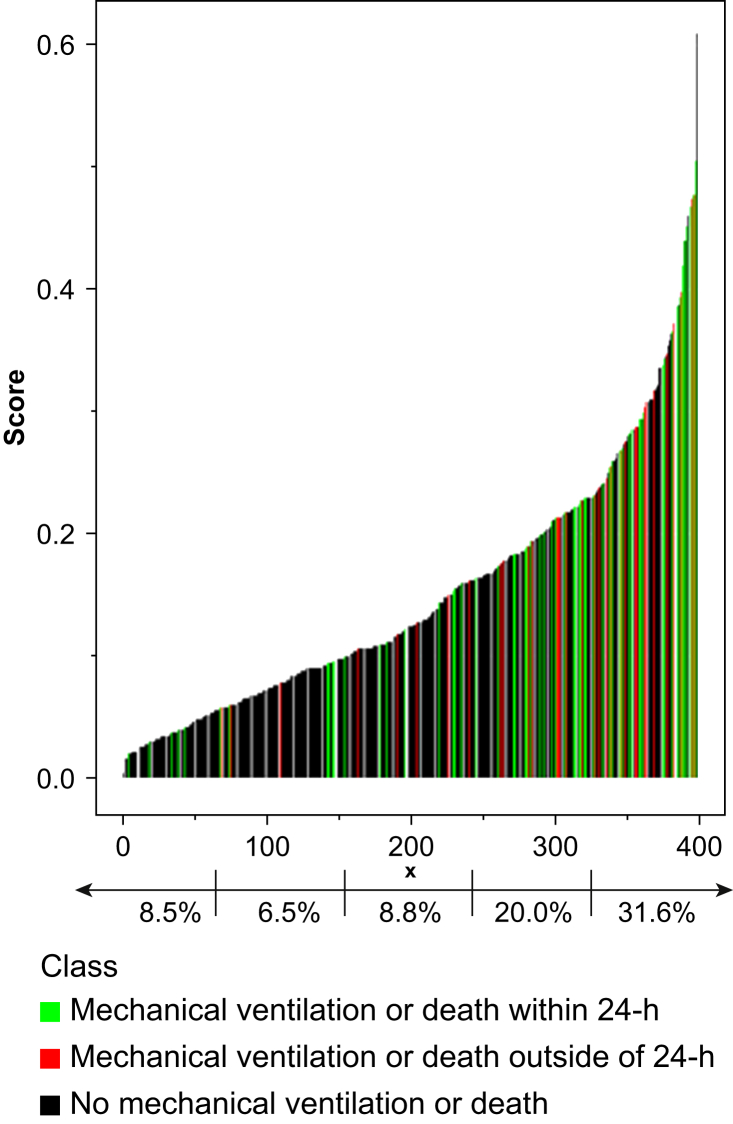
Next we graphed the maximum predicted score for each patient, along with their receipt of mechanical ventilation or death (Fig 2). By quintiles of machine learning scores, 8.5%, 6.5%, 8.8%, 20.0%, and 31.6% of patients (Fig 2) required mechanical ventilation or died within the subsequent 24 h.
Fig 2.
Maximum predicted score for each patient, along with ventilation requirement or death. Quintile 1, 8.5% died or required mechanical ventilation within 24 h. Quintile 2, 6.5% died or required mechanical ventilation within 24 h. Quintile 3, 8.8% died or required mechanical ventilation within 24 h. Quintile 4, 20.0% died or required mechanical ventilation within 24 h. Quintile 5, 31.6% died or required mechanical ventilation within 24 h.
