Abstract
Cholesterol 25-hydroxylase (CH25 H) is a key enzyme regulating cholesterol metabolism and also acts as a broad antiviral host restriction factor. Porcine deltacoronavirus (PDCoV) is an emerging swine enteropathogenic coronavirus that can cause vomiting, diarrhea, dehydration and even death in newborn piglets. In this study, we found that PDCoV infection significantly upregulated the expression of CH25H in IPI-FX cells, a cell line of porcine ileum epithelium. Overexpression of CH25H inhibited PDCoV replication, whereas CH25H silencing using RNA interference promoted PDCoV infection. Treatment with 25-hydroxycholesterol (25HC), the catalysate of cholesterol via CH25H, inhibited PDCoV proliferation by impairing viral invasion of IPI-FX cells. Furthermore, a mutant CH25H (CH25H-M) lacking hydroxylase activity also inhibited PDCoV infection to a lesser extent. Taken together, our data suggest that CH25H acts as a host restriction factor to inhibit the proliferation of PDCoV but this inhibitory effect is not completely dependent on its enzymatic activity.
Keywords: Porcine deltacoronavirus, Cholesterol 25-hydroxylase, 25-hydroxycholesterol, Viral entry
1. Introduction
Porcine deltacoronavirus (PDCoV) is a newly discovered porcine coronavirus belonging to the genus Deltacoronavirus, family Coronaviridea, and order Nidovirales (Chen et al., 2015; Ma et al., 2015; Woo et al., 2009). PDCoV has a single-stranded positive-sense RNA genome approximately 25.4 kb in length. Almost two-thirds of the genome consists of two open reading frames (ORFs), ORF1a and ORF1b, that encode two viral replicase polyproteins, pp1a and pplab; these are cleaved into 15 mature non-structural proteins by viral proteases. The other ORFs encode four structural proteins (spike [S], envelope [E], membrane [M], and nucleocapsid [N]), and three accessory proteins (NS6, NS7, and NS7a) (Fang et al., 2017, 2016). PDCoV was first identified from pig manure samples in 2009 (Woo et al., 2012) and the first outbreak of PDCoV on a farm occurred in the United States in 2014 (Homwong et al., 2016; Hu et al., 2015; Marthaler et al., 2014; Wang et al., 2014). Subsequently, PDCoV was reported in many other countries including China (Dong et al., 2015; Wang et al., 2015), Canada, South Korea (Lee et al., 2016), the Lao People’s Democratic Republic, Thailand and Vietnam (Janetanakit et al., 2016), causing severe economic losses for the pig industry. Because PDCoV is an emerging virus, little is known regarding its pathogenesis and virus-host interactions, and there are no effective drugs or vaccines to control the disease.
Cholesterol 25-hydroxylase (CH25 H) is a 31.6 kDa hydroxylase found in the endoplasmic reticulum whose main function is to regulate cholesterol metabolism in vivo (Lund et al., 1998). CH25H catalyzes the conversion of excess cholesterol to 25-hydroxycholesterol (25HC); 25HC can also negatively regulate cholesterol production, further inhibiting the accumulation of cholesterol (Janowski et al., 1999; Kandutsch et al., 1978). Cholesterol is an important component of cellular membranes and is crucial for viral invasion. Recent studies have found that the antiviral effects of CH25H are mediated by its product 25HC. 25HC can inhibit the replication of many viruses including Zika virus (ZIKV) (Li et al., 2017), human immunodeficiency virus (HIV) (Raleigh et al., 2018), hepatitis C virus (HCV) (Anggakusuma Romero-Brey et al., 2015; Chen et al., 2014; Xiang et al., 2015), Ebola virus (Liu et al., 2013), Nipah virus (Liu et al., 2013), porcine epidemic diarrhea virus (PEDV) (Zhang et al., 2019), porcine reproductive and respiratory syndrome virus (PRRSV) (Ke et al., 2017; Song et al., 2019, 2017), and Lassa virus (Shrivastava-Ranjan et al., 2016). Remarkably, CH25H-M, a mutant enzyme lacking hydroxylase activity, has antiviral effects against a variety of viruses including HCV (Chen et al., 2014), PRRSV (Ke et al., 2017), bovine parainfluenza virus type 3 (BPIV3) (Lv et al., 2019) and PEDV (Zhang et al., 2019). However, the antiviral effects and mechanisms of action of CH25H and 25HC against PDCoV remain unclear.
In this study, we investigated the expression and antiviral role of CH25H during PDCoV infection. We found that CH25H plays a negative role in the infection of PDCoV. Moreover, PDCoV infection significantly upregulated CH25H expression. The product of CH25H enzymatic activity, 25HC, inhibited PDCoV replication by blocking virus entry. CH25H-M retained the ability to inhibit PDCoV infection although the underlying mechanism remains unclear. These data may be helpful in the search for novel anti-PDCoV drugs and vaccines.
2. Materials and methods
2.1. Cell culture and viruses
LLC-PK1 cells were purchased from the American Type Culture Collection (ATCC CL-101) and maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen, Carlsbad, CA, USA) containing 10 % fetal bovine serum (Invitrogen). IPI-FX cells were derived from IPI-2I cells (porcine ileum epithelial cells) by sub-cloning through limited serial dilution (Wang et al., 2019). The cells were incubated at 37 °C under a humidified atmosphere containing 5 % CO2. PDCoV strain CHN-HN-2014 (GenBank accession number KT336560) was isolated from a piglet with severe diarrhea in China in 2014 and preserved in our laboratory (Dong et al., 2016).
2.2. Plasmids and small interfering RNA (siRNA) silencing
The coding sequences of porcine CH25H CH25H-M were cloned into pCAGGS-Flag. The mutant CH25H-M was engineered to lack catalytic activity by site-directed mutagenesis of histidine residues 242 and 243 to glutamine. The pCAGGS-CH25H-Flag and pCAGGS-CH25H-M-Flag plasmids were constructed and maintained in our laboratory (Ke et al., 2017). The siRNAs targeting pCH25H were designed and synthesized by GenScript (Ke et al., 2017). The primer sequences used were as follows: 5′-GGCAUGUGCUGCAUCACAATT-3′, 5′-UUGUGAUGCAGCACAUGCCTT-3′.
