Abstract
COVID-19 is one of the fatal pandemic throughout the world. For cellular fusion, its antigenic peptides are presented by major histocompatibility complex (MHC) in humans. Therefore, exploration into residual interaction details of CoV2 with MHCs shall be a promising point for instigating the vaccine development. Envelope (E) protein, the smallest outer surface protein from SARS-CoV2 genome was found to possess the highest antigenicity and is therefore used to identify B-cell and T-cell epitopes. Four novel mutations (T55S, V56F, E69R and G70del) were observed in E-protein of SARS-CoV2 after evolutionary analysis. It showed a coil➔helix transition in the protein conformation. Antigenic variability of the epitopes was also checked to explore the novel mutations in the epitope region. It was found that the interactions were more when SARS-CoV2 E-protein interacted with MHC-I than with MHC-II through several ionic and H-bonds. Tyr42 and Tyr57 played a predominant role upon interaction with MHC-I. The higher ΔG values with lesser dissociation constant values also affirm the stronger and spontaneous interaction by SARS-CoV2 proteins with MHCs. On comparison with the consensus E-protein, SARS-CoV2 E-protein showed stronger interaction with the MHCs with lesser solvent accessibility. E-protein can therefore be targeted as a potential vaccine target against SARS-CoV2.
Keywords: COVID-19, Epitope identification, Interactions with MHCs
1. Introduction
The novel severe acute respiratory syndrome coronavirus 2, also known as COVID-19 is an enveloped single-stranded RNA virus, which belongs to the family Coronavirdiae [1]. It was first identified at Wuhan in China in December 2019 [2], and since then it is spreading worldwide, resulting into deadly 2019–20 coronavirus pandemic (as declared by WHO in March 2020) [3] that affected millions worldwide. The virion comprises a nucleocapsid having phosphorylated nucleocapsid (N) protein and genomic RNA. The N-protein resides inside phospholipid bilayers [4]. It is enclosed by the spike glycoprotein trimer (S). The envelope (E) protein and membrane (M) protein (a type III transmembrane glycoprotein) reside between the S proteins on virus envelope [4]. E-protein is enigmatic and the smallest structural protein and resides on the outer surface of the virus. During replication, a minor portion of this protein gets integrated into the virion envelope and then expresses itself in the infected cell [5]. The remaining portion localizes at the site of intracellular trafficking. Here, it takes part into CoV assembly and budding, so it is essential for virus assembly and maturation [5]. The paramount cause for such a rapid spread of the virus is its alarmingly contagious nature with high reproductive rate and person-to-person transmission via respiratory droplets [6].
The progression of SARS-CoV-2 depends on the interaction between individual's immune system (include genetics (such as HLA genes), age, gender, nutritional status, neuroendocrine-immune regulation, and physical status) and the virus [7]. The clinical symptoms in patients affected with SARS-CoV-2 are highly unusual plus dry cough followed by fever, dyspnea, respiratory choking and viral pneumonia [7]. While fusing into the cells, its antigen will be presented to the antigen presentation cells (APC). Antigenic peptides are presented by major histocompatibility complex (MHC; or human leukocyte antigen (HLA) in humans) and then identified by virus-specific cytotoxic T lymphocytes (CTLs). However, SARS-CoV induces the generation of double-membrane vesicles where replication occurs. This hinders host detection of their dsRNA [8]. APCs can also be affected by coronavirus [9]. As the mechanism of entry is unknown for SARS-CoV2, therefore, to inhibit the immune illusion of SARS-CoV-2, it is essential to design its specific drug.
Absence of effective therapeutics and vaccines against this novel virus is further escalating the infection. Thus, it serves an immediate requirement to conquer this virus and protect the human race. The conventional approach to the production of vaccines relies heavily on antigen expression from in vitro culture models. On the other hand, the complete genome sequencing of the SARS-CoV2 or COVID-19 virus has been accomplished [10] (GenBank ID: MN908947.3). This has created an excellent scope to design novel subunit vaccines against SARS-CoV2 via reverse vaccinology approach. The more rapid in silico approach towards vaccine development would be extremely helpful in this urgent scenario.
The study focuses on utilizing different epitopes of the envelope protein of SARS-CoV2 to pave a pathway for vaccine development. The genome of SARS-CoV2 comprises nine protein coding genes, with three proteins (surface glycoprotein, envelope protein and membrane glycoprotein) present on the outer surface of the virus. The envelope protein has been identified to have the highest antigenicity in SARS-CoV2. The mutations and conformational changes in the SARS-CoV2 over the evolution were also studied. After identification of potential B-cell and T-cell epitopes, allergenicity testing, analysis of hydrophobicity, toxicity, host non-homology analysis and antigenic variation evaluation were done. The B- cell epitopes are those peptides that are recognized by surface immunoglobulins of B-cells, while T-cell epitopes are presented by MHC-I and MHC-II, which are recognized by CD8+ and CD4+ T-cells, respectively. These assessments would improve the performance of the selected vaccine candidates. The study also performed docking of human leukocyte antigens (HLA) with T cell epitopes to explore their interactions. The binding affinity values and dissociation constant values were also evaluated for SARS-CoV2 and MHCs complexes as well as for their consensus protein complexes. It, therefore, focuses on utilizing E-protein as a potential vaccine target and also provides cues towards subunit vaccine development. Therefore this study would instigate novel vaccine development against SARS-CoV2, which would, in turn, serve as a boon towards the survival of the human race.