Generalised linear modelling
Using GEE, nine features were found to be significantly associated with ventilation or death within 24 h (c-statistic=0.866; 95% CI, 0.863–0.869). The demographic features: age (adjusted odds ratio [aOR]=1.025; 95% CI, 1.008–1.043; P=0.005), male sex (aOR=2.817; 95% CI, 1.582–5.025; P<0.001), and BMI (aOR=1.035; 95% CI, 1.004–1.067; P=0.026) were all associated with mechanical ventilation or death. The laboratory findings of high sensitivity troponin (aOR=1.005; 95% CI, 1.001–1.010; P=0.014) and D-Dimer (aOR=0.983; 95% CI, 0.972–0.994; P=0.002) were also associated with our primary outcome. The vital signs – (1) previous ventilatory frequency (aOR=1.010; 95% CI, 1.003–1.017; P=0.004), (2) current ventilatory frequency (aOR=1.014; 95% CI, 1.007–1.021; P<0.001), (3) previous SpO2/FiO2 (aOR=0.999; 95% CI, 0.998–1.000; P=0.005), and (4) current SpO2/FiO2 (aOR=0.998; 95% CI, 0.998–0.999; P<0.001) – were also associated with our primary outcome. As the prediction window increased from 4 to 48 h, the discrimination remained similar (c-statistic: 4 h time window=0.865, 95% CI 0.862–0.868; 8 h window=0.854, 95% CI 0.850–0.856; 24 h window=0.866, 95% CI 0.863–0.869; 48 h window=0.840, 95% CI 0.837–0.843); and an increasing number of variables were selected (4 h: four significant variables, 8 h: five variables, 24 h: nine variables, 48 h: 11 variables). Sex, high-sensitivity troponin, previous ventilatory frequency, current ventilatory frequency, and previous SpO2/FiO2 and SpO2/FiO2 occurred consistently across multiple prediction windows. The full results of the GEE for each of the prediction windows can be seen in Supplementary Table S3.
Discussion
In the setting of COVID-19, the Random Forest algorithm is able to predict ventilation or death with high sensitivity (0.77) and specificity (0.80). Furthermore, we have very good discrimination (c-statistic=0.858; 95% CI, 0.841–0.874) for predicting our primary target (24 h prediction window), which improves as our prediction window narrows (4 h window, c-statistic=0.885; 95% CI, 0.858–0.924). [Interpretation of the c-statistic: 0.5–0.6 for a poor model, 0.6–0.7 for a good model, 0.8–0.9 for a very good model, and 0.9–1.0 for an excellent model.] Of the 10 features with the highest predictive value, nine are vital signs. By capturing the clinical trajectory, these dynamic features enable greater predictive utility to detect changes through the course of a hospitalisation. We have selected a list which can be easily and automatically extracted for potential integration into a clinical support system.21 In addition, we demonstrate consistent significance of key features (age, sex, BMI, high sensitivity troponin, blood glucose, SpO2/FiO2, and ventilatory frequency) across two independent modelling methodologies (Random Forest and GEE) and multiple prediction windows (4, 8, 24, and 48 h). This suggests a robust signal that can be leveraged for prediction of mechanical ventilation.
Concordance with previous results
Our highest utility predictor, SpO2/FiO2, has been used as a proxy for Pao2/FiO2 – which occurs in the diagnosis and grading of acute respiratory distress syndrome.22,23 As SpO2/FiO2 can be easily calculated, without the need for arterial blood draw and can be used to monitor continuously, this may represent a promising metric to assess for respiratory deterioration in general care patients, not just patients with COVID-19. Similar to other studies, we found that older,7 heavier,24 or male25 patients are more likely to require mechanical ventilation. Although other studies have found associations between renal failure, congestive heart failure, hypertension, diabetes, and cardiac arrhythmias critical illness or death,7,10,26 we found these to have only small utility in the machine learning algorithm and not associated with outcome in the GEE. Our lack of finding these previously reported associations may be attributable to different patient populations, different clinical practices, or to our more comprehensive list of potential factors. Both C-reactive protein24 and aspartate aminotransferase (AST)27 – which we have identified in our Random Forest model – have also been included in previous severity models. Tachypnoea is a well characterised clinical sign of respiratory decompensation.7 The discrimination of our ventilation model was also similar that reported in a critical illness model (c-statistic=0.88).10
Clinical decision making
Our algorithm can be integrated into a clinical support software with the ultimate goal of identifying patients before clinical decompensation.21 Our primary target (24 h prediction window) was selected to allow appropriate time for interventions, while still providing evidence of deterioration in dynamic features. The advantages of identifying potential respiratory failure 24 or 48 h in advance, include: (1) enrolment in clinical trials, (2) aggressive therapeutic interventions such as prone positioning or noninvasive mechanical ventilation, and (3) planning for appropriate ventilator allocation and utilisation. To identify the prediction window that optimises the trade-off between detection and potential intervention, we also quantified discrimination at 4, 8, and 48 h prediction windows. In our Random Forest model, we have the greatest discrimination to predict within 4 h (c-statistic=0.885; 95% CI, 0.858–0.924) and the lowest, but still very good, discrimination when predicting within 48 h (c-statistic=0.839; 95% CI, 0.825–0.854). This is expected because evidence of the imminent respiratory failure has likely started to manifest, improving the ability to predict, but a 4 h prediction window unfortunately allows the least opportunity for meaningful intervention. We have shown high discrimination (for the Random Forest Model) at 24 h. This can inform when the model is most useful. However, the utility of the model must account for both discrimination of the model and clinical actionability. In addition to the high discrimination, 24 h notice also allows the clinician an opportunity to make modifications in clinical care and preparation in resources for potential decompensation.