2.3. RNA isolation and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from treated cells using TRIzol reagent (Invitrogen) and reverse transcribed into cDNA using the Transcriptor First Strand cDNA Synthesis Kit (Roche, Basel, Switzerland) according to the manufacturer’s instructions. The following primers were used: PDCoV N (F 5′-CGCTTAACTCCGCCATCAA-3′, R 5′-TCTGGTGTAACGCAGCCAGTA-3′); pCH25H (F 5′- CAGTGCAACAGAGTCATCGG-3′, R 5′-GCCCTTCTTCCCAGTCTTCT-3′); and pGAPDH (F 5′- ACATGGCCTCCAAGGAGTAAGA-3′, R 5′-GATCGAGTTGGGGCTGTGACT-3′). qRT-PCR was performed using SYBR green PCR mix (Applied Biosystems, Foster City, CA, USA) and the ABI 7500 real-time PCR system (Applied Biosystems). Expression levels were normalized to those of glyceraldehyde 3-phosphate dehydrogenase. Absolute quantitative mRNA levels were calculated using standard curves.
2.4. Indirect immunofluorescence assay (IFA)
Following treatments, IPI-FX cells were washed three times with phosphate-buffered saline (PBS), then fixed with 4 % paraformaldehyde for 15 min and permeabilized with pre-chilled −20 °C methanol for 10 min. After washing three times with PBS, the cells were blocked with 5 % bovine serum albumin for 1 h and then incubated with primary antibodies for 1 h. The cells were washed with PBS and then incubated with Alexa Fluor-labeled secondary antibodies (Abbkine, Wuhan, China) for 45 min at 37 °C. Thereafter, nuclei were stained with 0.01 % 4′,6-diamidino-2-phenylindole (Beyotime, Shanghai, China) for 15 min. Fluorescent images were acquired using a confocal laser scanning microscope (Olympus FluoView v3.1; Olympus, Tokyo, Japan).
2.5. Western blotting
Treated cells were washed twice with cold PBS and harvested with lysis buffer (4 % sodium dodecyl sulfate, 3 % dithiothreitol, 0.065 mM Tris-HCl, pH 6.8, and 30 % glycerin) supplemented with a protease inhibitor cocktail (Sigma, St. Louis, MO, USA). The samples were centrifuged at 12,000 rpm for 5 min to remove cell debris. Equal volumes of samples were separated by 12 % SDS-PAGE and then proteins were electroblotted onto a polyvinylidene difluoride membrane (Millipore, Billerica, MA, USA). The membrane was blocked with Tris-buffered saline containing 5 % nonfat milk and 0.1 % Tween-20 and then treated with primary antibodies. The expression of CH25H was analyzed using a specific anti-mouse CH25H antibody (H00009023-M01, Novus Biologicals, Littleton, CO, USA). A specific monoclonal antibody against PDCoV N (diluted 1:1000) was used to detect the expression of N protein as described previously (Zhu et al., 2018). An anti-rabbit-β-actin monoclonal antibody (ABclonal, Wuhan, China) was used to assess the expression of β-actin and to confirm equal loading of protein samples.
2.6. Plaque assay for PDCoV titers
LLC-PK1 cells were grown as a monolayer in six-well tissue culture plates and infected for 1 h with 10-fold serial dilutions (1 mL each) of PDCoV-containing samples. After washing three times with PBS, the cells were covered with overlay medium (DMEM containing 1.8 % (w/v) Bacto agar and 2.5 μg/mL trypsin), incubated at 37 °C for 1.5 days, and examined using a plaque assay. The average plaque numbers and their standard deviations were calculated from three independent experiments. Viral titers were expressed as plaque-forming units/mL.
2.7. Statistical analysis
Differences between groups were assessed using two-tailed unpaired t tests and differences among multiple groups were assessed using analysis of variance. All statistical analyses were performed using GraphPad Prism 5.0 (GraphPad, San Diego, CA, USA). Values of P < 0.05 were considered statistically significant and indicated as follows in figures: *, P = 0.05; **, P = 0.01; ***, P = 0.001.
3. Results
3.1. PDCoV infection upregulates CH25H expression in IPI-FX cells
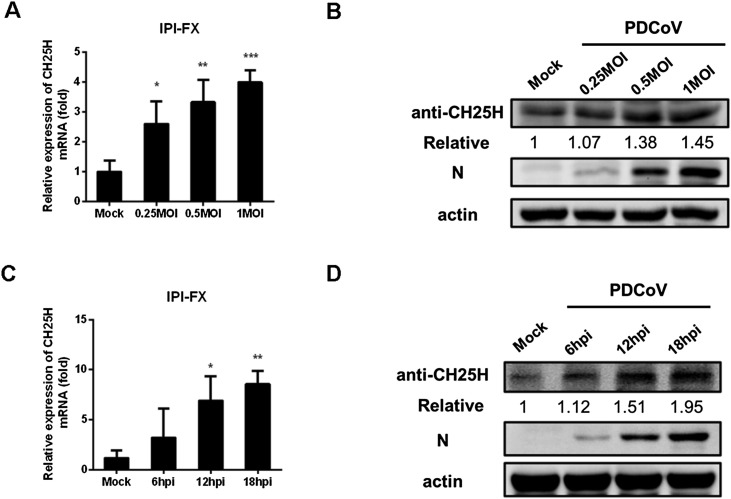
To assess the effects of PDCoV infection on the expression of CH25H, IPI-FX cells were infected with different doses of PDCoV (multiplicity of infection (MOI) 0.25, 0.5, and 1) for 12 h. CH25H mRNA and protein levels were assessed by qRT-PCR (Fig. 1 A) and western blotting (Fig. 1 B), respectively. Both mRNA and protein levels of CH25H were significantly up-regulated in IPI-FX cells infected with PDCoV compared with control cells. Subsequently, we assessed CH25H mRNA levels and protein levels at different times (6, 12, and 18 h post-infection (hpi)) in IPI-FX cells infected with PDCoV (MOI 1). There were no significant changes in the mRNA and protein levels of CH25H at 6 hpi, while both CH25H mRNA (Fig. 1 C) and protein levels (Fig. 1 D) were significantly upregulated at 12 and 18 hpi. These results indicated that CH25H expression was upregulated in IPI-FX cells infected with PDCoV in a time- and dose-dependent manner.
Fig. 1.
PDCoV infection upregulates CH25H expression in IPI-FX cells. (A, B) IPI-FX cells were infected with different doses of PDCoV (MOI 0.25, 0.5, and 1). Uninfected cells were used as a negative control. Samples were collected at 12 hpi and CH25H mRNA and protein levels were determined by qRT-PCR (A) and western blotting (B), respectively. (C, D) IPI-FX cells were inoculated with PDCoV (MOI 1) and samples were collected at 6, 12, and 18 hpi. CH25H mRNA and protein levels were assessed by qRT-PCR (C) and western blotting (D), respectively. PDCoV infection was verified by western blotting with an anti-PDCoV N protein antibody. β-actin used as a sample loading control. All data were presented as the means ± standard deviations of three independent experiments. *P < 0.05 and **P < 0.01.
3.2. CH25H inhibits PDCoV replication, but inhibition does not completely depend on its enzymatic activity
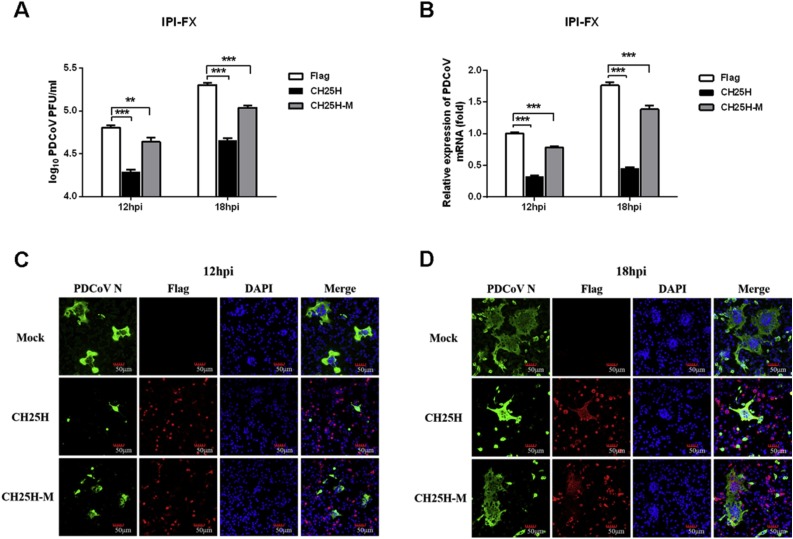
The histidine and glutamate-rich region of CH25H uses oxygen and diiron cofactors to catalyze the hydroxylation of cholesterol to yield 25HC. A previous study demonstrated that mutation of CH25H histidines 242 and 243 resulted in a mutant enzyme (CH25H-M) incapable of producing 25HC (Lund et al., 1998). To evaluate the effect of CH25H and its enzyme-activity mutant (CH25H-M) on PDCoV proliferation, IPI-FX cells were transiently transfected with the eukaryotic expression plasmids pCAGGS-Flag-CH25H or pCAGGS-Flag-CH25H-H242Q/H243Q (pCAGGS-Flag-CH25H-M). Subsequently, the cells were inoculated with PDCoV (MOI 0.5) at 28 h post-transfection. A plaque assay was performed to assess the inhibitory effect of CH25H and CH25H-M on PDCoV in LLC-PK1 cells. The viral titers in cells overexpressing CH25H or CH25H-M were significantly lower than those of control cells, however, the inhibitory effect of CH25H-M was weaker than that of CH25H (Fig. 2 A), suggesting that the antiviral effect of CH25H is not completely dependent on its enzymatic activity. Consistently, qRT-PCR (Fig. 2 B) and IFA (Fig. 2 C, D) demonstrated that PDCoV titers were suppressed in cells overexpressing CH25H or CH25H-M, with the inhibitory effect of CH25H being more pronounced than that of CH25H-M. These results showed that CH25H inhibited PDCoV proliferation but that inhibition did not totally depend on its enzymatic activity.
Fig. 2.
CH25H inhibits PDCoV replication and inhibition does not completely depend on enzymatic activity. (A, B) IPI-FX cells were transfected with CH25H and CH25H-M expression vectors or empty vector. At 28 h post-transfection, cells were infected with PDCoV (MOI 0.5). After another 12 or 18 h, cells were harvested for plaque assay (A) or qRT-PCR (B). (C, D) IPI-FX cells were transfected with CH25H and CH25H-M expression vectors or empty vector. At 28 h post-transfection, cells were infected with PDCoV (MOI 0.5). After another 12 (C) or 18 h (D), IFAs were conducted to detect PDCoV infection using an anti-PDCoV N protein antibody. Scale bar =50 μm. All data are presented as the means ± standard deviations of three independent experiments. *P < 0.05 and **P < 0.01.