2. Materials and methods
2.1. Identification of most antigenic outer protein of SARS-CoV2 and sequence analysis
COVID19 genome has GenBank ID: MN908947.3. Out of essential three proteins (E, M and S protein) that constitute the outer surface of the virus, E protein was identified as most antigenic executing VaxiJen v2.0 [11] server (VaxiJen score: 0.6025), while M and S protein has lower VaxiJen score of 0.5102 and 0.4646, respectively. Thus E protein is the most powerful antigen and has potential to elicit highest immune response. Through the usage of Virus-mPLoc [12], the localization site in the host cell where the viral protein resides after its fusion, was analyzed. To analyze the evolutionary divergence of E-protein of SARS-CoV2 with that of other CoV strains, the homologous sequences (having sequence identity >50% and query coverage >90%) were retrieved utilizing PSI-BLAST [13]. All the E protein sequences other than envelope protein of SARS-CoV2 were used to build a consensus sequence utilizing Unipro UGENE software [14]. The pair-wise alignment using EMBOSS WATER [15] was performed between the E-protein of SARS-CoV2 and the consensus sequence of E-proteins of closely related viruses to reveal the amino acid changes that accumulated in E-protein of SARS-CoV2. Phylogenetic tree comprising all the homologous proteins was constructed using Phylogeny.fr, where the alignment was achieved through MUSCLE and the tree was constructed by PhyML program using maximum likelihood algorithm [16].
2.2. Modeling modeling of SARS-CoV-2 E-protein and the consensus E-protein
For building the three-dimensional structures of SARS-CoV-2 E-protein and the consensus E-protein, first, Swiss Model [17] and Phyre2 [18] were utilized for homology modeling and fold recognition processes, respectively. These two methods failed to predict the complete 3D structure, so the ab-initio method using QUARK [19] was utilized. The ab-initio molecular modeling relies on de novo prediction of protein's 3D structure from its amino acid sequence without using template information. [19].
2.3. Energy minimization and molecular dynamics of modeled protein
The modeled proteins were then subjected for energy minimization using ModRefiner to remove the distorted geometries and the most interactive state was tried to achieve in terms of backbone topology, hydrogen bonding, atomic orientation etc. [20]. Distortions in the loop regions were further optimized using ModLoop to achieve proper conformation of ψ-φ angles [21].
3DRobot [22] was utilized to generate conformations (decoys) of the modeled structures. It uses TM-align to select templates from the PDB library [22]. Replica exchange Monte Carlo simulation was utilized to generate decoys using the templates [22].An atomic-level two-step iterative energy minimization procedure was utilized to remove steric clashes. Thus, improved hydrogen bonding network was achieved by using this procedure [22]. In this study, 9 decoys were generated from each of the two models using RMSD cut-off 12 Å.
2.4. Selection of the modeled structure
Protein models were checked to have no residues in the disallowed region in Ramachandran plot through PROCHECK [23] and the ERRAT value was also analyzed [24]. Through Protein Quality Predictor (ProQ) server, overall protein quality was checked through LG score and MaxSub score [25]. For a model to have satisfying quality, the LG score>1.5 and MaxSub score>0.1 is desired [25]. Z-score was also evaluated by ProSA for supporting the proper protein quality [26]. The best models were selected based on the aforementioned parameters.
2.5. Conformation analysis
The extent of conformational deviation between E-protein of SARS-CoV-2 and other related viruses were predicted through comparison of different structural units like helices, 310 helices, coils, strands etc. were calculated through STRIDE [27]. This would help to understand how the E-protein of SARS-CoV-2 has been structurally evolved from other related viruses.
2.6. B-cell and T-cell epitope identification and analysis
ABCpred server [28] using neural network algorithm helped to predict the B-cell epitopes, keeping 0.5 as threshold and amino acid length as 10. The T-cell epitopes of MHC-I and MHC-II were predicted using two IEDB servers through neural network algorithm [29,30]. These servers also identified the most probable HLAs where the epitopes bind [30,31]. MHC-1 binding T-cell epitopes were also predicted by NetCTL1.2 server [32] and the consensus predicted epitopes by two servers were considered. The antigenicity of the individual predicted epitopes was then carried out by VaxiJen v2.0 [11]. The AllerTOP v.2.0 server [33] was utilized to predict which epitope peptides are probable allergens. The epitopes that were found to be non-allergens were considered for further analysis. The prediction of net toxicity and hydrophobicity of epitopes were carried out through ToxinPred server [34]. To avoid epitopes demonstrating cross reactivity against the host's own proteins, host non-homology assessment was performed by subjecting the epitope sequence to BLASTp [35] against human proteome. The e-value cutoff of 0.01 was set. No epitopes must be homologous to human's own protein to avoid autoimmune responses.