The Random Forest model has a sensitivity of 0.77 and a specificity of 0.80. Determination of the optimal identification threshold should weigh the risk of falsely identifying a patient as at risk for mechanical ventilation (increased monitoring and resource utilisation, aggressive intervention) vs failing to identify a patient who is susceptible to future deterioration (missed opportunity to alter clinical trajectory and a delay in recognising the need for increased acuity of care). The number of patients needed to identify (NNI) is 3.2 for the highest quintile and 5.0 for the second highest quintile, which are reasonable numbers that limit false positives while identifying patients in need of life-saving, but invasive, therapy.
Clinical correlates
For additional insight into patient characteristics our algorithm is likely to misclassify, we reviewed the patients with the lowest predictive score, who ultimately required mechanical ventilation within 24 h (‘false negatives’), and patients with high predictive scores who never required ventilation (‘false positives’). Patients the algorithm failed to identify were disproportionately missing data for highly predictive features, such as Pao2/FiO2, ventilatory frequency, heart rate, and SpO2. Specifically, seven of the 10 patients with the lowest predicted scores, who received mechanical ventilation within 24 h (i.e. the false negatives), were found to be missing data for key features. Our algorithm was programmed to overcome this pitfall, by propagating values from the previous time window, when no new values are recorded. Therefore, these false negative cases skew early in their hospital course, where no prior values are recorded and missing values are imputed to population mean. As with any predictive metric, our algorithm is inherently limited by the quality of data recorded. Furthermore, the absence of regularly recorded vital signs may be associated with unrecognised decompensation, because of lower prioritisation of medical documentation in an emergency situation or as a reflection of the medical care team's attentiveness. Because of inherent limitations secondary to incomplete data, we have characterised missing data in Supplementary Table S4. Static variables (e.g. age, height, weight, and comorbidities such as chronic pulmonary disease) have no missing values across our dataset. This contrasts with dynamic variables which have some missing values. For laboratory values and vital signs, this likely reflects how often they were clinically indicated. For example, SpO2, which is missing in 47% of our 4 h prediction windows, may be typically checked less frequently than every 4 h in a stable, general care patient; however, we do not have the reasons why SpO2 was not recorded. Future studies may benefit from including absence or presence of a value as part of the algorithm.
We also reviewed the patients with the highest predictive scores who did not require ventilation within 24 h (‘false positive’). Five of the 10 patients with the highest predictive scores ultimately required mechanical ventilation during their hospital course, suggesting our algorithm was successful in detecting future respiratory decline, but not within the pre-specified prediction window.
To assess the utility of our predictions on a patient level, we quantified the percentage of patients in each risk quintile requiring ventilation or dying within 24 h of their maximum risk score (Fig 2). Patients in risk quintiles 1, 2, and 3 had an 8.5%, 6.5%, and 8.8% risk, respectively. This compares with 20.0% risk in the fourth quintile and 31.6% risk for a patient in the fifth quintile. Even though a patient in the highest risk quintile still has less than a 1 in 3 chance of requiring mechanical ventilation within the next 24 h, the clinical provider may decide that because of the high mortality in patients requiring mechanical ventilation, the increased patient risk (31.6% compared with <10% in the three lowest risk quintiles or 15.1% in our overall cohort) merits closer attention or more aggressive care.