3.3. Silencing of CH25H promotes PDCoV proliferation
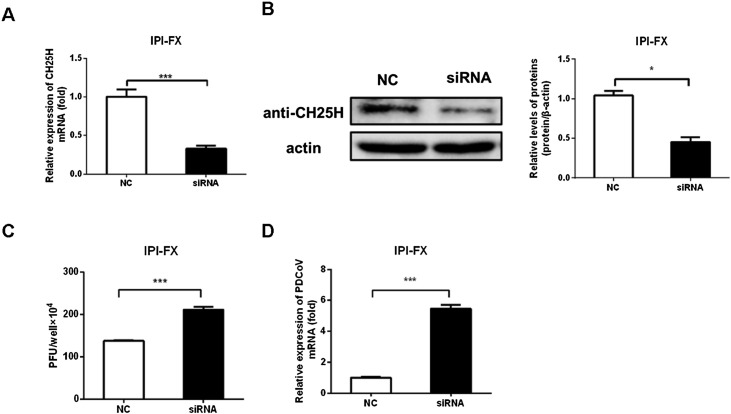
To further assess whether CH25H was a host restriction factor against PDCoV infection, IPI-FX cells were transfected with specific siRNAs targeting porcine CH25H to silence endogenous expression of CH25H. At 36 h post-transfection, CH25H mRNA and protein levels were assessed by qRT-PCR (Fig. 3 A) and western blotting (Fig. 3 B). Treatment with siRNA had clear knockdown effects on CH25H expression (Fig. 3 B). IPI-FX cells were transfected with the siRNA and infected with PDCoV (MOI 0.5) at 36 h post-transfection. PDCoV titers and PDCoV N gene copy number were determined by plaque assay and qRT-PCR analysis, respectively. As shown in Fig. 3 C, viral titers in cells silenced for CH25H expression were significantly higher than those in cells transfected with a negative-control siRNA. Consistently, qRT-PCR demonstrated that PDCoV N mRNA levels were increased in conjunction with downregulation of CH25H expression (Fig. 3 D). Based on the results of overexpression and knockdown assays, we concluded that CH25H was a host restriction factor against PDCoV infection.
Fig. 3.
Silencing of CH25H using siRNA promotes PDCoV proliferation. (A, B) IPI-FX cells in 24-well plates were transfected with CH25H-specific siRNAs or a negative-control (NC) siRNA. At 36 h post-transfection, cells were harvested and knockdown efficiency was determined by qRT-PCR (A) and western blotting (B, left panel). The histogram (B, right panel) showed the silence efficiency based on the results of western blotting. (C, D) IPI-FX cells in 24-well plates were transfected with CH25H-specific siRNAs or a negative control (NC) siRNA. At 36 h post-transfection, cells were infected with PDCoV (MOI 0.5). Cells were harvested at 18 hpi to analyze PDCoV titers and mRNA levels by plaque assay (C) and qRT-PCR (D), respectively. All data are presented as the means ± standard deviations of three independent experiments. *P < 0.05 and **P < 0.01.
3.4. 25HC inhibits PDCoV infection
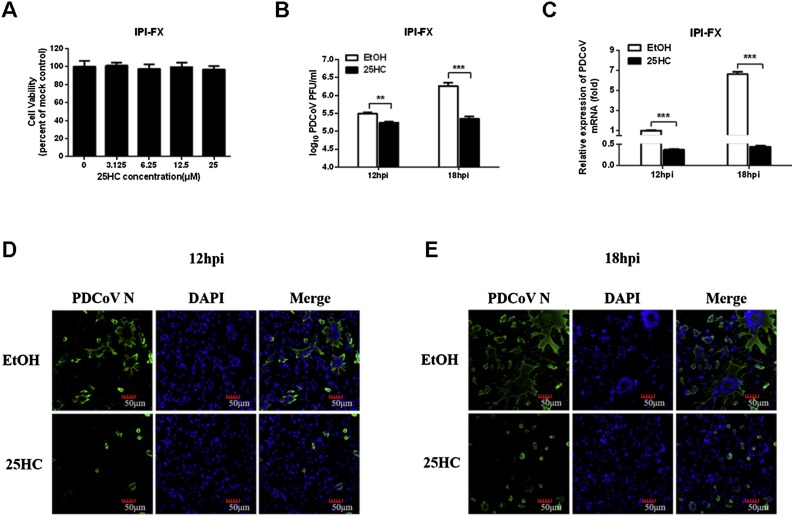
Previous studies suggested that CH25H exerts its antiviral activity by catalyzing the conversion of cholesterol to 25HC (Shawli et al., 2019; Xie et al., 2019; Yuan et al., 2019). To further investigate the inhibitory effect of 25HC on PDCoV infection, the cytotoxicity of 25HC was first assessed in IPI-FX cells using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. To this end, IPI-FX cells were cultured in 96-well plates and treated with various concentrations of 25HC (3.125, 6.25, 12.5, and 25 μM) or EtOH for 24 h. As shown in Fig. 4 A, treatment with 12.5 μM 25HC had no effect on cell viability. Then, IPI-FX cells were pretreated with 25HC (12.5 μM) for 8 h, followed infection with PDCoV (MOI 0.5). Viral titers were determined by plaque assay in LLC-PK1 cells. The results showed that treatment with 25HC significantly inhibited PDCoV production (Fig. 4 B). As expected, qRT-PCR and IFA also showed that treatment with 25HC reduced PDCoV N mRNA levels and infection by PDCoV (Fig. 4 C–E).
Fig. 4.
25HC inhibits PDCoV proliferation. (A) Assessment of the cytotoxicity of 25HC against IPI-FX cells. IPI-FX cells were incubated with different concentrations of 25HC or EtOH for 24 h and then evaluated using a MTT assay. (B–E) IPI-FX cells were pretreated with 25HC at the indicated concentrations for 8 h prior to PDCoV infection in the presence of 2.5 μg/mL trypsin (MOI 0.5). The infected cells were cultured in the presence of 25HC and harvested at 12 or 18 hpi for the plaque assay (B), qRT-PCR (C) or IFA (D, E). IFA was performed using a specific antibody against PDCoV N protein. Scale bar =50 μm. All data are presented as the means ± standard deviations of three independent experiments. *P < 0.05 and **P < 0.01.
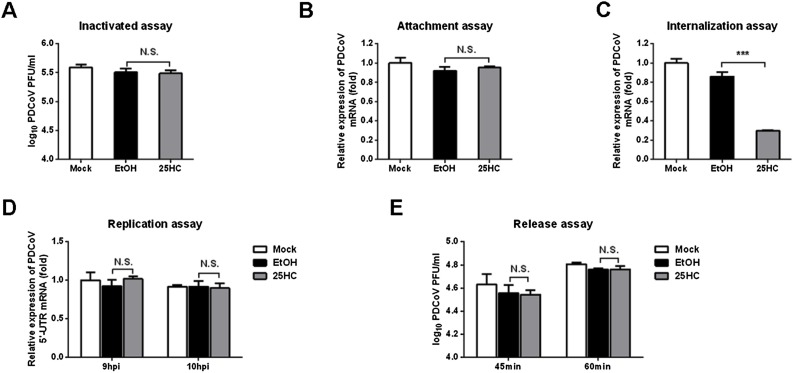
3.5. 25HC inhibits PDCoV invasion
To further investigate which stages (adsorption, invasion, replication or release) were targeted by 25HC during the PDCoV replication cycle, we first tested whether 25HC could directly inactivate PDCoV particles. To this end, PDCoV was mixed with 25HC (12.5 μM) or EtOH and incubated at 37 °C for 3 h. Viral titers were assessed by plaque assay in LLC-PK1 cells. As shown in Fig. 5 A, 25HC treatment did not have any direct effect on PDCoV particles.
Fig. 5.
25HC inhibits PDCoV invasion. (A) Viral inactivation assay. PDCoV and 25HC were incubated at 37 °C for 3 h. LLC-PK1 cells cultured in six-well plates were prechilled at 4 °C for 1 h. The growth medium was replaced with a mixture of 25HC (12.5 μM) or EtOH and PDCoV. After incubation at 4 °C for another 2 h, the cells were washed with precooled PBS, covered with overlay medium, incubated at 37 °C for a further 1–2 days, and assessed using a plaque assay. (B) Adsorption assay. IPI-FX cells cultured in 24-well plates were pretreated with 25HC (12.5 μM) or EtOH for 1 h at 37 °C and then prechilled at 4 °C for 1 h. The growth medium was then replaced with a mixture of 25HC (10 μM) or EtOH and PDCoV (MOI 0.5) in the presence of 2.5 μg/mL trypsin for 1 h at 4 °C. After washing three times with prechilled PBS, PDCoV N protein mRNA levels were measured by qRT-PCR. (C) Penetration assay. IPI-FX cells cultured in 24-well plates, prechilled at 4 °C for 1 h and then infected with PDCoV (MOI 0.5) in the presence of 2.5 μg/mL trypsin for 1 h at 4 °C. The virus-containing medium was replaced with fresh medium containing 25HC (12.5 μM) or EtOH. The temperature was raised to 37 °C for 3 h and then the cells were washed with PBS, pH 3. PDCoV N protein mRNA levels were assessed by qRT-PCR. (D) Replication assay. IPI-FX cells were infected with PDCoV (MOI 0.5) in the presence of 2.5 μg/mL trypsin at 37 °C for 8 h. Cell-free virus particles were removed. Cells were treated with fresh medium containing 25HC (12.5 μM) or EtOH for 9 or 10 h. The cells were collected for qRT-PCR to assess levels of negative-sense PDCoV RNA. (E) Release assay. IPI-FX cells were inoculated with PDCoV (MOI 0.5) at 4 °C for 1 h. At 12 hpi, the inoculum was replaced with fresh medium containing 25HC (12.5 μM). Cell supernatants were harvested at 45 and 60 min and titrated using a plaque assay. All data are presented as the means ± standard deviations of three independent experiments. *P < 0.05 and **P < 0.01.
To assess the effect of 25HC on PDCoV adsorption, IPI-FX cells were pretreated with 25HC (12.5 μM) or EtOH at 37 °C for 1 h. The cells were then were prechilled at 4 °C for 1 h. The culture medium was replaced with a mixture of 25HC (12.5 μM) or EtOH and PDCoV (MOI 0.5). After incubation at 4 °C for another 1 h, the cells were washed with precooled PBS and then PDCoV N mRNA levels were determined by qRT-PCR. As shown in Fig. 5 B, 25HC treatment did not block viral attachment to IPI-FX cells.
To assess viral penetration, IPI-FX cells were prechilled at 4 °C for 1 h and then incubated with PDCoV (MOI 0.5) at 4 °C for another 1 h. Next, virus-containing medium was replaced with fresh medium containing 25HC (12.5 μM) or EtOH and incubated at 37 °C for 3 h. The cells were washed with PBS, pH 3. Levels of PDCoV N mRNA was measured by qRT-PCR. As shown in Fig. 5 C, 25HC blocked PDCoV internalization.
To investigate whether 25HC affects PDCoV replication, IPI-FX cells were incubated with PDCoV (MOI 0.5) for 8 h. The virus-containing medium was replaced with fresh medium containing 25HC (12.5 μM) or EtOH. At 9 and 10 hpi, the infected cells were collected and PDCoV negative-sense RNA levels were assessed by qRT-PCR using primers targeting the viral 5′ untranslated region. As shown in Fig. 5 D, 25HC treatment did not perturb PDCoV RNA replication.
Finally, we investigated whether 25HC affects release of PDCoV using a plaque assay. IPI-FX cells were infected with PDCoV (MOI 0.5). Virus-containing medium was replaced with fresh medium containing 25HC (12.5 μM) or EtOH at 12 hpi. The cell supernatant was harvested at 45 and 60 min after medium replacement for the plaque assay. As shown in Fig. 5 E, 25HC did not inhibit the release of PDCoV. Taken together, these results indicated that 25HC inhibited PDCoV proliferation by blocking viral entry but had no effect on the adsorption, replication, or release stages of the viral life cycle. Moreover, 25HC did not directly inactivate PDCoV in vitro.