2.7. Antigenic variability of SARS-COV-2
The antigenic variability was determined using ρepitope analysis [36]. It is calculated from:
ρepitope = (number of amino acid differences in the dominant epitope) ÷ (total number of amino acids in the dominant epitope). The ρepitope calculated was based on residual differences between epitope regions of SARS-COV-2 E-protein and the corresponding residues of the E-protein of the consensus sequence.
2.8. Molecular docking and interaction studies
The docking of the E-proteins (SARS-CoV2 and consensus) with the predicted most probable binding HLAs was carried out using PatchDock [37] to analyze their interactions and binding affinity. PatchDock has shown very high efficiency for protein-protein or protein-ligand docking that uses an algorithm verified by benchmark 0.0 and Critical Assessment of PRediction of Interactions (CAPRI) for antigen-antibody and other protein complexes [37]. It performs docking based on geometric shape complementarity in proteins, where molecules are divided into flat, convex and concave surfaces and only surface with shape complementarity will be docked [37]. The algorithm used by PatchDock ensures wide interface areas with negligible steric clashes and was observed to create complexes in near-native state for most of the cases.
The selected T-cell epitope regions were used binding sites during molecular docking. Out of 20 solution complexes, solution 1 model was chosen having the best geometric shape complementarity score. The interaction and binding patterns in the complexes were then analyzed using Protein Interaction Calculator (P.I.C), where the longer residue protein was considered as receptor, while the other one as ligand [38]. Hydrophobic bonds, H-bonds and ionic (cation-pi) bonds were focused upon. The interactions were also analyzed through PyMOL [39] for consensus results. Presence of ionic interaction [40] is well documented to strengthen the protein complex interaction. Net solvent accessibility was evaluated for each of the residues in the epitope regions as well for the individual epitopes after their respective interactions [41].
2.9. Binding affinity and KD calculation of the docked complexes
PRODIGY (PROtein binDIng enerGY prediction) was utilized to calculate the binding affinities and dissociation constants for the docked complexes of MHC-I and MHC-II [42]. It uses intermolecular contacts to find out binding affinities in docked complexes. Total six docked complexes were used to find out the binding affinity and KD values. Binding affinity and dissociation constant were expressed in terms of ΔG value (kcal/mol) and Molar (M), respectively. The temperature set for the calculation was 37 degree centigrade.
3. Results and discussion
3.1. Evolutionary divergence of SARS-CoV2 envelope protein
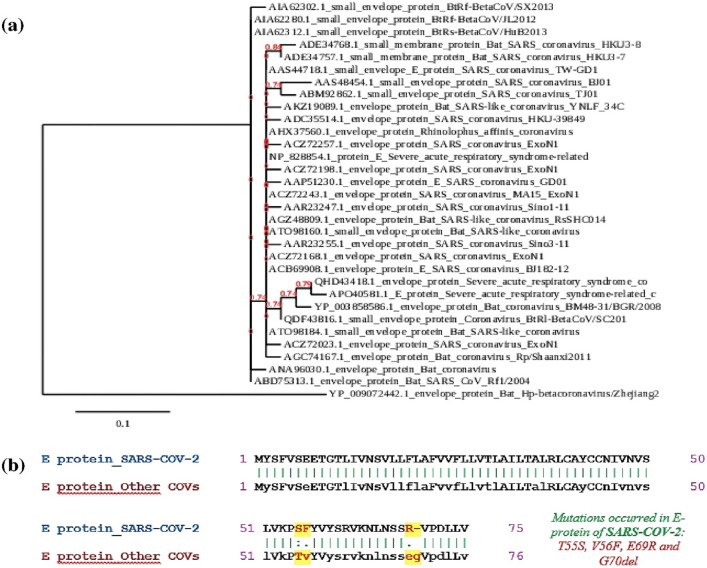
The PSI-BLAST [13] against E-protein of SARS-CoV2 revealed 32 homologous envelope proteins from other CoV strains, having e-value <0.005, query coverage >90% and sequence identity >50%. The evolutionary relationships were depicted by the construction of a phylogenetic tree, which has been demonstrated in Fig. 1 (a). The E-protein of SARS-CoV-2 (QHD43418.1) is the most closely related one to E protein of “Severe acute respiratory syndrome-related coronavirus” (APO40581.1). This virus was known to cause a large-scale epidemic in China in 2002–2003 with high mortality rate [43]. On the other hand, E-protein of SARS-CoV-2 is the most distantly related to that of BtRf-BetaCoV/SX2013 (AIA62302.1), BtRs-BetaCoV/HuB2013 (AIA62312.1) and BtRf-BetaCoV/JL2012 (AIA62280.1) strains.
Fig. 1.
(a) Phylogenetic tree depicting the evolutionary relationship of SARS-CoV-2 with other CoV strains. (b) The pairwise alignment between SARS-CoV-2 E-protein and consensus E-protein of other related CoV strains. [The amino acid differences in pairwise alignment were highlighted yellow.]