In our highest risk cohort, the NNI a single new case of mechanical ventilation was 3.2, and for our second risk quintile the NNI was 5.0. This means that for every three patients our algorithm identifies as being in the highest risk group (or five in the second quintile), we will correctly detect one new case requiring mechanical ventilation in the next 24 h. Given the high mortality associated with mechanical ventilation,7, 8, 9 an NNI <11 may be considered reasonable, particularly if the intervention is low-risk or low-cost. The intervention may be as low-risk and low-cost as using continuous monitoring with SpO2 rather than intermittent monitoring, thus detecting a decrease in the SpO2/FiO2 ratio, our strongest indicator of risk for mechanical ventilation or death. If desired, the desired threshold can be adjusted up or down based on type of intervention and availability of resources.
The Random Forest identified initiation of intravenous heparin (Z-score=1.60) and hydroxychloroquine (1.37) in the algorithm. Other pharmacologic agents, such as tocilizumab (0.85), remdesivir (0.15), and hydrocortisone (0), had very low association. No pharmacologic agents were selected in the GEE models. Potential reasons include inadequate statistical power, differences in patient population, or a reflection of pharmacologic utility.
High-sensitivity troponin was included in the Random Forest Model (4.59) and was selected in multiple GEE models. Although the mechanism of respiratory deterioration remains unresolved, the association between myocardial injury, myocarditis, myocardial infarction, and thromboembolic events has been previously described and merits further study and incorporation into predictive models.28
Strengths and limitations
Our study used two very dissimilar techniques (Random Forest and GEE) for analysing the data and found similar discrimination and similar factors being associated with mechanical ventilation and death. Our study possessed several limitations. First, we were unable to account for all predictive features that may contribute to pending respiratory failure. In our study, we included some features, such as SpO2/FiO2, which had not been previously characterised in the progression of COVID-19, and included basic relationships between features (change in values); however, other features and more complex relationships were potentially missed by our methodology. The lack of institutional criteria for intubation also introduces heterogeneity in our primary outcome, although the variability in provider practice likely also increases the generalisability of our model.
Additional limitations to our study include those inherent to our single-centre, observational study design: our conclusions require prospective multicentre validation. We also failed to explore the causal relationship between our predictive features and the outcome. In addition, the model's positive predictive value is a function of outcome incidence. As the pandemic has progressed, the fraction of infected individuals who require mechanical ventilation or die has decreased.29 This means the positive predictive value will be lower and the NNI will be higher if the model were applied to the current, less critically ill patient population as compared with the patients in our dataset.
Overfitting was another potential concern. This was addressed through our selection of generalised linear modelling, which adjusts standard error estimates by an estimated overfitting parameter. To mitigate this potential issue within our Random Forest model, cross-validation was independent, with all time points corresponding to a single patient restricted to the same fold.
Although we demonstrated that tachypnoea, hypotension, and hypoxia are associated with impending respiratory decline, we do not address whether addressing these homeostatic imbalances through vasopressors or supplemental oxygen mitigate progression of respiratory decline. A final limitation is lack of external validation of our models. To mitigate this intrinsic issue, independent cross-validation was performed. Randomly dividing all the time points to different folds would result in time points from the same patient in many different folds. We would like to estimate how well the model generalises to completely independent samples. To ensure a conservative estimate of how well the model generalises during cross-validation, we have ensured that all time points from the same patient are restricted to the same fold.
Another limitation of this study is that the rapidly evolving understanding of COVID-19 and advances in clinical management, necessitate re-calibration of the machine learning model at regular time intervals. This is an important consideration when applying this model to new data and an additional limitation of this study. For example, even though hydroxychloroquine was associated with the outcome in the Random Forest Model, that association probably does not hold today because of evolving practice patterns.30
Conclusions
A Random Forest Machine learning approach and a GEE approach, using demographic data, vital signs, medication records, laboratory studies, and medical comorbidities can be leveraged to predict which patients with COVID-19 are likely to require mechanical ventilation. Of the 10 features with highest predictive value, nine are vital signs. SpO2/FiO2 can be easily estimated and monitored continuously, providing a promising metric to assess for respiratory collapse in patients with COVID-19. Future studies will (1) validate the algorithm on a larger number of patients across additional healthcare systems, (2) integrate the complexity of the model within clinician workflow, and (3) assess if clinical features identified by the algorithm may provide targets for medical intervention to alter the clinical course.