4. Discussion
25HC has been described as having potent antiviral activity against a panel of enveloped and nonenveloped viruses via multiple mechanisms that depend on the virus-host context. As a cholesterol inhibitor, 25HC may have multiple antiviral mechanisms including blockade of viral entry by modifying cell membranes, disruption of viral replication via membranous web formation, or interference with the circulation of cholesterol between organelles. For instance, 25HC showed inhibitory effects on PEDV (Zhang et al., 2019), rabies virus (Yuan et al., 2019), ZIKV (Li et al., 2017), PRRSV (Ke et al., 2017; Song et al., 2019) and HIV entry (Saulle et al., 2020); 25HC suppressed HCV proliferation by disrupting membranous web formation (Anggakusuma Romero-Brey et al., 2015); and 25HC inhibited human rotavirus infection by interfering with recycling of cholesterol between the endoplasmic reticulum and late endosomes, a process that is exploited by rotavirus to penetrate cells (Civra et al., 2018). Recently, Jeon and Lee found that treatment of swine testicle cells or virions with methyl-β-cyclodextrin hindered PDCoV entry, and that addition of exogenous cholesterol restored PDCoV infectivity (Jeon and Lee, 2018). Furthermore, they also found that cholesterol in both the target cell membrane and the viral envelope was required for PDCoV entry (Jeon and Lee, 2018). In our present study, treatment with 25HC did not affect the infectivity of PDCoV particles, suggesting that 25HC does not remove cholesterol from virions. However, treating IPI-FX cells with 25HC significantly inhibited PDCoV proliferation by blocking viral entry, indicating that 25HC inhibited PDCoV replication by affecting the distribution of cholesterol within cell membranes. We cannot rule out the possibility that the 25HC and viral cholesterol competitively bind to a receptor to attenuate PDCoV entry. The key functional receptor of PDCoV has not yet been identified, and thus we could not determine its cholesterol dependence. Cholesterol in the cellular membrane plays an important role in interactions between the severe acute respiratory syndrome coronavirus (SARS-CoV) spike protein and the cellular receptor angiotensin converting enzyme 2 (ACE2) (Glende et al., 2008). Most molecules described as receptors for dengue virus are present within cholesterol-rich complexes (Cambi et al., 2004; Reyes-Del Valle et al., 2005; Wan et al., 2012). Future studies should address whether cholesterol promotes PDCoV entry through the interaction between viral spike protein and cellular receptors. In addition, whether the expressions of CH25H and its products 25HC are up-regulated in small intestinal tissues of PDCoV-infected pigs needs further investigations.
There is limited information on the mechanism through which a CH25H mutant (CH25H-M) lacking hydroxylase activity inhibits virus production. However, several studies have shown that the antiviral activity of CH25H does not entirely depend on its enzymatic activity. CH25H-M interacts with HCV NS5A to disrupt the formation of the NS5A dimer, which is necessary for HCV replication (Chen et al., 2014). Similarly, CH25H-M inhibits PRRSV production by degrading PRRSV nsp1α with diminished antiviral activity (Ke et al., 2017). CH25H-M also restricted PEDV (Zhang et al., 2019) and BPIV3 (Lv et al., 2019) infection, although the underlying mechanisms are currently unclear. In the present study, overexpression of CH25H-M also inhibited the proliferation of PDCoV in IPI-FX cells. The mechanism through which CH25H-M inhibits PDCoV proliferation remains to be further studied. We speculate that CH25H may interact with PDCoV proteins to affect replication and assembly of PDCoV, and that CH25H-M retains this function.
In summary, we demonstrated for the first time that CH25H inhibits PDCoV proliferation. We also showed that 25HC inhibits infection by PDCoV, supporting the view that 25HC has broad antiviral activity. In addition, we found that CH25H-M had some antiviral effect, suggesting that the ability of CH25H to inhibit PDCoV proliferation did not absolutely depend on its enzymatic activity. These findings may inform the development of anti-PDCoV drugs.
5. Author statement
The work described in this manuscript has not been submitted elsewhere for publication and is under consideration by any other journal. All the author listed have approved to submit this manuscript to Virus Research. If accepted for publication, we assure that it will not be published elsewhere without the consent of the Publisher.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31730095, U1704231, 31902247), and the Special Project for Technology Innovation of Hubei Province (2017ABA138).
References
- Anggakusuma Romero-Brey I., Berger C., Colpitts C.C., Boldanova T., Engelmann M., Todt D., Perin P.M., Behrendt P., Vondran F.W., Xu S., Goffinet C., Schang L.M., Heim M.H., Bartenschlager R., Pietschmann T., Steinmann E. Interferon-inducible cholesterol-25-hydroxylase restricts hepatitis C virus replication through blockage of membranous web formation. Hepatology. 2015;62(3):702–714. doi: 10.1002/hep.27913. [DOI] [PubMed] [Google Scholar]
- Cambi A., de Lange F., van Maarseveen N.M., Nijhuis M., Joosten B., van Dijk E.M., de Bakker B.I., Fransen J.A., Bovee-Geurts P.H., van Leeuwen F.N., Van Hulst N.F., Figdor C.G. Microdomains of the C-type lectin DC-SIGN are portals for virus entry into dendritic cells. J. Cell Biol. 2004;164(1):145–155. doi: 10.1083/jcb.200306112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Wang S., Yi Z., Tian H., Aliyari R., Li Y., Chen G., Liu P., Zhong J., Chen X., Du P., Su L., Qin F.X., Deng H., Cheng G. Interferon-inducible cholesterol-25-hydroxylase inhibits hepatitis C virus replication via distinct mechanisms. Sci. Rep. 2014;4:7242. doi: 10.1038/srep07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Gauger P., Stafne M., Thomas J., Arruda P., Burrough E., Madson D., Brodie J., Magstadt D., Derscheid R., Welch M., Zhang J. Pathogenicity and pathogenesis of a United States porcine deltacoronavirus cell culture isolate in 5-day-old neonatal piglets. Virology. 2015;482:51–59. doi: 10.1016/j.virol.2015.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civra A., Francese R., Gamba P., Testa G., Cagno V., Poli G., Lembo D. 25-Hydroxycholesterol and 27-hydroxycholesterol inhibit human rotavirus infection by sequestering viral particles into late endosomes. Redox Biol. 2018;19:318–330. doi: 10.1016/j.redox.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N., Fang L., Zeng S., Sun Q., Chen H., Xiao S. Porcine deltacoronavirus in Mainland China. Emerg Infect Dis. 2015;21(12):2254–2255. doi: 10.3201/eid2112.150283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N., Fang L., Yang H., Liu H., Du T., Fang P., Wang D., Chen H., Xiao S. Isolation, genomic characterization, and pathogenicity of a Chinese porcine deltacoronavirus strain CHN-HN-2014. Vet. Microbiol. 2016;196:98–106. doi: 10.1016/j.vetmic.2016.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P., Fang L., Liu X., Hong Y., Wang Y., Dong N., Ma P., Bi J., Wang D., Xiao S. Identification and subcellular localization of porcine deltacoronavirus accessory protein NS6. Virology. 2016;499:170–177. doi: 10.1016/j.virol.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P., Fang L., Hong Y., Liu X., Dong N., Ma P., Bi J., Wang D., Xiao S. Discovery of a novel accessory protein NS7a encoded by porcine deltacoronavirus. J. Gen. Virol. 2017;98(2):173–178. doi: 10.1099/jgv.0.000690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glende J., Schwegmann-Wessels C., Al-Falah M., Pfefferle S., Qu X., Deng H., Drosten C., Naim H.Y., Herrler G. Importance of cholesterol-rich membrane microdomains in the interaction of the S protein of SARS-coronavirus with the cellular receptor angiotensin-converting enzyme 2. Virology. 2008;381(2):215–221. doi: 10.1016/j.virol.2008.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homwong N., Jarvis M.C., Lam H.C., Diaz A., Rovira A., Nelson M., Marthaler D. Characterization and evolution of porcine deltacoronavirus in the United States. Prev. Vet. Med. 2016;123:168–174. doi: 10.1016/j.prevetmed.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Jung K., Vlasova A.N., Chepngeno J., Lu Z., Wang Q., Saif L.J. Isolation and characterization of porcine deltacoronavirus from pigs with diarrhea in the United States. J. Clin. Microbiol. 2015;53(5):1537–1548. doi: 10.1128/JCM.00031-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janetanakit T., Lumyai M., Bunpapong N., Boonyapisitsopa S., Chaiyawong S., Nonthabenjawan N., Kesdaengsakonwut S., Amonsin A. Porcine Deltacoronavirus, Thailand, 2015. Emerg Infect Dis. 2016;22(4):757–759. doi: 10.3201/eid2204.151852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski B.A., Grogan M.J., Jones S.A., Wisely G.B., Kliewer S.A., Corey E.J., Mangelsdorf D.J. Structural requirements of ligands for the oxysterol liver X receptors LXRalpha and LXRbeta. Proc. Natl. Acad. Sci. U. S. A. 1999;96(1):266–271. doi: 10.1073/pnas.96.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J.H., Lee C. Cholesterol is important for the entry process of porcine deltacoronavirus. Arch. Virol. 2018;163(11):3119–3124. doi: 10.1007/s00705-018-3967-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandutsch A.A., Chen H.W., Heiniger H.J. Biological activity of some oxygenated sterols. Science. 1978;201(4355):498–501. doi: 10.1126/science.663671. [DOI] [PubMed] [Google Scholar]
- Ke W., Fang L., Jing H., Tao R., Wang T., Li Y., Long S., Wang D., Xiao S. Cholesterol 25-hydroxylase inhibits porcine reproductive and respiratory syndrome virus replication through enzyme activity-dependent and -independent mechanisms. J. Virol. 2017;91 doi: 10.1128/JVI.00827-17. 00827-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Chung H.C., Nguyen V.G., Moon H.J., Kim H.K., Park S.J., Lee C.H., Lee G.E., Park B.K. Detection and phylogenetic analysis of porcine deltacoronavirus in Korean swine farms, 2015. Transbound. Emerg. Dis. 2016;63(3):248–252. doi: 10.1111/tbed.12490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Deng Y.Q., Wang S., Ma F., Aliyari R., Huang X.Y., Zhang N.N., Watanabe M., Dong H.L., Liu P., Li X.F., Ye Q., Tian M., Hong S., Fan J., Zhao H., Li L., Vishlaghi N., Buth J.E., Au C., Liu Y., Lu N., Du P., Qin F.X., Zhang B., Gong D., Dai X., Sun R., Novitch B.G., Xu Z., Qin C.F., Cheng G. 25-hydroxycholesterol protects host against Zika virus infection and its associated microcephaly in a mouse model. Immunity. 2017;46(3):446–456. doi: 10.1016/j.immuni.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.Y., Aliyari R., Chikere K., Li G., Marsden M.D., Smith J.K., Pernet O., Guo H., Nusbaum R., Zack J.A., Freiberg A.N., Su L., Lee B., Cheng G. Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity. 2013;38(1):92–105. doi: 10.1016/j.immuni.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E.G., Kerr T.A., Sakai J., Li W.P., Russell D.W. cDNA cloning of mouse and human cholesterol 25-hydroxylases, polytopic membrane proteins that synthesize a potent oxysterol regulator of lipid metabolism. J. Biol. Chem. 1998;273(51):34316–34327. doi: 10.1074/jbc.273.51.34316. [DOI] [PubMed] [Google Scholar]
- Lv L., Zhao G., Wang H., He H. Cholesterol 25-Hydroxylase inhibits bovine parainfluenza virus type 3 replication through enzyme activity-dependent and -independent ways. Vet. Microbiol. 2019;239:108456. doi: 10.1016/j.vetmic.2019.108456. [DOI] [PubMed] [Google Scholar]
- Ma Y., Zhang Y., Liang X., Lou F., Oglesbee M., Krakowka S., Li J. Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. mBio. 2015;6(2):e00064. doi: 10.1128/mBio.00064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marthaler D., Raymond L., Jiang Y., Collins J., Rossow K., Rovira A. Rapid detection, complete genome sequencing, and phylogenetic analysis of porcine deltacoronavirus. Emerg Infect Dis. 2014;20(8):1347–1350. doi: 10.3201/eid2008.140526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raleigh D.R., Sever N., Choksi P.K., Sigg M.A., Hines K.M., Thompson B.M., Elnatan D., Jaishankar P., Bisignano P., Garcia-Gonzalo F.R., Krup A.L., Eberl M., Byrne E.F.X., Siebold C., Wong S.Y., Renslo A.R., Grabe M., McDonald J.G., Xu L., Beachy P.A., Reiter J.F. Cilia-associated oxysterols activate smoothened. Mol. Cell. 2018;72(2):316–327. doi: 10.1016/j.molcel.2018.08.034. e315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Del Valle J., Chavez-Salinas S., Medina F., Del Angel R.M. Heat shock protein 90 and heat shock protein 70 are components of dengue virus receptor complex in human cells. J. Virol. 2005;79(8):4557–4567. doi: 10.1128/JVI.79.8.4557-4567.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saulle I., Ibba S.V., Vittori C., Fenizia C., Mercurio V., Vichi F., Caputo S.L., Trabattoni D., Clerici M., Biasin M. AIDS; 2020. Sterol Metabolism Modulates Susceptibility to HIV-1 Infection. [DOI] [PubMed] [Google Scholar]
- Shawli G.T., Adeyemi O.O., Stonehouse N.J., Herod M.R. The oxysterol 25-Hydroxycholesterol inhibits replication of murine norovirus. Viruses. 2019;11(2) doi: 10.3390/v11020097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava-Ranjan P., Bergeron E., Chakrabarti A.K., Albarino C.G., Flint M., Nichol S.T., Spiropoulou C.F. 25-hydroxycholesterol inhibition of lassa virus infection through aberrant GP1 glycosylation. mBio MBio. 2016;7:01808–01816. doi: 10.1128/mBio.01808-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z., Zhang Q., Liu X., Bai J., Zhao Y., Wang X., Jiang P. Cholesterol 25-hydroxylase is an interferon-inducible factor that protects against porcine reproductive and respiratory syndrome virus infection. Vet. Microbiol. 2017;210:153–161. doi: 10.1016/j.vetmic.2017.09.011. [DOI] [PubMed] [Google Scholar]
- Song Z., Bai J., Nauwynck H., Lin L., Liu X., Yu J., Jiang P. 25-Hydroxycholesterol provides antiviral protection against highly pathogenic porcine reproductive and respiratory syndrome virus in swine. Vet. Microbiol. 2019;231:63–70. doi: 10.1016/j.vetmic.2019.02.035. [DOI] [PubMed] [Google Scholar]
- Wan S.W., Lin C.F., Lu Y.T., Lei H.Y., Anderson R., Lin Y.S. Endothelial cell surface expression of protein disulfide isomerase activates beta1 and beta3 integrins and facilitates dengue virus infection. J. Cell. Biochem. 2012;113(5):1681–1691. doi: 10.1002/jcb.24037. [DOI] [PubMed] [Google Scholar]
- Wang L., Byrum B., Zhang Y. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014. Emerg Infect Dis. 2014;20(7):1227–1230. doi: 10.3201/eid2007.140296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.W., Yue H., Fang W., Huang Y.W. Complete genome sequence of porcine deltacoronavirus strain CH/Sichuan/S27/2012 from Mainland China. Genome Announc. 2015;3(5) doi: 10.1128/genomeA.00945-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Fang L., Liu S., Ke W., Wang D., Peng G., Xiao S. Susceptibility of porcine IPI-2I intestinal epithelial cells to infection with swine enteric coronaviruses. Vet. Microbiol. 2019;233:21–27. doi: 10.1016/j.vetmic.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Huang Y., Yuen K.Y. Coronavirus diversity, phylogeny and interspecies jumping. Exp. Biol. Med. (Maywood) 2009;234(10):1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86(7):3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Tang J.J., Tao W., Cao X., Song B.L., Zhong J. Identification of cholesterol 25-hydroxylase as a novel host restriction factor and a part of the primary innate immune responses against hepatitis C virus infection. J. Virol. 2015;89(13):6805–6816. doi: 10.1128/JVI.00587-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T., Feng M., Dai M., Mo G., Ruan Z., Wang G., Shi M., Zhang X. Cholesterol-25-hydroxylase is a chicken ISG that restricts ALV-J infection by producing 25-hydroxycholesterol. Viruses. 2019;11(6) doi: 10.3390/v11060498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Wang Z., Tian B., Zhou M., Fu Z.F., Zhao L. Cholesterol 25-hydroxylase suppresses rabies virus infection by inhibiting viral entry. Arch. Virol. 2019;164(12):2963–2974. doi: 10.1007/s00705-019-04415-6. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Song Z., Wang M., Lan M., Zhang K., Jiang P., Li Y., Bai J., Wang X. Cholesterol 25-hydroxylase negatively regulates porcine intestinal coronavirus replication by the production of 25-hydroxycholesterol. Vet. Microbiol. 2019;231:129–138. doi: 10.1016/j.vetmic.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Liu S., Wang X., Luo Z., Shi Y., Wang D., Peng G., Chen H., Fang L., Xiao S. Contribution of porcine aminopeptidase N to porcine deltacoronavirus infection. Emerg. Microbes Infect. 2018;7(1):65. doi: 10.1038/s41426-018-0068-3. [DOI] [PMC free article] [PubMed] [Google Scholar]