3.2. Analysis of mutations in SARS-CoV2 envelope protein
In order to analyze the extent of mutations in the E-protein of SARS-CoV-2, the amino acid changes between E-protein of SARS-CoV-2 and the consensus E-protein of other closely related CoV strains were compared. The consensus sequence was achieved through MSA, based on which amino acids are likely to be present in most of the sequences. The consensus sequence, therefore, gave the best representation of all the homologous sequences. To detect the amino acid mutations accumulated in E-protein of SARS-CoV-2, the pairwise alignment between SARS-CoV-2 E-protein and consensus E-protein was accomplished as depicted in Fig. 1(b). From the pair-wise alignment, three point mutations (T55S, V56F and E69R) and one amino acid deletion (G70del) were observed to occur in E-protein of SARS-CoV-2. Presence of any such mutations in the protein's epitope region would affect the protein's overall epitope's structure, antigenicity and in turn vaccine development, which was described in later sections.
3.3. Analysis of conformational changes between SARS-CoV-2 E-protein and consensus E-protein of other CoV strains
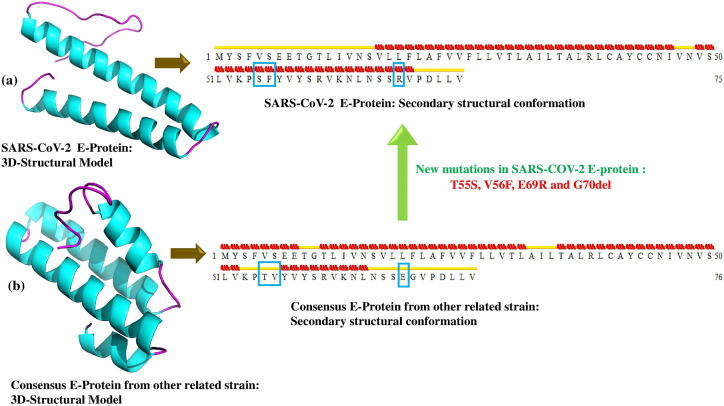
The best model of E-protein and its respective consensus were chosen based on their maximum satisfaction of the stereo-chemical parameters (Suppl.Table 1) with no residues in the disallowed regions of the Ramachandran plot. The 5th model and the 2nd model were observed to be the best models for E-protein and consensus, respectively (Suppl. Table 1). Significant differences in the structures of the two models (Fig. 2 ) were observed. The exploration of its secondary structure revealed a net loss of helices in N-terminal and its gain in the C-terminal in SARS-CoV-2 E-protein compared to the consensus E-protein structure. Two point mutations (T55S, V56F) caused coil➔helix transformation and mutation E69R and G70del causes turn➔helix transformation in that particular location. For both the cases, the protein is acquiring more rigid and structured conformations.
Fig. 2.
Demonstration of 3D and secondary structure of (a) SARS-CoV-2 E-protein and (b) Consensus E-protein of other related CoV strains.
3.4. B-cell and T-cell epitope identification and analysis
A total of five B-cell epitopes (Suppl.Table 2) were identified in the protein that has the potential to bind to the surface immunoglobulin of the B-cells and trigger production of antibodies. MHC-I binding and MHC-II binding T-cell epitopes were identified to be three and seven, respectively (Suppl.Table 3–4). These epitopes bind to MHC-I and MHC-II respectively and are presented to T-cells. The most probable binding HLAs (one with lowest percentile rank) for each epitope were also revealed (Suppl.Table 3–4). The prediction of antigenicity of the individual epitopes demonstrated that the epitopes have high antigenicity value except one B-cell and one MHC-II binding T-cell epitope with low antigenicity value (VaxiJen score<0.4) (Suppl.Tables 2 and 4). These epitopes, therefore, were excluded. The assessment of allergenicity revealed that one MHC-I binding T-cell epitope and two B-cell epitopes are probable allergens, thus have the potential to provoke type-I hypersensitivity reactions. One B-cell and one MHC-II binding T-cell epitope was seen to have the potential of showing toxicity (Suppl.Table 2–3). These epitopes, therefore, must not be considered during vaccine development. Thus only the epitopes identified as probable non-allergens were considered for further analysis. Host non-homology assessment revealed that no selected epitopes have significant homology with human proteins with an e-value cutoff of 0.01. Thus the epitope peptides are pathogen specific and thus would generate minimum autoimmune response. The prediction of hydrophilicity and hydrophobicity demonstrated a net hydrophobic nature in most of the predicted epitopes (Suppl.Table 2–4). However, for an epitope to be properly exposed to the aqueous environment, lesser hydrophobicity is preferred. B-cell epitope, satisfying all above criteria like “ILTALRLCAY” and MHC-I binding T-cell epitopes like “LTALRLCAY” and “VSLVKPSFY” would be a better candidate in terms of lowest hydrophobicity (Table 1 ). MHC-II binding T-cell epitope ‘LAFVVFLLVTLAILT’ have the highest hydrophilicity value (Table 1) and thus, is more probable to be exposed for better interaction with immune cells (Suppl.Table 4). This epitope also had highest immunogenicity score and thus was considered as best MHC-II binding epitope and thus selected for further studies. All the selected SARS-CoV-2 E-protein epitopes used in further studies were listed in Table 1.