Authors' contributions
Study conception: NJD, CBD, MCE
Study design: NJD, CBD, MRM, CP, KKT, MCE
Data interpretation: NJD, CBD, GM, MRM, CP, KKT, MCE
Data analysis (Random Forest Model): CBD
Data analysis (logistic regression and GEE models): GM
Developing the initial and final drafts of the manuscript: NJD, MCE
Assimilation of intellectual content from all co-authors: NJD, MCE
Critical revision of the work for important intellectual content: CBD, GM, MRM, CP, KKT
Acknowledgements
The authors acknowledge Erin O. Kaleba (Data Office for Clinical and Translational Research, University of Michigan Medical School, Ann Arbor, MI, USA) for help with data acquisition.
Handling editor: Michael Avidan
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bja.2020.11.034.
Declarations of interest
National Institutes of Health (K01-HL141701, to MRM); Foundation for Anesthesia Education and Research (to NJD). CBD is a paid consultant for Thrive Earlier Detection. He is also an inventor on various technologies unrelated to the work described in this manuscript. Some of the licenses are or will be associated with equity or royalty payments. The terms of all these arrangements are being managed by Johns Hopkins University in accordance with its conflict of interest policies. KKT is a founder and equity holder in AlertWatch Inc, a University of Michigan Software Startup Company. All other authors declare no competing interests.
Funding
National Institutes of Health (K01-HL141701 to MRM) and Foundation for Anesthesia Education and Research (to NJD).
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.World Health Organization . 2020. Naming the coronavirus disease (COVID-19) and the virus that causes it.https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it.2020 Available from: accessed 23 April 2020. [Google Scholar]
- 2.Deng Y., Liu W., Liu K. Clinical characteristics of fatal and recovered cases of coronavirus disease 2019 in Wuhan, China: a retrospective study. Chin Med J. 2020;133:1261–1267. doi: 10.1097/CM9.0000000000000824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N., Zhang D., Wang W. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spiezia L., Boscolo A., Poletto F. COVID-19-related severe hypercoagulability in patients admitted to intensive care unit for acute respiratory failure. Thromb Haemost. 2020;120:998–1000. doi: 10.1055/s-0040-1710018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mao L., Wang M., Chen S. Neurological manifestations of hospitalized patients with COVID-19 in Wuhan, China: a retrospective case series study. JAMA Neurol. 2020;77:683–690. doi: 10.1001/jamaneurol.2020.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J.-J., Dong X., Cao Y.-Y. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy. 2020;75:1730–1741. doi: 10.1111/all.14238. [DOI] [PubMed] [Google Scholar]
- 7.Zhou F., Yu T., Du R. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang X., Yu Y., Xu J. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richardson S., Hirsch J.S., Narasimhan M. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City area. JAMA. 2020;323:2052–2059. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang W., Liang H., Ou L. Development and validation of a clinical risk score to predict the occurrence of critical illness in hospitalized patients with COVID-19. JAMA Intern Med. 2020;180:1081–1089. doi: 10.1001/jamainternmed.2020.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ji D., Zhang D., Xu J. Prediction for progression risk in patients with COVID-19 pneumonia: the CALL score. Clin Infect Dis. 2020;71:1393–1399. doi: 10.1093/cid/ciaa414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gong J., Ou J., Qiu X. A tool to early predict severe corona virus disease 2019 (COVID-19): a multicenter study using the risk nomogram in Wuhan and Guangdong, China. Clin Infect Dis. 2020;71:833–840. doi: 10.1093/cid/ciaa443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang X., Coffee M., Bari A. Towards an artificial intelligence framework for data-driven prediction of coronavirus clinical severity. CMC Comput Mater Con. 2020;63:537–551. [Google Scholar]