Table 1.
List of selected B-cell and T-cell epitopes.
| B-cell epitopes |
T-cell epitopes |
|||
|---|---|---|---|---|
| Epitope | Position | Epitope | Position | Most probable binding HLA |
| MHC-I binding | ||||
| VFLLVTLAIL | 25–34 | LTALRLCAY (T1) | 34–42 | HLA-A*01:01 (PDB ID: 6AT9_A) |
| ILTALRLCAY | 33–42 | VSLVKPSFY (T2) | 49–57 | HLA-A*30:02 (PDB ID: 6J2A_A) |
| LLFLAFVVFL | 18–37 | MHC-II binding (Best epitope) | ||
| LAFVVFLLVTLAILT (T3) (Core peptide: VFLLVTLAI) | 21–35 | HLA-DRB1:15:01 (PDB ID: 5V4M_C,F,I,L) | ||
The epitope positions were found to be same for the SARS-CoV2 E-protein and consensus E-protein.
3.5. Antigenic variability of the identified epitopes
The B-cell and T-cell epitopes were analyzed for their antigenic variability. It highlighted the epitopes to have the residues that got specifically mutated in SARS-CoV-2 E-protein when compared to the consensus sequence. The antigenic distances of all the epitopes are zero except in two (one MHC-1 Binding “VSLVKPSFY” and another MHC-II binding core peptide: ‘FYVYSRVKN’) with antigenic distance of 0.22 and 0.11, respectively. The production of vaccine targeted epitopes with no antigenic distance (antigenically conserved) is longer lasting and successful, even when the pathogen mutates into a more powerful strain. However, epitopes having non-zero antigenic distance helps to explore some novel mutations in SARS-CoV-2. These epitopes contain two important point mutations at amino acid level (T➔S and V➔F) in novel SARS-CoV-2 (Fig. 1 (b)). Comparing the conformation of this epitope region in E-protein of SARS-CoV-2 with consensus E-protein, as described in Section 3.3, there is a notable transition from coil➔helix in this epitope region. This might change the epitope's overall interaction with their corresponding HLAs.
3.6. Interactions and binding patterns
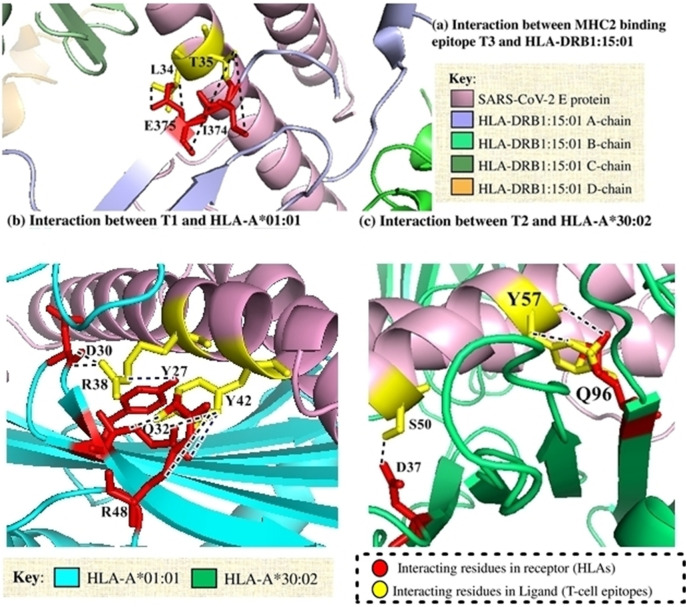
Docking of the selected T-cell epitopes with the respective and most probable binding HLAs (Table 1) was done to analyze their binding patterns and residues involved. Interaction with MHC-I was observed to get accomplished through two different epitope regions namely T1 and T2 of SARS-CoV-2 E protein and consensus E-protein. Asp38 and Tyr42 from T1 epitope from SARS-CoV-2 E-protein played a major role in H-bond. Apart from H-bond interactions, Tyr42 was also involved in cation-pi interaction. Majority of non-covalent interactions were done by Tyr57 from T2 with Gln96 from A chain of MHC-I. No ionic interactions were observed from MHC-II protein while 32nd, 34th and 35th residues from T3 epitope of E-protein interacted in majority to form the H-bonds with MHC-II (Suppl. Table 5-7).
Mainly, A-chain of MHC-I and MHC-II was observed to play a chief role. Altogether, the interactions through MHC-I was found to be more dominating for forming stronger binding patterns with numerous side chain- side chain and ionic interactions. The ionic interactions created a charged environment for the partner protein. All the major interactions have been depicted in Fig. 3 . On the contrary, from the consensus sequence of E-protein, only Cys40 and Ala41 from T1 epitope were observed to form three and two H-bond interactions with Gln115 and Asp122, respectively of MHC-I (Suppl. Table 8). 52nd, 53rd and 57th residues played a dominant role from T2 from the consensus E-protein with MHC-I. Tyr57 got involved in cation-pi interactions along with hydrophobic interactions (Suppl. Table 9). On the other hand, only few residues in H-bonded non-covalent interactions were observed in consensus T3 epitope with MHC-II (Suppl. Table 10).
Fig. 3.
Major hydrogen-bonding (black-dots) and ionic (blue-dots) interactions between different T-cell epitopes and their corresponding HLAs [F, C, I, L chains of PDB ID: 5V4M (HLA-DRB1:15:01) was renamed to A, B, C and D chains respectively by Discovery Studio].
3.7. Evaluation of solvent accessible surface area (ASA)
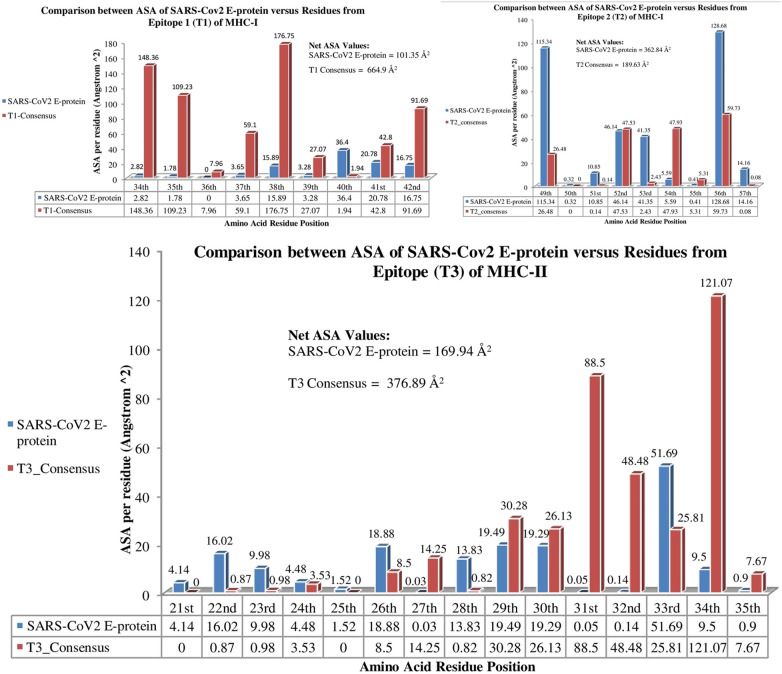
Lesser solvent accessible surface area (ASA) was observed for either of the MHCs when interacted with SARS-CoV2 E-protein in comparison to their interaction with consensus E-protein. This also supported the interaction results (Suppl. Table 5–10). The net solvent accessibility value after T1 and T2 from SARS-CoV2 E-protein interacted with MHC-I was observed to be 464.19 Å2 while that after consensus E-protein interacted with MHC-I was found to be 854.53 Å2 (Fig. 4 ). Similarly, 169.94 Å2 and 376.89 Å2 were the solvent accessibilities when MHC-II interacted with SARS-CoV2 E-protein and consensus E-protein, respectively (Fig. 4). This implies that better interaction have taken part from the evolutionarily evolved SARS-CoV2 E-protein in comparison to the consensus E-protein. Fig. 4 gives the detailed evaluation for the residue based solvent accessibility along with the net solvent accessibility from the epitopes with MHCs.
Fig. 4.
Comparative analysis for residue wise solvent accessibility values for SARS-CoV2 E-protein and consensus E-protein after they interact with their respective MHCs.
3.8. Analysis of binding affinities for the docked complexes
The binding affinities for MHC-I and MHC-II complexes bound individually with SARS-CoV2 protein complex were observed to have higher ΔG values and lower dissociation constants, in comparison to their respective complexes with consensus E-protein (Table 2 ). Therefore, SARS-CoV2 forms a spontaneous interaction with MHCs. This, once more affirms the stronger interaction as well.
Table 2.
Calculation of binding affinities and dissociation constants.