- 14.Luo W., Phung D., Tran T. Guidelines for developing and reporting machine learning predictive models in biomedical research: a multidisciplinary view. J Med Internet Res. 2016;18:e323. doi: 10.2196/jmir.5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.University of Michigan – Anesthesia clinical research committee (ACRC) 2020. https://anes.med.umich.edu/research/acrc.html Available from: accessed 20 May 2020. [Google Scholar]
- 16.Quan H., Sundararajan V., Halfon P. Coding algorithms for defining comorbidities in ICD-9-CM and ICD-10 administrative data. Med Care. 2005;43:1130–1139. doi: 10.1097/01.mlr.0000182534.19832.83. [DOI] [PubMed] [Google Scholar]
- 17.Douville N.J., Jewell E.S., Duggal N. Association of intraoperative ventilator management with postoperative oxygenation, pulmonary complications, and mortality. Anesth Analg. 2020;130:165–175. doi: 10.1213/ANE.0000000000004191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Reilly Nugent A., Kelly P.T., Stanton J., Swanney M.P., Graham B., Beckert L. Measurement of oxygen concentration delivered via nasal cannulae by tracheal sampling. Respirology. 2014;19:538–543. doi: 10.1111/resp.12268. [DOI] [PubMed] [Google Scholar]
- 19.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 20.Team RC . R Foundation for Statistical Computing; Vienna, Austria: 2018. R: a language and environment for statistical computing.https://www.R-project.org/ [Google Scholar]
- 21.Tremper K.K., Mace J.J., Gombert J.M., Tremper T.T., Adams J.F., Bagian J.P. Design of a novel multifunction decision support display for anesthesia care: AlertWatch® OR. BMC Anesthesiol. 2018;18:16. doi: 10.1186/s12871-018-0478-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pandharipande P.P., Shintani A.K., Hagerman H.E. Derivation and validation of SpO2/FiO2 ratio to impute for PaO2/FiO2 ratio in the respiratory component of the sequential organ failure assessment (SOFA) score. Crit Care Med. 2009;37:1317. doi: 10.1097/CCM.0b013e31819cefa9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen W., Janz D.R., Shaver C.M., Bernard G.R., Bastarache J.A., Ware L.B. Clinical characteristics and outcomes are similar in ARDS diagnosed by oxygen saturation/FiO2 ratio compared with PaO2/FiO2 ratio. Chest. 2015;148:1477–1483. doi: 10.1378/chest.15-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petrilli C.M., Jones S.A., Yang J. Factors associated with hospitalization and critical illness among 4,103 patients with COVID-19 disease in New York City. BMJ. 2020;369:m1966. doi: 10.1136/bmj.m1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nepogodiev D., Bhangu A., Glasbey J.C. Mortality and pulmonary complications in patients undergoing surgery with perioperative SARS-CoV-2 infection: an international cohort study. Lancet. 2020;396:27–38. doi: 10.1016/S0140-6736(20)31182-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jehi L., Ji X., Milinovich A. Development and validation of a model for individualized prediction of hospitalization risk in 4,536 patients with COVID-19. PLoS One. 2020;15 doi: 10.1371/journal.pone.0237419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L., He W., Yu X. Coronavirus disease 2019 in elderly patients: characteristics and prognostic factors based on 4-week follow-up. J Infect. 2020;80:639–645. doi: 10.1016/j.jinf.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Long B., Brady W.J., Koyfman A., Gottlieb M. Cardiovascular complications in COVID-19. Am J Emerg Med. 2020;38:1504–1507. doi: 10.1016/j.ajem.2020.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.CDC . 2020. COVIDView: A weekly surveillance summary of U.S. COVID-19 activity.https://www.cdc.gov/coronavirus/2019-ncov/covid-data/covidview/index.html Available from: accessed 23 April 2020. [Google Scholar]
- 30.Skipper C.P., Pastick K.A., Engen N.W. Hydroxychloroquine in nonhospitalized adults with early COVID-19: a randomized trial. Ann Intern Med. 2020;173:623–631. doi: 10.7326/M20-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.