| Complex name/ parameters | Binding affinity (ΔG in kcal/mol) |
Dissociation constant in molar units (Kd) |
|---|---|---|
| MHC-I_SARS-CoV2 E protein complex | −19.35 | 3.25E-14 |
| MHC-I_Consensus E protein complex | −18.15 | 3.60E-12 |
| MHC-II_SARS-CoV2 E protein complex | −22.8 | 8.00E-17 |
| MHC-II_Consensus E protein complex | −17.9 | 2.40E-13 |
4. Conclusion and future scope
The outer SARS-CoV-2 envelope (E) protein was studied to be most antigenic and thus would allow better interaction with immune cells. This protein is also the smallest outer surface protein, therefore easier for recombinant production. Through an evolutionary analysis of SARS-CoV-2 E-protein, four novel mutations (T55S, V56F, E69R and G70del) were observed to occur in E-protein of SARS-CoV-2 when compared to the consensus E-protein of other related viruses. These mutations altered lead to a massive conformational shift from more flexible coil/turn to more structured, stable and rigid helix, at the mutation site. This study also focused on designing potential B-cell and T-cell epitopes that could be used for the development of subunit vaccines. Those epitopes were also computationally screened for unwanted effects to host such that they should not show allergenicity, toxicity or cross-reactivity when administered to an individual. Three B-cell epitopes (VFLLVTLAIL, ILTALRLCAY and LLFLAFVVFL), two MHC-I binding (LTALRLCAY, VSLVKPSFY) and an MHC-II binding (VFLLVTLAI) were identified as the best epitopes for development of future subunit vaccine. The epitopes were also analyzed for their antigenic variability to check the epitope that had residues which got specifically mutated in SARS-CoV-2 E-protein when compared to the consensus sequence of all other strains. All the epitopes were observed to be highly conserved except ‘VSLVKPSFY’ that have two SARS-CoV-2 specific point mutations: T➔S and V➔F that caused a coil➔helix transition in its structure. Conserved epitopes would serve as long lasting vaccines, while this epitope with novel mutations would provide further scope to study the virus evolution and understand their effects on vaccine development. This study also analyzed the non-covalent interactions with the T-cell epitopes and their corresponding binding HLAs through molecular docking. It was observed that the predominant ionic and side chain- side chain interactions were numerous and were giving a more stable and strengthening interaction through MHC-I epitopes in comparison to MHC-II epitope, when interacted with E-protein. Gln96 (MHC-I) and Tyr57 (E-protein) was found to play a predominant role with several interactions. Tyr42 from T1 epitope of E-protein formed several H-bond interactions as well as ionic interaction with Arg48 from MHC-I. Leu34 and Thr35 from E-protein formed several H-bond interactions with MHC-II. Moreover, T-cell epitopes in SARS-CoV-2 E protein showed more interactions with HLAs than the consensus E-protein. Moreover, the stronger and spontaneous interaction by SARS-CoV2 with the MHCs was also supported through the evaluation of higher ΔG values and lower dissociation constants, on contrary to the consensus E-protein involved complexes. These stronger interactions would trigger higher levels of T-cell activation, which in turn might cause higher immune response. The net solvent accessibility for SARS-CoV2 E-protein after interaction with its respective MHCs showed a stronger and better interaction in comparison to the consensus E-protein. This might serve SARS-CoV-2 E protein as a better vaccine target. This residue-level analysis helped to explore the detailed structural evolution of SARS-CoV-2 E-protein and therefore paves the way towards using E-protein for vaccine development against SARS-CoV-2. Vaccine development and clinical trials would serve to be effective only after analyzing the residual and structural details of a particular protein and there stands this study to be novel and worthily paramount.
CRediT authorship contribution statement
Shreya Bhattacharya, Arundhati Banerjee, Sujay Ray conceived of the presented idea. Shreya Bhattacharya, Arundhati Banerjee, did introduction result and discussion as well as reference formatting. Conformational analysis and binding affinity analysis specifically carried out by Sujay Ray.Some specific portion of materials of method was done by Sujay Ray. Molecular Docking portion was carried out by Shreya Bhattacharya. Arundhati Banerjee investigated some specific area sush as SASA and interaction pattern. Figures and Tables were constructed by Shreya Bhattacharya, Arundhati Banerjee, Sujay Ray. The abstract and discussion portion was specifically oriented by Arundhati Banerjee. Response to reviewer was made by Shreya Bhattacharya with help of Sujay Ray. Total design of the work was done by Sujay Ray and supervised the findings of this work. Some critical calculations were made by Sujay Ray. The data validation was carried out by Sujay Ray. All authors discussed the results and contributed to the final manuscript.
Declaration of competing interest
None.
Acknowledgments
Acknowledgement
Authors would like to thank Department of Biosciences and Bioengineering,IITG, Department of Biochemistry and Biophysics, University of Kalyani, India and Amity Institute of Bioechnology, Amity University, Kolkata, India for their support. No funding support was received for this research work.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijbiomac.2020.12.192.
Appendix A. Supplementary data
Supplementary tables
References
- 1.Gorbalenya A.E., Baker S.C., Baric R.S. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hui D., Azhar E.I., Madani T., Ntoumi F., Kock R., Dar O., et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health — the latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO Director-general's opening remarks at the media briefing on COVID-19 - 11 March 2020, Who.Int. 2020. https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020
- 4.Li G., et al. Coronavirus infections and immune responses. J. Med. Virol. 2020;92:424–432. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16 doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y., Gayle A., Wilder-Smith A., Rocklöv J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. Journal Of Travel Medicine. 2020;27 doi: 10.1093/jtm/taaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Xiaowei, Geng Manman, Peng Yizhao, Meng Liesu, Lu Shemin. Molecular immune pathogenesis and diagnosis of COVID-19. J. Pharm. Anal. 2020;10(2):102–108. doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Snijder E.J., van der Meer Y., Zevenhoven-Dobbe J., et al. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 2006;80:5927e5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.V.D. Menachery, A. Schafer, K.E. Burnum-Johnson, et al., MERS-CoV and H5N1 influenza virus antagonize antigen presentation by altering the epigenetic landscape, Proc. Natl. Acad. Sci. U. S. A. 115 (2018) E1012eE1021. [DOI] [PMC free article] [PubMed]
- 10.Wu F., Zhao S., Yu B., Chen Y., Wang W., Song Z., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doytchinova I., Flower D. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics. 2007;8 doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen H., Chou K. Virus-mPLoc: a fusion classifier for viral protein subcellular location prediction by incorporating multiple sites. J. Biomol. Struct. Dyn. 2010;28:175–186. doi: 10.1080/07391102.2010.10507351. [DOI] [PubMed] [Google Scholar]
- 13.Altschul S. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okonechnikov K., Golosova O., Fursov M. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 2012;28:1166–1167. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- 15.Rice P., Longden I., Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 16.Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwede T. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelley L., Mezulis S., Yates C., Wass M., Sternberg M. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu D., Zhang Y. Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins: Structure, Function, And Bioinformatics. 2012;80:1715–1735. doi: 10.1002/prot.24065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu D., Zhang Y. Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys. J. 2011;101:2525–2534. doi: 10.1016/j.bpj.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fiser A., Sali A. ModLoop: automated modeling of loops in protein structures. Bioinformatics. 2003;19:2500–2501. doi: 10.1093/bioinformatics/btg362. [DOI] [PubMed] [Google Scholar]
- 22.Deng H., Jia Y., Zhang Y. 3DRobot: automated generation of diverse and well-packed protein structure decoys. Bioinformatics. 2015;32:378–387. doi: 10.1093/bioinformatics/btv601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laskowski R., MacArthur M., Moss D., Thornton J. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. doi: 10.1107/s0021889892009944. [DOI] [Google Scholar]
- 24.Colovos C., Yeates T. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2:1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wallner B., Elofsson A. Can correct protein models be identified? Protein Sci. 2003;12:1073–1086. doi: 10.1110/ps.0236803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wiederstein M., Sippl M. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frishman D., Argos P. Knowledge-based protein secondary structure assignment. Proteins Struct. Funct. Genet. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 28.Saha S., Raghava G. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins: Structure, Function, And Bioinformatics. 2006;65:40–48. doi: 10.1002/prot.21078. [DOI] [PubMed] [Google Scholar]
- 29.Dhanda S., Karosiene E., Edwards L., Grifoni A., Paul S., Andreatta M., et al. Predicting HLA CD4 immunogenicity in human populations. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.01369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trolle T., Metushi I., Greenbaum J., Kim Y., Sidney J., Lund O., et al. Automated benchmarking of peptide-MHC class I binding predictions. Bioinformatics. 2015;31:2174–2181. doi: 10.1093/bioinformatics/btv123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paul S., Lindestam Arlehamn C., Scriba T., Dillon M., Oseroff C., Hinz D., et al. Development and validation of a broad scheme for prediction of HLA class II restricted T cell epitopes. J. Immunol. Methods. 2015;422:28–34. doi: 10.1016/j.jim.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larsen M., Lundegaard C., Lamberth K., Buus S., Lund O., Nielsen M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinformatics. 2007;8 doi: 10.1186/1471-2105-8-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dimitrov I., Bangov I., Flower D., Doytchinova I. AllerTOP v.2—a server for in silico prediction of allergens. J. Mol. Model. 2014;20 doi: 10.1007/s00894-014-2278-5. [DOI] [PubMed] [Google Scholar]
- 34.Gupta S., Kapoor P., Chaudhary K., Gautam A., Kumar R., Raghava G. In Silico approach for predicting toxicity of peptides and proteins. PLoS One. 2013;8 doi: 10.1371/journal.pone.0073957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altschul S., Gish W., Miller W., Myers E., Lipman D. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/s0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 36.Zhou R., Pophale R.S., Deem M.W. 2009. Computer-assisted Vaccine Design. arXiv preprint arXiv:0904.4705. [Google Scholar]
- 37.Schneidman-Duhovny D., Inbar Y., Nussinov R., Wolfson H. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:W363–W367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tina K.G., Bhadra R., Srinivasan N. PIC: protein interactions calculator. Nucleic Acids Res. 2007;35:W473–W476. doi: 10.1093/nar/gkm423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeLano W.L. PyMOL: an open-source molecular graphics tool. CCP4 Newsletter on protein crystallography. 2002;40:82–92. [Google Scholar]
- 40.Baldwin R.L. How Hofmeister ion interactions affect protein stability. Biophys. J. 1996;71(4):2056–2063. doi: 10.1016/S0006-3495(96)79404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fraczkiewicz R., Braun W. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J. Comput. Chem. 1998;19:319–333. [Google Scholar]
- 42.Xue L., Rodrigues J., Kastritis P., Bonvin A.M.J.J., Vangone A. PRODIGY: a web-server for predicting the binding affinity in protein-protein complexes. Bioinformatics. 2016 doi: 10.1093/bioinformatics/btw514. [DOI] [PubMed] [Google Scholar]
- 43.Cheng V., Lau S., Woo P., Yuen K. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin. Microbiol. Rev. 2007;20:660–694. doi: 10.1128/cmr.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables