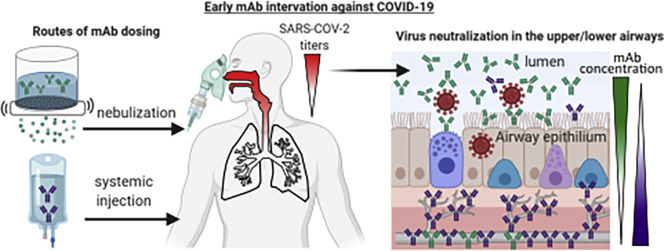
Abstract
To address the COVID-19 pandemic, there has been an unprecedented global effort to advance potent neutralizing mAbs against SARS-CoV-2 as therapeutics. However, historical efforts to advance antiviral monoclonal antibodies (mAbs) for the treatment of other respiratory infections have been met with categorical failures in the clinic. By investigating the mechanism by which SARS-CoV-2 and similar viruses spread within the lung, along with available biodistribution data for systemically injected mAb, we highlight the challenges faced by current antiviral mAbs for COVID-19. We summarize some of the leading mAbs currently in development, and present the evidence supporting inhaled delivery of antiviral mAb as an early intervention against COVID-19 that could prevent important pulmonary morbidities associated with the infection.
Graphical abstract
1. Introduction
Severe Acute Respiratory Disease Syndrome Coronavirus 2 (SARS-CoV-2), the etiologic agent of coronavirus disease 2019 (COVID-19), has caused a global pandemic at a scale not seen for nearly 100 years. In response, historic efforts bridging across governments, industry, and academia have leveraged massive financial investments and scientific efforts to advance potential vaccines and therapeutics into the clinic at a pace never before witnessed.
SARS-CoV-2 can be classified under a group of viruses that cause acute respiratory infections (ARIs), characterized by their respiratory tropism and predominant spread within the airways until infection of the deep lung. Thus, we believe there are important lessons to be gained from studying efforts in developing vaccines and therapeutics for other ARIs. Despite the large number of common ARIs, including influenza, respiratory syncytial virus (RSV), parainfluenza virus (PIV), metapneumovirus, rhinoviruses, and seasonal coronaviruses, there remains no vaccine or treatment for virtually any ARIs, with the exception of influenza and, now, SARS-CoV-2. Even then, for influenza vaccines, the efficacy remains modest, ranging from a low of 19% to a high of 60% between 2009 and 2019 in the U.S. [1]. While the poor efficacy is frequently attributed to the difficulty in accurately predicting which influenza strains will circulate in different communities during an upcoming flu season [2], the actual reasons are likely multifold. There are important subpopulations that generally do not respond well to vaccines, including infants, the elderly, and immunocompromised adults. For instance, infants 1–2 months old have been shown to only rarely produce a humoral response against the viral surface glycoproteins in response to vaccination, in contrast with older children [3]. This finding is in line with studies of antibody titers observed in children under 6 months who are hospitalized with RSV infection, among whom fewer than 50% develop a neutralizing antibody response [4]. Influenza vaccination also suffers rapid intraseason waning of protective immunity, with effectiveness declining ~16% every 28 days [5], possibly due to decline of influenza vaccine–induced human bone marrow plasma cells [6]. Recently results from the Pfizer/BioNTech and Moderna Phase III trials suggest that they may provide >90% protection against symptomatic disease during the immediate weeks following the second (booster) injection, compared to saline control. This has led the FDA to issue emergency use authorization (EUA) for both vaccines. The efficacy of another vaccine (AstraZeneca/Oxford) was reported as ~62% compared to the meningococcal conjugate vaccine control. In all cases, the durability of the immunity will only be revealed in the coming months and years. Likewise, since the Phase III study enrolled participants without history of allergic response or otherwise underlying conditions, the safety profile of the vaccines in these individuals remains to be determined. Even with highly effective vaccines, however, there will likely be patient subgroups who fail to benefit comparably (e.g. immunocompromised adults) as well as those who are not vaccinated and become sick. Thus, there will almost certainly be a strong demand for effective treatment options for COVID-19 in the coming years despite the increasing availability of safe and effective vaccines.
Among the various potential therapeutic interventions, monoclonal antibodies (mAbs) represent one of the most promising classes of molecules due to their longstanding track record of safety in humans, their exceptional specificity to the virus (which minimizes risk of off-target effects), and their ability to coordinate the immune defense in the fight against infection. Technological advances over the past two decades in sequencing and single cell screening, as well as manufacturing, have positioned mAbs to quickly respond to the COVID-19 pandemic. In this review, first we highlighted important pathophysiology associated with SARS-CoV-2, protective functions of antibodies in mucus, and some of the leading mAb under development for SARS-CoV-2. We then examined the track record of mAbs that have been advanced to address other ARIs in the past and used the insights to identify opportunities where the potential efficacy of antiviral mAbs for COVID-19 can be improved.
2. Mechanism of spread of many acute respiratory infections (ARIs)
2.1. Predominant apical infection and shedding
The human respiratory system can be broadly divided into (i) the upper respiratory tract (URT) encompassing the nasal cavity and pharynx, (ii) the lower respiratory tract (LRT) that begins at the trachea and extends all the way down to bronchioles, and (iii) deep lung, i.e. the alveolae. Within the lung, there are two distinctive epithelia: a ciliated epithelium capable of secreting and clearing mucus that lines the conducting airways, and a specialized epithelium that lines the alveolus. The alveolar epithelium is dominated by pulmonary alveolar type I (AT1) cells, which are covered by no more than ~200 nm of liquid (termed epithelial lining fluid) [7] and make up more than 95% of the alveolar surface area, where they play a central role in gas exchange [8]. In contrast, the airway epithelium possesses a unique morphology and function, including a polarized epithelium with tight junctions, formation of cilia, and the secretion of mucins that form a viscoelastic airway mucus (AM) gel overlaying the epithelium. These cellular features critically impact how viruses infect and propagate within the lung, but such features cannot be accurately recapitulated by in vitro cultures of airway epithelial cells that are fully submerged in liquid media, per standard culture methods.
The most rigorous model capturing this complex pulmonary physiology is created by culturing human nasal or tracheobronchial epithelial cells, collected from airway brushings or from cadaver airway tissue, on semi-permeable membranes for at least 20–25 days. This model, commonly referred to as well-differentiated human airway epithelial (WD-HAE) culture, exposes cells to air in the apical compartment but provides essential nutrients through direct contact with the culture media in the basal compartment. This air-liquid interface culture results in a polarized, well-differentiated, ciliated airway epithelium with a secreted mucus layer [[9], [10], [11]].
Unlike studies using submerged liquid cultures with non-polarized epithelial cells [12], studies using WD-HAE cultures revealed that many viruses responsible for common ARIs almost always exclusively infect via the apical (luminal) side of the airway epithelium, with little to no productive infection when viruses are introduced into the basal (serosal) compartment (Fig. 1 ). More importantly, infected cells appear to predominantly, if not exclusively, shed progeny viruses back into the apical compartment (i.e. into AM secretions), with limited to no shedding of virus into the basal compartment. For example, influenza infects WD-HAE cultures apically, and almost exclusively sheds progeny virus into AM with little basolateral viral shedding [[13], [14], [15], [16]]. This apical shedding phenomenon is consistent with prior findings that Rab11, a GTP-binding protein related to endocytic recycling, is both crucial for the budding of influenza and is exclusively trafficked to the apical membrane in polarized epithelial cells [17,18]. Similar apical infection and shedding has been confirmed for other common viruses responsible for common ARIs, including RSV [[19], [20], [21], [22], [23]], PIV [24], and the betacoronavirus HKU1 [25].
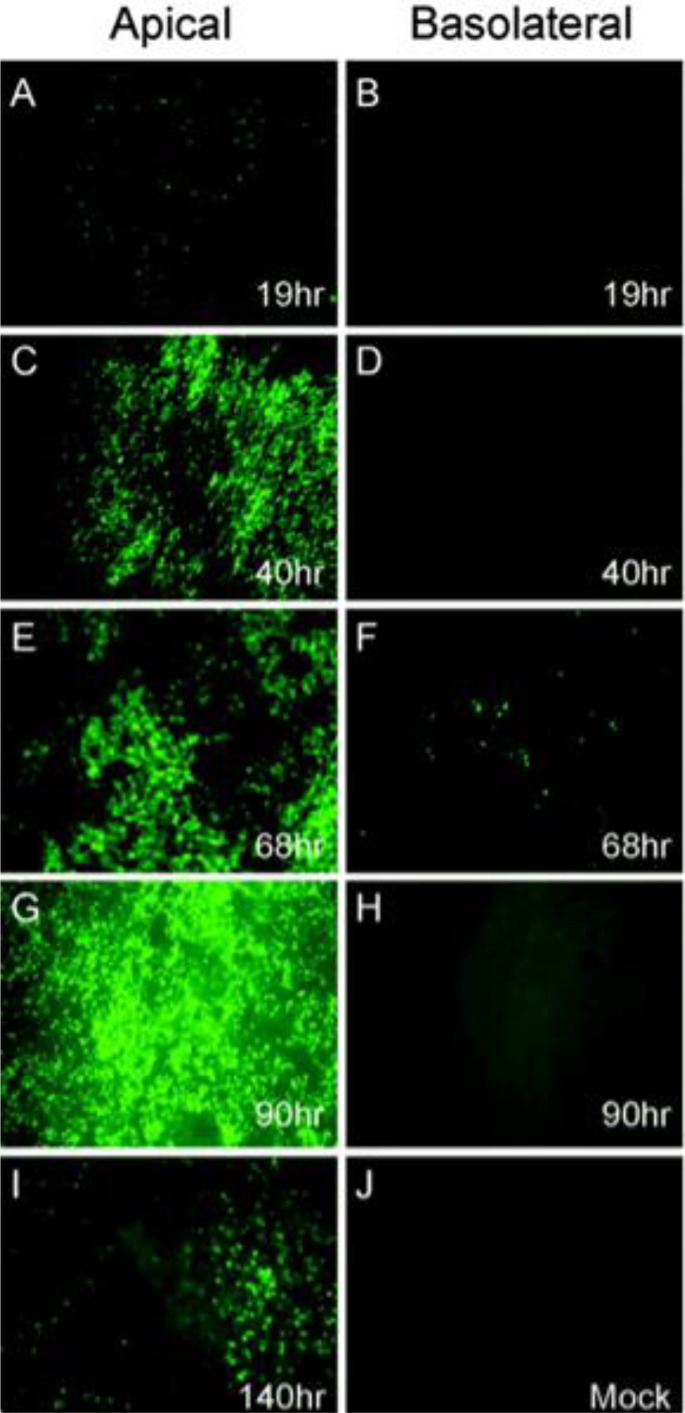
Fig. 1.
Infection and spread of SARS-CoV GFP infection in HAE over time after apical or basolateral inoculation. HAE were inoculated via the apical (A, C, E, G, and I) or basolateral (B, D, F, and H) compartments with SARS-CoV GFP and GFP-positive cells and assessed over time (1 to 5 days post-infection). Apical inoculation leads to progressive increase in GFP fluorescence over time, with significant infection after 40 h (C) and maximum fluorescence after 90 h (G). Basolateral infection is not effective, only a small proportion of cells are GFP positive 68 h after infection. Results are representative of three repeats. Adapted from [26] with permission from American Society for Microbiology, Journal of Virology.
SARS-CoV-1 and SARS-CoV-2 bind the same receptor for cellular entry: Angiotensin-converting enzyme 2 (ACE2). Similar to influenza and RSV, SARS-CoV-1 only productively infects WD-HAE cultures when the virus is inoculated apically, with no appreciable infection when the same amount of virus is inoculated basally (Fig. 1) [26]. Equally importantly, there is ~1000-fold greater virus shed into the apical compartment relative to the basal compartment [26]. The near exclusive apical infection and shedding of SARS-CoV-1 is consistent with the trafficking of ACE2 to the apical membrane of the human airway epithelium in vivo and in WD-HAE cultures in vitro [[26], [27], [28]]. Given that SARS-CoV-2 binds the same ACE2 receptor for cellular entry, it is not surprising that SARS-CoV-2 undergoes the same preferential apical infection and shedding [[26], [27], [28]], and that apical infection of polarized cells in vitro leads to substantially more virus than basolateral infection [29]. Immunofluorescent staining of airway biopsy tissues found that ACE2 was detected exclusively on the apical surface of cells [30].
2.2. Apical infection and shedding consistent with clinical features of ARIs
2.2.1. Influenza and RSV
Based on studies using WD-HAE cultures, apical shedding of virus and subsequent reinfection appears to be the primary route responsible for the spread of influenza and RSV from the URT to the LRT, before eventually infecting the deep lung. In other words, the infection does not spread to the deep lung either through the systemic circulation or by direct cell-to-cell transmission. This stepwise propagation of the infection from the URT to LRT and, finally, the deep lung is consistent with the clinical hallmarks of common ARIs. Infections with influenza and RSV first result in symptoms in the URT such as cough, congestion, and sore throat. These symptoms create opportunities for molecular diagnosis of the infection while the infection is still primarily in the URT, before the virus extensively infects the LRT and finally the deep lung. There is generally at least several days between the onset of URT symptoms and virus-induced bronchiolitis and pneumonia that necessitates medical attention, including hospitalization in a small fraction of individuals, primarily those with immature or compromised immune systems such as infants, immunocompromised adults, and the elderly. This time frame between the first development of symptoms and progression to severe disease, which varies depending on the virus, represents a prime window of opportunity to prevent LRT infections and pathologies, as exemplified by the current guidelines for prescribing antivirals such as oral oseltamivir, approved by the FDA for treatment of acute uncomplicated influenza, within 2 days of onset of illness.
This mechanism of apical shedding and propagation of many ARIs is also consistent with current standards of diagnosing these infections using either nasal or nasopharyngeal swabs, rather than blood sampling. Indeed, nasopharyngeal swab is the collection method for the commonly used respiratory pathogen panels, tests used to simultaneously diagnose or rule-out dozens of respiratory pathogens, including the four seasonal coronaviruses, adenovirus, metapneumovirus, influenza A and B, parainfluenza, RSV, rhinovirus, and bacteria such as Chlamydia pneumoniae, Mycoplasma pneumoniae, Bordatella pertussis, and others [31,32]. In contrast, analysis of blood from patients infected with ARIs typically shows low-to-no systemic viremia, including those infected by influenza [33], RSV [34], and MPV [35]. Detectable titers of infectious viruses generally only start entering the systemic circulation when the infection has reached the deep lung, where viruses can more easily penetrate the much-thinner epithelial lining, and where infection-induced inflammation can more readily compromises epithelial barrier function [[36], [37], [38]].
2.2.2. SARS-CoV-1 and SARS-CoV-2
Boucher and colleagues conducted RNA mapping of ACE2 expression in various regions throughout the respiratory tract to identify key sites for SARS-CoV-2 infection [36]. Their quantitative assessment revealed relatively high ACE2 expression in the trachea and bronchi, lower ACE2 expression in the bronchioles and alveoli, and relatively uniform expression of Transmembrane Serine Protease 2 in both the URT and LRT [36]. Differential ACE2 expression correlates with greater infectivity of SARS-CoV-2 in polarized cultures of cells from different parts of the respiratory tract [36]. These findings are consistent with the hypothesis that SARS-CoV-2 infections first establish in the nose and URT – sites that are most exposed and most susceptible to transmission – followed by spread through the LRT before eventually infecting the deep lung. Apical shedding and propagation also explains why clinical reports to date indicate relatively limited viremia of SARS-CoV-2 (i.e. infectious viruses in the blood) before the disease has progressed to more severe infection, hyper-inflammation, and lung injury to the more fragile alveoli [36].
For SARS-CoV-1, a summary WHO report [39] described the incubation period as typically being from 2 to 7 days, potentially as long as 10 days after exposure. Then, ~3–7 days following the first development of symptoms, a lower respiratory phase begins, including dyspnea that sometimes leads to hypoxia and ultimately requires mechanical ventilation in 10–20% of infected patients [39]. Thus, the clinical course of SARS mirrors our current understanding of the pathogenesis of ARIs, in which early disease predominantly affects the URT, and clinical deterioration typically occurrs following progression to the LRT and the deep lung. Given that SARS-CoV-2 targets the same ACE2 receptor, it is not surprising that the clinical presentation and progression of COVID-19 is also quite similar. There is generally a substantial lag between the first symptoms of SARS-CoV-2 infection in the URT to when these patients begin to experience dyspnea (5–7 days after symptoms), and a delay of ~9.5 days between symptoms and ICU admission [[40], [41], [42]]. Thus, the first few days following the development of symptoms represents a crucial window for interventions aimed to prevent progression to LRT disease. Case series and contact tracing studies following patients with COVID-19 have suggested an incubation period that varies across individuals but is typically ~6 days between exposure and the development of symptoms [43].
3. Antibody function in airway mucus (AM)
Antibodies play a major role in the body's adaptive immunity to infections. Indeed, greater levels of anti-flu mAb in the nasal mucosa have been shown to be correlated with more rapid elimination of the virus in humans [44]. Nearly all vaccines seek to elicit effective antibody titers against viruses of interest; passive immunization or therapy with antiviral mAbs seek to bypass the need for the immune system to learn to produce neutralizing antibodies by directly producing and delivering the antibodies needed. Given the apical infection and shedding of SARS-CoV-2 that concentrates the viruses in the AM overlaying the respiratory epithelium, it is imperative to understand the characteristics of endogenous antibodies in AM, particularly their functions in blocking the spread of the infection, in order to ensure we can recapitulate these essential functions through either vaccine-elicited antibody response or administered mAb.
3.1. Antibody isotype, abundance, and source of antibodies in AM
Unlike the gastrointestinal tract, where sIgA is the predominant antibody isotype, and unlike cervicovaginal mucus lining the female reproductive tract where IgG is the predominant antibody, there is an abundance of both sIgA and IgG in the AM lining the airways in the URT and LRT.
IgA has two subclasses, IgA1 and IgA2, and is the classic antibody isotype associated with mucosal protection. Dimeric IgA (dIgA) is produced by plasma cells in the lamina propria and comprises two IgA molecules linked by a J-chain protein. The dIgA is then bound by secretory component (SC), a protein found in the plasma membrane on the basolateral surfaces of a sub-population of mucosal epithelial cells. This dIgA-SC complex on the plasma membrane is then transcytosed and released into the AM as secretory IgA (sIgA) [45,46]. While ~90% of IgA in the systemic circulation is monomeric, ~50% of IgA in AM is dimeric, i.e., sIgA [47,48]. Since current manufacturing technologies are unable to produce and deliver large quantities of sIgA, the rest of this review will focus on the role of IgG in mucosal protection.
Abundant quantities of IgG have been found in AM and nasal lavages [49], including all four subtypes of IgG. In general, the ratio of IgG to sIgA is in the range of 1:1 to 1:3 [50,51]; early work by Deuschl and Johansson showed that comparable levels of both IgG (~0.12 mg/mL) and sIgA (~0.14 mg/mL) are detected in tracheo-bronchial lavages; the actual concentrations in undiluted AM are most likely substantially higher, possibly exceeding 1 mg/mL. Similarly, there are comparable levels of total IgG (~240 μg/mL) and IgA (~337 μg/mL) in induced sputum [52].
At the gas-exchange surface of the alveoli, the barrier to the blood is thinnest; thus, a fraction of immunoglobulins in the blood can enter via passive transudation [53]. Along the conducting airways, plasma cells associated with local lymphoid tissue can directly secrete immunoglobulins [54]. IgA2 represents ~20% of total serum IgA, but ~30% of total lung IgA [55]; these differences reflect considerable local production of sIgA in the lung, with some estimates suggesting >80% of sIgA in tracheobronchial secretions comes from local plasma cell production [51]. Plasma cells producing IgG are also located in the bronchial mucosa [56,57]. By analyzing the ratio of albumin to different IgGs in induced sputum, it has been estimated that local plasma cell production accounts for over 50–60% of the IgG1 and IgG2 in the lung, and as much as ~80–90% of the IgG3 and IgG4 [58].
3.2. Mechanisms of antibody-mediated protection
3.2.1. Neutralization
The Fab domains of IgG can bind with exquisite specificity to epitopes on the viral surface [59]. When the antibodies bind viral epitopes that are responsible for binding the host cell receptor, IgG can directly inhibit cellular entry and infection [60]. In addition, some virus-bound antibodies can interfere with cellular processes essential for productive infection without interfering with viral entry into the cells. Collectively, antibodies that can directly neutralize the virus without the aid of other immune cells or immune factors are referred to as neutralizing antibodies. Unsurprisingly, following vaccination, the generation of neutralizing antibodies is one of the best predictors of protection against future infections [61]. Nevertheless, not all induced antibodies produced in response to vaccination or infection can neutralize the virus directly; instead, the Fc domain of virion-bound IgG can facilitate other effector functions that enhance protection against viral infections.
3.2.2. Classical Fc effector functions
In addition to neutralization, IgGs in the lung can facilitate effector functions such as complement activation, opsonization, and antibody-dependent cellular cytotoxicity (ADCC) [62,63]. When IgG has opsonized a target pathogen, they can initiate complement-dependent cytotoxicity (CDC) [64], a chain of events that begins when the Fc of antibodies bound to a pathogen or pathogen-infected-cell attract complement protein C1q, leading to a cascade of events that ultimately cause direct disruption to the membrane (through the formation of a membrane attack complex) and the release of C3a and C5a, potent anaphylatoxins that mediate vasodilation, chemotaxis of leukocytes, and other inflammatory effector functions that aid the innate response to infection [62].
ADCC is a similar process that is initiated when IgG binds viral proteins on an infected cell's surface. However, instead of interacting with C1q, the Fc domain of the attached IgG interacts with Fc receptors on leukocytes, inducing the release of cytotoxic cargo that directly kill the infected cell [62]. For example, the infusion of anti-simian immunodeficiency virus antibodies to animals infected with SIV demonstrated a subsequent decrease in viremia with a profile suggesting that the ADCC led to direct killing of virus-infected cells [65]. In the lung, alveolar macrophages carry out ADCC [66]: ~25% of alveolar macrophages can bind IgG3, and ~ 10% bind IgG4, with little binding IgG1 or IgG2 [67]. Interestingly, IgG can interact with lung-specific surfactant protein A (SP-A) to enhance some of its effector functions. For instance, SP-A bound to IgG-opsonized pathogens enhances phagocytosis relative to opsonization with IgG alone [68]. Importantly, IgG that are paired with SP-A are still capable of forming immune complexes and binding to C1q to initiate the classical complement cascade [63,68].
3.2.3. Fc effector functions specific to mucus: muco-trapping via mucin-crosslinking
A recently discovered yet little recognized effector function of virus-specific IgG is to crosslink viruses to the mucin mesh [[69], [70], [71], [72], [73], [74]], leading to their immobilization in AM (Fig. 2 ). By trapping viruses in AM, shed progeny viruses are unable to readily diffuse through AM to spread the infection within the lung. Trapped viruses are then quickly eliminated from the lung through natural mucociliary or cough-driven clearance, thereby offering a mechanism of direct physical clearance of viruses from infected lungs.
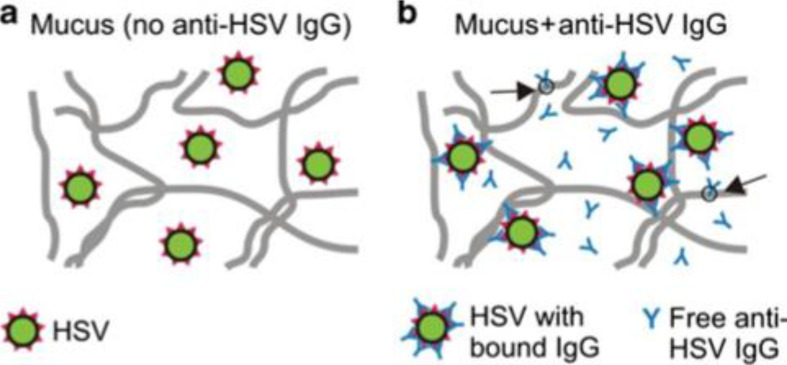
Fig. 2.
Proposed mechanism of antibody (Ab)-mediated trapping of viruses in mucus. Schematic showing (a) herpes simplex virus (HSV) readily penetrating native cervicovaginal mucus (CVM) with little-to-no endogenous HSV immunoglobulin G (IgG), and (b) anti-HSV IgG trapping HSV in CVM by multiple transient, low affinity bonds in mucins. By forming only short-lived, low-affinity bonds with mucus, free Ab, such as IgG, are able to diffuse rapidly through mucus and bind to viruses. As IgG molecules accumulate on the virus surface, they form multiple low-affinity bonds between the virus and mucus gel. A sufficient number of these transient low-affinity bonds ensure viruses are effectively trapped in mucus at any given time, thereby reducing the flux of infectious virions that can reach target cells. Adapted from [69] with permission from Springer Nature.
This unique mechanism of mucosal antibody protection was long overlooked because the affinity of individual antibody molecules to mucins was thought to be far too weak to directly crosslink pathogens to mucins. Indeed, the diffusion coefficient of IgG and IgA antibodies in human mucus is only slowed ~10% compared to in water [75,76], indicating that any bond between antibodies and mucins is exceedingly transient (on the order of seconds or fractions of a second) and readily broken up by thermal excitation [75]. Nevertheless, multiple IgGs can bind the same virus or bacteria, and the resulting array of bound antibodies on any individual pathogen/antibody complex can form multivalent interactions with the mucin mesh, providing avidity sufficient to trap individual pathogens nearly permanently.
This concept was first illustrated with herpes simplex virus (HSV), where HSV-specific IgG mediated effective trapping of HSV in human cervicovaginal mucus and protected against vaginal HSV transmission in mice [69]. Extension of this concept to AM was recently illustrated with influenza virus, where mobility was directly correlated with the presence of endogenous influenza-binding antibodies in AM, even for influenza virus-like particles (VLPs) that lack the ability to bind sialic acids on mucins [77].
IgG can effectively purge non-infectious, Ebola virus-like particles from the lungs of mice within just 30 min of intranasal mAb dosing [71] (Fig. 3 ). Unpublished work from our group suggests nebulized muco-trapping IgG into RSV-infected lambs can effectively reduce RSV infectious viral load in the lung to near non-detectable levels within just 3 days, even when administered at near peak viral titer in the lung, leading to greatly reduced bronchiolitis in the treated animals.
Fig. 3.
Ebola pseudovirus distribution in the mouse lung airways. A–D, Representative transverse 50-μm-thick frozen tissue sections showing the distribution of Ebola pseudovirus in the mouse trachea treated with phosphate-buffered saline (PBS) (A, B) or ZMapp (C, D). Red corresponds to Ebola pseudovirus, and blue corresponds to 4′,6-diamidino-2-phenylindole (DAPI)-stained cell nuclei. Arrows indicate the inner lining of the trachea. E, Quantification of Ebola pseudovirus signal in mouse trachea treated with PBS (control) or ZMapp compared with blank tissue. Data represent n = 3 mice per group with, on average, 10 tissue sections quantified per mouse. Error bars represent standard error of the mean. * indicates a statistically significant difference (P < 0.05) based on a two-tailed Student's t-test assuming unequal variance. Adapted from [71] with permission from Oxford Academic Press, Journal of Infectious Diseases.
The crosslinking of IgG-Fc with mucins appears to be mediated by specific N-glycans on IgG-Fc; removing either the Fc or the N-glycans greatly reduced muco-trapping [69]. There is likely a goldilocks range for the affinity between individual IgG and mucins that optimizes this effector function [72]: if the affinity is too weak, many IgG molecules must be bound to an individual pathogen to generate sufficient avidity to trap. In contrast, if the affinity is too strong, IgG lose their ability to undergo rapid diffusion in mucus and quickly accumulate on the pathogen surface. The ability for Fab domains on IgG to bind viruses with high specificity, coupled with IgG-Fc that interact with mucins, effectively transform AM overlaying the airway epithelium into a potent adhesive filter that can quickly clear viruses from the lung with exceptional specificity and potency [72].
4. mAb against SARS-CoV-2
In this section we summarize the current Ab development efforts against SARS-CoV-2, including information about their epitopes and also highlight some other approaches using non-Ab scaffolds including results from animal models where applicable. Further information about the current status of SARS-CoV-2 Abs in clinical trials is also discussed.
4.1. Spike protein as antiviral mAb target
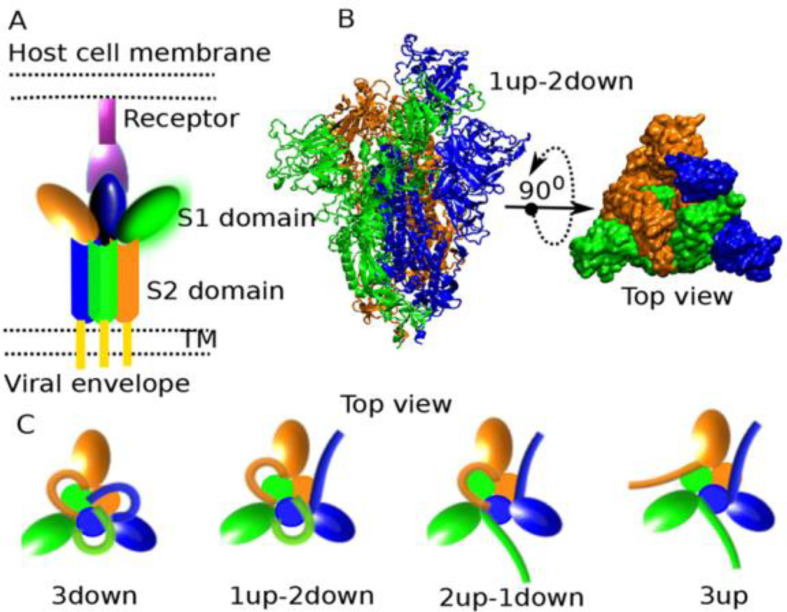
Coronaviruses infect host cells by engaging key host-cell receptors through its trimeric Spike (S) glycoprotein. A monomer of S glycoprotein of SARS-CoV-2 (~180 kDa) has two subunits: (i) S1, which contains an N terminal domain (NTD) and the Receptor-Binding Domain (RBD) responsible for binding to host-cell receptors, and (ii) S2, which contains a fusion peptide (FP), heptapeptide repeat sequences (HR1, HR2), a transmembrane domain (TM) and a cytoplasmic domain, promotes fusion of the viral and cellular membranes [78,79]. The S protein exists as a trimer on the viral envelope. Cryo-EM imaging of SARS-CoV-1 reveals each virion possesses ~50–100 spike trimers, with an average distance between spikes of ~15 nm [80]. The sequence of SARS-CoV-2 is about 70% homologous to SARS-CoV-1, and the two share about 80% sequence identity in the RBD and the requirement of ACE2 for entry [[81], [82], [83], [84]], suggesting SARS-CoV-2 may have similar a presentation of spikes on the viral surface.
Structural characterization of the SARS-CoV-2 S protein by Cryo-EM [81,85,86] shows that each of the S1 on the trimeric spike can independently assume either an open (“up”) conformation that exposes the RBD, or in the closed (“down”) conformation, whereby the surface of RBD that engages ACE2 is buried inside the trimer and thus not accessible for receptor binding (Fig. 4 ). The binding of S protein to ACE2 has been extensively characterized [81,85,[87], [88], [89], [90], [91]]. Binding of host ACE2 to exposed RBD leads to a 3-up conformation that is unstable, leading to S1 shedding and S2 refolding to promote membrane fusion [81,92]. In structural studies, the proportion of S1 RBD in the up conformation for SARS-CoV-1 prefusion spikes (i.e. 1-up or 2-up) in both unbound and ACE2-bound state was found to be about 55%, but almost no spikes had 3-up conformation, and the 2-up conformation was the most common [93,94]. Trypsin cleavage of SARS-CoV-1 spike results in about 95% of spikes in the up conformation that are capable of binding ACE2 [94].
Fig. 4.
Conformations of SARS-CoV-2 spike protein. A) Schematic depicting S protein bound to ACE2 receptor, including S1 and S2 subunits. B) Top and side view of S protein with one S1 subunit in up conformation. C) Movement of the S1 subunit makes it possible for spike protein to assume conformations where alL S1 units are down, one S1 unit is up, two S1 units are up, or all S1 units are up. Reproduced from [95] with permission from ACS, https://pubs.acs.org/doi/10.1021/acs.jpclett.0c01431. Further permission related to the material excerpted should be directed to the ACS.
Not surprisingly, given the structural homology between their spike proteins, some SARS-COV-1 binding mAbs also bind SARS-CoV-2, including CR30222 [96,97], S309 [98], and, similarly, some mAbs isolated from B-cells of some patients who have recovered from SARS-CoV-2 infection are capable of binding the spike protein of SARS-CoV-1 [99]. While a majority of potent neutralizing mAbs bind the RBD and block ACE2 binding, neutralizing mAbs can also bind other parts of the spike, including NTD or the S2 subunit. The precise mechanisms for how non-RBD binding mAbs can effectively neutralize SARS-CoV-2 remain unclear. While ACE2 can only bind RBD when at least one S1 is in the up conformation, structural alignment of CR3022–SARS-CoV-2 RBD suggests that the binding of CR3022 to RBD can be sterically hindered if the RBD on an adjacent protomer adopted a down conformation [96]. Thus, mAbs capable of binding recombinant RBD in vitro may still fail to neutralize SARS-CoV-2, depending on the epitope availability resulting from dynamic fluctuation of the S protein. Indeed, there are significant differences in neutralization potency of RBD binding mAbs with similar binding affinity [99,100].
4.2. Human mAbs isolated from convalescent blood
Since the onset of the pandemic, there has been an unprecedented global effort spanning across large pharma, startup companies, and academic researchers to discover and engineer mAbs against SARS-CoV-2 that can potently neutralize the virus. Much of this effort has centered on isolating potent neutralizing Ab against SARS-CoV-2 from B-Cells of patients who have recovered from the infection [[98], [99], [100], [101], [102], [103], [104], [105], [106], [107], [108], [109], [110], [111], [112], [113], [114]], with the notion that their recovery may be at least partially attributed to protection offered by specific antibodies produced by their immune system. More importantly, unlike in vitro affinity maturation by phage/yeast display that may create unnatural Abs with poor stability and pharmacokinetics in a complex biological environment, B-cell secreted Abs are already pre-selected for stability and activity. The COVID-19 pandemic has shown that, with the advances in single cell sequencing, deep sequencing, microfluidics, and cell sorting that have occurred over the past 2 decades, it is now possible to isolate individual clones of potent neutralizing mAbs on the order of days to weeks.
To isolate mAbs that can effectively neutralize SARS-CoV-2, the most frequently exploited approach is to identify B-cells with B-cell receptors (BCR) that bind the RBD fragment of the S protein with high affinity, since such binding likely sterically inhibits interactions between RBD and ACE2. In good agreement with this expectation, many of the isolated mAbs [[98], [99], [100], [101], [102], [103], [104], [105], [106], [107], [108], [109], [110], [111], [112], [113], [114]] bind S proteins with picomolar affinity. For example mAbs C121, C144, and C135 isolated by the Nussenzweig group have respective IC50 values of 1.64, 2.55, and 2.98 ng/mL against SARS-CoV-2 [99]. This in turn translates to exceptional neutralization potency against SARS-COV-2 [115]: many of them possess IC50 below 15 ng/mL against pseudoviruses, with comparable potencies against live virus (Table 1 ).
Table 1.
Select human or humanized mAb against SARS-CoV-2.
| mAb | Group | Antigen | IC50 (pseudovirus) | IC50 (live virus) | Refs |
|---|---|---|---|---|---|
| S309 | Veesler/Corti | S-Protein | – | 69 ng/mL | [98] |
| C121 C135 C144 |
Nussenzweig | RBD | – | 1.6 ng/mL 2.98 ng/mL 2.55 ng/mL |
[99] |
| COV2-2196 | Crowe | S-protein | 0.7 ng/mL | 15 ng/mL | [100] |
| REGN10933 REGN10987 |
Regeneron | S-protein + RBD boost | 6.42 ng/mL 6.09 ng/mL |
5.61 ng/mL 6.32 ng/mL |
[105] |
| P2C-1F11 | Zhang | RBD | 30 ng/mL | 30 ng/mL | [113] |
| B38 | Liu | RBD | – | 177 ng/mL | [112] |
| ADI-56046 | Walker | S-Protein | 40 ng/mL | 76 ng/mL | [114] |
| 2–15 | Ho | S-Protein | 5 ng/mL | 0.7 ng/mL | [104] |
| CT-P59 | Celltrion | RBD | 8.4 ng/mL | [116] | |
| CB6 | Junshi Bioscience | RBD | 33.3 ng/mL | 36 ng/mL | [109] |
| CC12.1 | Burton | S-Protein | 19 ng/mL | 22 ng/mL | [107] |
| BD-368-2 | Xie | S-Protein + RBD | 1.2 ng/mL | 15 ng/mL | [103] |
Despite highly varying neutralizing titers among patients' convalescent plasma [117], potent neutralizing Ab was isolated in vast majority of patients [99]. Interestingly, many of the most potent mAbs appeared to form with only modest levels of somatic hypermutation, as reflected by the small number of mutations compared to their closest germline sequences [118,119]. Very little somatic hypermutation or clonal B cell expansion was observed in five patients for up to 2.5 months after SARS-CoV-2 transmission [106]. The precise immunological mechanisms responsible for the limited somatic hypermutation or clonal expansion are unclear; it may be possible that the high affinity of near-germline IgG antibodies against SARS-CoV-2 simply limits antigen access to the germinal center. Nevertheless, the limited need for somatic hypermutations to generate high affinity neutralizing mAbs against SARS-CoV-2 is consistent with the highly comparable neutralization potencies (2–10 ng/mL range) among the numerous mAbs generated independently by different groups.
Beyond isolating mAbs from recovered patients, it is possible to employ other methods to isolate human/humanized mAbs against SARS-CoV-2. One such approach is Regeneron's high-throughput isolation and screening of antibodies from their proprietary, humanized VelocImmune® mice and VelociGene® technologies [120]. The same platform was used to generate Regeneron's anti MERS-CoV mAbs, REGN 3051 and REGN 3048, which possess affinities between 40 and 48 pM. Most recently, Regeneron applied its VelocImmune® mice and sequencing of B-cells from convalescent COVID-19 patients to isolate potent SARS-COV-2 neutralizing antibodies [105]. The isolated mAbs had affinities for spike proteins between 37.1 and 42.8 pM, and IC50 against live virus of 37.4 to 42.1 pM (5.61 to 6.30 ng/mL). A cocktail of two antibodies, REGN10933 and REGN10987, is currently in clinical trials for treating hospitalized (NCT04426695) or ambulatory (NCT04425629) adult patients. Other humanized mouse technologies for isolation of monoclonal exist, and a review of different monoclonal antibodies isolated with these platforms can be found elsewhere [121].
A number of non-RBD binding yet neutralizing mAb have also been described [98,104,110]; these mAbs do not block ACE2 binding, but still neutralize infection. For example, in a study by Liu et al, NTD binding mAbs 2–17, 5–24 and 4–8 had respective IC50 values of 7, 8 and 9 ng/mL, and mAbs 2–43 and 2–51 that bind neither RBD nor NTD had IC50 values of 3 and 7 ng/mL for neutralization of live SARS-CoV-2 virus infection, comparable to some of the best RBD-binding mAbs [104]. Although many of the mAbs that block ACE2 interaction are only able to bind the RBD on S proteins in the up conformations, other non-RBD-binding mAb are also able to bind S in the down conformation [99,100,104,105].
4.3. Non-human mAb
4.3.1. Nanobodies
Camelid sera contain unique heavy chain antibodies (VHH) that do not incorporate a light chain [122]. Similarly, cartilaginous fishes have heavy-chain antibodies (IgNAR) from which antigen domain antibodies VNAR can be obtained [123]. These antigen binding domains, commonly referred to as single-domain antibodies or nanobodies, can be expressed recombinantly in non-mammalian cultures, and do not require proper heavy-light chain pairing. While their small size and stability profile is thought to offer potential advantages over traditional IgG-based mAbs, they also pose some limitations, including short serum half-life, an inability to facilitate effector functions, and immunogenicity. Some of these can be addressed by the addition of an IgG Fc, which would necessitate production using mammalian cultures. Nanobodies are currently in clinical trials for a diverse array of human diseases [124]; Caplacizumab was the first nanobody approved by the FDA in 2019 [125]. Clinical development of ALX-0171, an inhaled nanobody against RSV, was recently discontinued [126]. Potent nanobodies have been developed against SARS-CoV-2 [[127], [128], [129], [130], [131], [132]], with some like mNB6tri [130] possessing picomolar IC50s (2.3 ng/mL) in live SARS-CoV-2 infection assays.
4.3.2. Other biological scaffolds
Other scaffolds have also been used to develop potent inhibitors of SARS-CoV-2 infection. Linksy et al. [133] and Cao et al. [134] developed molecules by de novo design using helices capable of binding RBDs on S proteins; the best molecule, LCB1, had IC50 of 24 pM (0.16 ng/mL) in live SARS-CoV-2 infection assays [134]. Human VH domain based molecules (VH-Fc) capable of neutralizing SARS-CoV-2 infection have also been developed by various groups [133,135,136]. Ab8 had an IC50 of 40 ng/mL for neutralization of live SARS-CoV-2 [133].
4.3.3. ACE2 decoys and derivatives
Viral escape mutants can readily develop against specific mAb, with escape mutants still retaining ACE2 binding [137]. Thus, in contrast to mAb discovery or de novo design, a number of research groups have focused on developing ACE2 decoys, including recombinant ACE2-Fc (fusion of ACE2 and IgG1-Fc). ACE2-Fc can block infections by SARS-CoV-1 [138] and SARS-CoV2 [136,139]. Nevertheless, the potency of recombinant WT ACE2 is 2–3 logs worse than the best neutralizing mAbs. To improve the potency of ACE2 decoys, a number of groups have engineered variants of ACE2 using structural knowledge and screening of mutant libraries [[140], [141], [142]]. Some of these mutants exhibit significantly improved affinity to SARS-CoV-2 RBD, with neutralization potency that rivals some of the human mAbs.
Other studies have attempted to increase the valency of ACE2 or ACE2 mutants by linking them to a trimerization domain instead of an IgG1 Fc to generate mutant ACE2 trimers, demonstrating, in some cases, better potency than ACE2-Fc dimers [143,144]. Another approach linked an NTD binding mAb to ACE2-Fc, resulting in ~100 fold improvement over ACE2-Fc in binding and neutralization [145].
4.4. Efficacy in animal models to date
Alongside the current intense efforts for SARS-CoV-2 mAb discovery, many groups have been exploring suitable animal models for COVID-19. Various animal models have been developed for SARS-CoV-2 infection studies, including the use of transgenic mice with hACE2, Syrian hamsters, ferrets, and non-human primate models in rhesus macaques, cynomolgus macaques, and African green monkeys. Several reviews have recently summarized these and other animal models and findings [146,147].
A number of studies have focused on the use of mAbs for passive immunoprophylaxis, given to animals prior to viral challenge to assess protection against infection. Viral RNA copy numbers and infectious virus titers were reduced by 4–6 logs in golden Syrian hamsters intraperitoneally administered a dose of 1.5 mg/kg of a mAb 2–15, 24 h prior to intranasal challenge with 105 PFU of SARS-CoV-2 [104]. In Syrian hamsters given 0.9 mg/kg – 16.5 mg/kg of the mAb CC12.1, 24 h prior to intranasal virus challenge, reduced viral RNA copy numbers by ~2–3 logs [107].
A small number of in vivo studies have assessed both prophylaxis and therapeutic efficacy. In rhesus macaques, viral titers from throat swabs were reduced by 6 logs and 3 logs in animals receiving 50 mg/kg of CB6(LALA) intravenously 24 h prior-to or 1 and 3 days after infection, respectively [109]. Using mice expressing human ACE2, mAb COV2-2196, mAb COV2-2381, or a cocktail of mAb COV2-2196 and mAbCOV2-2130 demonstrated 3 logs reduction in lung viral titers at 200 μg/mouse administered 24 h before or 12 h after intranasal challenge with 4 × 105 PFU SARS-CoV-2 [100]. Additionally, there were 2–3 logs reduction in viral RNA copies in nasal swabs of macaques dosed with 50 mg/kg mAb COV2-2196 or mAb COV2-2381, 3 days before challenge with 1.1 × 104 PFU SARS-CoV-2 [100]. Greater prophylactic efficacy vs. therapeutic efficacy was also observed with other studies. In hACE2 transgenic mouse, 20 mg/kg injection of the mAb BD-368-2 reduced 105 TCID50 SARS-CoV-2 lung viral titers by 6 and 3 logs when given 24 h before and 2 h after infection, respectively [113]. Similarly, again using hACE2 transgenic mouse, 25 mg/kg mAb B38 or mAb H4 given intravenously 12 h after intranasal infection with 5 × 105 TCID50 COVID19 virus reduced lung viral RNA copies by ~3 logs, with reduced lung damage [99].
4.5. Anti-SARS-CoV-2 mAbs in the clinic
Due to the considerable costs associated with advancing a mAb into the clinic, only a small fraction of the discovered mAbs described above have reached the clinic over the first 6 months of the pandemic (as of Nov 2020). A number of other human mAbs, nanobodies, and ACE2-decoys will likely initiate clinical studies over the next 6 months.
The first clinical trial of a mAb to treat COVID-19 was Eli Lilly mAb LY-CoV555 (partnered with AbCellera) initiated on 5/28/2020 (NCT04411628). It is a human IgG1 targeting the spike protein derived from human B cells from convalescent patients. It was well tolerated at all doses tested, with no serious drug-related severe adverse events (SAEs) reported to date. There are currently multiple ongoing Phase 3 trials underway for LY-CoV555 for inpatients (NCT04501978) and outpatients (NCT04518410) with COVID-19, as well as in nursing home residents and staff (NCT04497987). Additionally a Phase 3 trial of combination of LY-CoV555 and LY-CoV016 for participants with mild to moderate COVID-19 Illness (NCT04427501) is also underway [148]. Recent data released from the ongoing studies of LY-CoV555 have shown that treatment was associated with a trend toward decreased hospitalization among patients who received any dose of mAb (relative to placebo), as well as a slight decrease in symptom severity up until day 6 (but not after). Surprisingly, only the 2800 mg (medium dose) group in the LY-CoV555 study resulted in a statistically significant reduction in viral load by day 11 relative to placebo, whereas the higher dose (7000 mg) did not [149]. Despite these confounding results, LY-CoV555 received emergency used authorization (EUA) for treatment of recently diagnosed COVID-19 patients at risk of developing severe disease by the FDA on November 9, 2020.
Regeneron initiated Phase 1 trial for its REGN-COV2 therapy (a combination of mAbs REGN10933 and REGN10987) on 6/10/2020 (NCT04425629, NCT04426695), and is currently in phase 3 (NCT04452318). REGN-COV2 was granted emergency use authorization for the treatment of early COVID-19 by the FDA on November 21, 2020. REGN-COV is not authorized for use for those who are hospitalized or require supplemental oxygen therapy. This EUA was based on initial data from the REGN-COV2 trials that found that treatment appeared to reduce viral load as well as reduce the likelihood of COVID-19-related medical visits by 57% through day 29, relative to placebo, but found no apparent benefit when administered to patients with more advanced disease [150]. Celltrion initiated a trial for their mAb CT-P59 on 6/18/2020 (NCT04525079) and has not seen any drug-related serious adverse events at any dose tested to date. Vir Biotechnology and GSK initiated a trial for their mAb VIR-7831 for the Early Treatment of Coronavirus Disease 2019 (COVID-19) in non-hospitalized patients on 8/27/2020. Many more mAbs isolated from SARS-CoV-2 patients' B-cells are initiating clinical trials and can be followed using the COVID-19 Antibody Therapeutics Tracker website [151].
A number of companies have advanced or are advancing a single mAb for COVID-19 (e.g. Eli Lilly, Celltrion), and others are advancing a cocktail of two mAbs (e.g. Regeneron). A key distinction between single mAb vs. mAb cocktail is the risk of viral escape with mutations such as N439K [149] and Y453F [149]. Studies with a replicating VSV-SARS-CoV-2-S virus have shown that multiple independent viral escape mutants can be readily generated to each of the individual antibodies tested [137,152]. In contrast, viral escape mutants are not selected in presence of non-competing or partially competing mAb cocktails. Thus, the use of a pair of non-competing mAbs may be highly desirable to overcome the possibility of viral escape against specific mAbs.
5. Clinical efficacy of mAbs for the treatment or prevention of ARIs
5.1. Previous antiviral mAb in clinical trial for ARIs
The efficacy of influenza vaccines are highly variable, and only moderately reduce the rates of hospitalization, especially in high risk groups: among adults ≥65, those who receive the flu vaccine, have a 14–43% reduced risk of hospitalization relative to unvaccinated patients [153]. Furthermore, there are many ARIs for which no vaccine or effective therapies are available, including RSV, MPV, PIV, adenovirus, seasonal coronaviruses (e.g. NL63-CoV), rhinoviruses, and others. Collectively, these ARIs affect tens of millions each year in the U.S. alone. Given the substantial annual morbidity and mortality caused by these ARIs, it is no surprise that significant efforts have been made to develop mAb-based therapeutics for respiratory infections [154]. The majority to date have focused on RSV and influenza, which represent the two largest areas of unmet medical need among ARIs.
Unfortunately, nearly all such efforts to develop mAb as therapy for ARIs have been met with disappointing results. Some human or humanized mAbs developed as antivirals that have advanced past Phase 1 studies include MEDI-8852 (influenza; IC50 ~ 41–4050 ng/mL; 750 or 3000 mg) [155,156], MHAA4549 (influenza; IC50 ~ 195–6765 ng/mL; 3600 or 8400 mg) [44,157], CR8020 (influenza; IC50 ~ 9–500 ng/mL; 30 mg/kg) [158], CT-P27 (influenza; IC50 ~ 15 μg/mL; 10–20 mg/kg) [136], diridavumab (influenza; IC50 ~ 18–2200 ng/mL; 30 mg/kg) [159,160], VIS-410 (influenza; IC50 ~ 30–7000 ng/mL; 2000 or 4000 mg) [161,162], Motavizumab (RSV; IC50 ~ 20 ng/mL; 15 mg/kg [163]), and palivizumab (RSV; IC50 ~ 163–360 ng/mL; 15 mg/kg) [[163], [164], [165]]. None of these mAbs were noted to have major safety concerns, which supports the anticipated safe use of mAbs against SARS-CoV-2. Unfortunately, they also did not show appreciable efficacy as a therapeutic, even when administered at very high doses (e.g. 3000 mg per patient for MEDI-8852 and 8400 mg per patient for MHAA4549). Only one - palivizumab, also known as Synagis® - has received approval as a prophylaxis; however, its modest efficacy and high costs have limited its clinical use to only the most severely premature infants.
5.2. Potential mechanisms for prior failures
Given the immense efforts to advance antiviral mAbs as a treatment for COVID-19, we believe it is important to examine why similar efforts to development treatments for other similar ARIs in the past have all failed, in order to better guide current development.
A common feature of all therapeutic antiviral mAbs listed in Table 1 is the universal focus on systemic delivery, either by intramuscular (IM), subcutaneous (SC), or intravenous (IV) injections. As noted above, SARS-CoV-2, just like other common viruses responsible for ARIs, preferentially infects the airway epithelium via the apical membrane and shed progeny viruses back into the airway lumen as the infection spreads from the URT to the LRT. This unique pathophysiology implies that therapeutic concentrations of mAbs must be achieved in the AM secretions where viruses concentrate in order to effectively neutralize viruses that are actively shed into the AM and limit the continued spread of the infection within the lung. However, non-human primate studies have shown that the concentration of mepolizumab in broncheoalveolar lavage fluid (BALF), following IV injection, is ~500-fold lower than the concentration of mepolizumab in plasma [166]. Even greater differences in BALF vs. plasma concentration were observed in biodistribution studies of motavizumab (anti-RSV mAb) in cynomolgus monkeys, where BALF and plasma concentrations of ~100 ng/mL and ~ 200,000 ng/mL, respectively, were measured 4 days following an IV dose at 30 mg/kg [167]. We have observed comparable magnitude difference in BALF vs. plasma levels of palivizumab injected in neonatal lambs (unpublished observations).
The limited concentrations of mAb measured in BALF following IV delivery imply that the failures of many antiviral mAb therapies in the past may simply be attributed to a failure in achieving therapeutic concentrations of mAb in the lung airways early enough in the course of disease progression. Indeed, near complete neutralization of viruses requires mAb concentrations well in excess of their IC50 (e.g. 5–10-fold excess). With many of the mAbs that were advanced into clinical studies in the past, neutralization potencies were in the low nanomolar affinity (i.e. 20–1000 ng/mL) range. Thus, in nearly all cases, the mAb dosing was not likely to have achieved effective inhibitive concentrations at the site of infection. It should be noted that the mAb levels needed to effectively prevent infection by passive immunization may be substantially less than the mAb levels needed to treat an active infection, since infections are typically initiated with low titers of viruses. Even with the highly inefficient pulmonary distribution of systemically dosed mAb, it may be possible to prevent infection by a small number of viruses, as reflected by the immunoprophylactic efficacy of palivizumab, motavizumab, and MEDI-8897 in reducing RSV hospitalization in clinical studies [[168], [169], [170]]. However, in the context of a much-greater viral load seen in an active, established infection of the respiratory tract, much higher levels of mAb are needed to quickly and effectively limit further viral spread. This accentuates the need for any COVID-19 mAb therapy to efficiently distribute the administered mAb into the lung airways in order to quickly block the progression of the infection from the URT and LRT into the deep lung.
Another likely factor in the poor clinical efficacy of treating ARIs using antiviral mAbs to date is the timing of mAb administration into the patients. Infusions are typically done in a hospitalized setting; not surprisingly, with most of the previous clinical trials, patients infected with RSV and influenza are often given the mAb only as they are hospitalized. By that point, the viral load with many ARIs is already beginning to wane, and most morbidities in this later stage of disease are driven by infection-related inflammation (e.g., through inappropriate bradykinin signaling following depletion of ACE2 [171] or through a feed-forward cycle of inflammation driven by IL-6 and other innate mediators [172,173]. The delayed initiation of mAb dosing is further complicated by the potentially slow distribution of systemically dosed therapies into the respiratory tract, leading to substantial delays before reaching Cmax in the lung. This effectively allows additional time for viruses to replicate and spread, resulting in further inflammation until inhibitory concentrations of the drug are finally reached in the lung. For instance, it takes 3 days of administering osteltamivir, an anti-influenza therapeutic, twice per day before achieving steady-state drug concentrations in the lung [174]. The time it takes IgGs, which are large, hydrophilic molecules with no mechanism of active transport, to effectively distribute into the lungs after systemic administration is likely to be equally slow.
6. Strategies to overcome past failures
6.1. Advancing more potent mAb
mAbs distribute very poorly from the systemic circulation into the lung [175], with only ~1% of the intravenously dosed mAb reaching the lung tissue [176]. One obvious way to increase the likelihood of efficacy is to improve the neutralization potency (i.e. IC50) of the mAb; assuming the same amount of mAb can reach the lung, a more potent mAb will more effectively inhibit viral replication locally in the lung than a less potent mAb. Indeed, the majority of the current anti-SARS-COV-2 mAbs in active preclinical and clinical development have exceptional potencies (IC50 values <10 ng/mL range) (Table 1). These potencies substantially exceed nearly all of the antiviral mAbs that were previously advanced into clinical studies and failed, including palivizumab against RSV (Synagis; IC50 ~ 163–360 ng/mL [[163], [164], [165]]), MEDI8852 against influenza (IC50 ~ 41–4050 ng/mL [155,156]), and MHAA4549 against influenza (IC50 ~ 195–6765 ng/mL [44,157]). Theoretically, assuming the concentrations of these anti-SARS-CoV-2 mAbs in BALF can reach ~0.1–0.2% of the mAb concentration in plasma, consistent with prior non-human primate studies, the local concentration should substantially exceed the IC50 of these mAbs. Indeed, ~100 ng/mL mAb concentration in BALF, arising from 200,000 ng/mL concentration in plasma [167], would correspond to 10 times greater mAb than an IC50 of 10 ng/mL).
While it may seem promising to simply dose more potent mAb to enhance efficacy, it should be noted that other antiviral mAbs with comparable neutralization potency in vitro have been advanced to clinical studies only to fail to provide meaningful benefit. For instance, IV infusion of the anti-RSV mAb motavizumab (IC50 ~ 20 ng/mL [163]) was not effective as a treatment for RSV [177]. Indeed, despite clear theoretical benefit, the extent to which greater potencies of mAbs can translate to better outcomes in the clinical remains unclear. For instance, there did not appear to be an appreciable difference in the prophylactic effectiveness of MEDI-8897 vs. motavizumab in early clinical studies [178,179] despite ~5-fold greater neutralization potency [180] and 9-fold better activity in a cotton rat model of RSV infection [181]. As it stands, it seems that greater neutralization potency in vitro may not predict effectiveness in vivo, as exemplified by an exceptionally potent mAb against Ebola in vitro affording no efficacy in vivo despite no evidence of neutralization escape [182].
Another approach is to simply dose mAb at higher doses per patient. For instance, in primate studies with motavizumab possessing YTE mutations in its Fc that lead to greater quantities of motavizumab in the systemic circulation, there appeared to be a comparable increase in motavizumab in the BALF. Nevertheless, in vitro studies suggest the rates of IgG transfer across monolayers of primary cultured alveolar epithelial cells may be saturated with higher levels of IgG [183]. In Eli Lilly's double-blind, placebo-controlled Phase 2 study evaluating LY-CoV555 at 700 mg, 2800 mg, and 7000 mg injected per patient, only the middle dose, but not the highest dose, met its prespecified primary endpoint of change from baseline in viral load at Day 11. Due to the small clinical trial size, it remains to be determined whether higher dosing of LY-CoV555 would translate to greater clinical efficacy. Many ongoing clinical studies are similarly testing mAb dosing at levels that greatly exceed what was tested with prior mAb therapies for ARIs in the past. Unfortunately, dosing at exceptionally high doses per patient is not an ideal solution when access to treatment is already expected to be limited by mAb availability and production capacity.
6.2. Initiating mAb therapy earlier
As discussed in Section 2.2.2, SARS-CoV-2 infections initiate in the URT before progressing through the LRT to the deep lung. In severe cases of deep lung infections, a hyperinflammatory milieu associated with dramatically increased expression of IL-6, CRP, and other inflammatory markers, is associated with worse prognoses [172]. The hyperinflammatory responses frequently trigger acute respiratory distress syndrome (ARDS); a recent survey showed that ~33% of hospitalized COVID-19 patients develop ARDS, ~26% are transferred to the ICU, and ~ 16% die [184]. By this stage of the infection, the positive feedback loop of inflammatory signaling can proceed in the absence of innate sensing of viral pathogen-associated molecular patterns [36,172,185], as evidenced by the fact that at least some patients with critical disease who receive convalescent plasma treatment can fully clear virus within 3 days (per RT-PCR), but still experience worsening decompensation and eventually death [186]. Inflammation in the deep lung can also can lead to further life-threatening conditions such as organ failure, cerebrovascular disease, and sepsis [42], as well as debilitating, life-long pulmonary fibrosis [187,188]. These realities highlight the need for early intervention to prevent the spread of infection to the LRT prior to the initiation of a hyperinflammatory response by the innate immune system.
COVID-19 patients are hospitalized due to morbidities associated with the inflammation caused by SARS-CoV-2 infections, rather than the presence of the virus itself. In our opinion, administering antiviral mAb to hospitalized patients with LRT infections is akin to turning off the gas supply that fuels a raging fire: while undoubtedly important, the rest of the fire (i.e. inflammation) must be put out to save the home. Indeed, it is unlikely that, in late stages of infection, antiviral mAb alone can quickly dampen the inflammation when administered after the infection reaches this terminal hyperinflammatory phase. Earlier treatment also implies there are lower viral titers in the lung at the time of initiating mAb therapy, making the spread of the viruses easier to stop and allowing for more time for the infused mAb to reach the lung before the viruses potentially spread into the LRT. Thus, the ideal antiviral therapy should be initiated early during the course infection, as soon as infection can be practically diagnosed, in order to prevent the associated pulmonary morbidities associated with infections in the deep lung. If caught early, the fire likely goes out on its own; URT symptoms typically resolve without requiring further medical attention.
Fortunately, this opportunity exists with COVID-19. Several studies have shown that the median time from first symptoms to dyspnea and potential hospitalization is another ~5–7 days [40,41], and ~ 8–12 days to ARDS [89]. This underscores a prime window whereby antiviral mAb can be dosed in patients after the onset of symptoms, with adequate time for the mAb to prevent the virus from spreading past the LRT into the deep lung. Reflecting the paradigm shift to initiating mAb therapies earlier, Eli Lilly has recently completed a Phase 2 clinical trial to determine the safety and efficacy of LY-CoV555 and LY-CoV016 for treatment of COVID-19 in an outpatient setting (NCT04427501). Eli Lilly announced in an interim report that treatment with any dose of mAb (in a pooled analysis) led to a hospitalization or ER visit incidence rate of 1.7% (5/302), much lower than the 6% (9/150) in the placebo group, despite only the middle dose group achieving its primary endpoint of viral titer reduction [148]. In parallel, Eli Lilly has started a phase 3 trial to evaluate whether LY-CoV555 stops the residents of nursing homes from developing COVID-19, creating customized mobile research units to run the clinical trial at nursing homes (NCT04497987). Regeneron is also conducting Phase 1/2/3 trials testing the safety and efficacy of antibody cocktail REGN-COV2 in infected ambulatory patients (NCT04425629), as well as prevention of infection in asymptomatic patients who are household contacts of individuals infected by SARS-CoV-2 (NCT04452318). Both LY-CoV555 (bamlanivimab) and REGN-COV2 (casirivimab and imdevimab) have received emergency used authorization from the FDA to treat recently diagnosed COVID-19 patients in outpatient settings immediately after diagnosis, but are not indicated for patients already on supplemental oxygen [189,190].
6.3. Direct delivery to the lung
Virtually all mAb therapeutics currently on the market, and notably all antiviral mAbs that were previously advanced into clinical studies for ARIs and failed, are administered systemically either via intravenous, subcutaneous, or intramuscular injections. The failures of systemically dosed antiviral mAb for ARIs is particularly notable, since nearly all antiviral mAbs currently under clinical development for COVID-19 are also administered systemically. As discussed above, systemic mAb dosing results in far lower levels of mAb in the lung relative to the overall dose of mAb administered, thereby limiting efficacy. Methods that can deliver greater amounts of mAb more quickly to where the viruses are concentrated will likely more effectively limit the spread of SARS-CoV-2 to the deep lung.
Inhaled delivery of mAb offers a number of distinct advantages over systemic mAb delivery. First, unlike systemic dosing, where the concentration of mAb in the lung is usually reduced 500–2000+ fold compared to the concentration in the circulation (see Section 5.2), a much larger fraction of the dosed mAb would reach the site where the viruses are concentrated: the AM. In a non-human primate study, mAb concentrations in the lung measured by in vivo microdialysis reached 1 μg/mL shortly after nebulization [191]; the authors had unexpectedly poor delivery likely due to an incompatible mouthpiece. In another macaque study, nebulized IgG, IgA, and IgM in the epithelial fluid of the lung reached concentrations between 500 and 1000 μg/mL immediately after delivery [192]. In this study, 1.33 mg/mouse of IgG administered by intranasal deposition was required to prevent bacterial pneumonia. In a Phase I trial for inhaled omalizumab that was self-administered using a jet nebulizer, for the 10 mg dose cohort, 273 to 2380 μg of omalizumab was estimated to have been delivered to the lung [193]. Given the typical neutralization potencies of antiviral mAbs (IC50 in the 10–300 ng/mL range [163]), it is likely that inhaled delivery of mAb on this scale would easily result in pulmonary mAb concentrations that are well in excess of the IC50. Achieving and sustaining mAb titers above its IC50 should translate to more effective inhibition of infections in the lung. For instance, in a mouse influenza challenge model, neutralizing mAb dosed intranasally achieved superior efficacy than the same mAb administered intraperitoneally, even at lower doses, when the treatment is initiated later in the infection [194]. Intranasal dosing of neutralizing mAb also reduced the dose of mAb needed in a RSV challenge cotton rat model by orders of magnitude; 4 g/ kg of IVIG delivered by i.p injection were required to achieve the same level of protection as 0.025 g/kg delivered by intranasal deposition [195]. Topical delivery of neutralizing mAbs into the lung has effectively suppressed viral replication in other animal models [[194], [195], [196]].
Another advantage with inhaled delivery of mAb is the time it takes to reach local Cmax. When inhaled, the maximum concentration of mAb in local lung tissues is naturally highest immediately following inhalation. In contrast, limited extravasation from systemic circulation into the lung means that systemically dosed drugs typically take many hours if not days to reach a local Cmax. Indeed, steady state concentrations of osteltamivir in the lung can only be achieve after 3 days of twice-daily dosing [174]. Poor biodistribution of mAb into the respiratory tract was reflected by a mouse study showing that, whereas 100% of nebulized cetuximab was present in the lung 2 h after nebulization, only ~1% of cetuximab was localized to the lung 2 h after i.v. injection [176]. Even after 48 h, nebulization leads to a much greater fraction of mAb in the lung (>50-fold excess) compared to the same dose of mAb dosed systemically.
6.3.1. Methods for inhaled delivery
Generally, mAb can be delivered in the lung in two forms: either as micronized dry powder aerosol with suitable aerodynamic diameter to enable efficient inhaled delivery through the use of dry powder inhalers (DPI), or as a liquid that can be aerosolized through the use of nebulizers. DPIs have been used to deliver small drugs into the lungs for asthma and COPD for decades. They are simple to use, require no electrical power, and are highly portable. Vectura, in partnership with UCB, previously advanced VR942 (an anti-IL13 mAb fragment for treatment of asthma) into Phase 1 and 2 clinical studies, demonstrating good safety in the phase I clinical trial (NCT02473939) [197]. More recently, Novartis investigated CSJ117, an anti-thymic stromal lymphopoietin antigen binding antibody fragment formulated into DPI, in a double-blind, placebo-controlled study in 28 patients with mild, atopic asthma, and observed no treatment-emergent serious adverse events [198]. Nevertheless, DPIs also have a number of important disadvantages relevant to mAb delivery. First, DPIs result in a large fraction of the drug deposited in the mouth and throat and have limited delivery to the upper respiratory tract (i.e., sinuses). Second, some patients, particularly elderly and pediatric patients, have trouble using DPIs as intended. Finally, the formulation of complex biomacromolecules such as mAb into DPIs require extensive development effort, with timelines that may not be compatible with the urgencies needed to quickly respond to a pandemic. For these reasons, ~75% of inhaled therapeutics in clinical development have pursued the nebulization of liquid protein preparations [199].
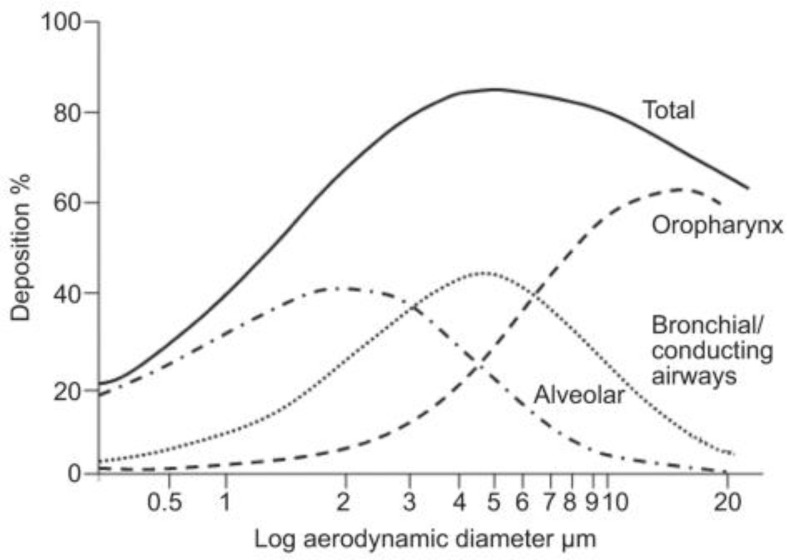
There are 3 major classes of nebulizers: jet, ultrasonic, and mesh nebulizers; they differ with respect to the physical process utilized to generate aerosols, but they typically each generate particles in the 2 to 5 μM range [200]. For optimal deposition into the conducting lung airways as well as the deep lung, the median mass aerodynamic diameter of the droplets should generally be in the 4–5 μm range [[201], [202], [203]]. As shown in Fig. 5 , the distribution of droplet sizes generated from a nebulizer can enable effective delivery to all parts of the respiratory tract, from the URT to LRT to the deep lung [204,205]. For instance, intrapulmonary deposition of a nebulized radiolabeled drug with median droplet diameter of 2.9 μm (89.2% of droplets had a diameter < 5 μm) ranged from 33 to 40%, while peripheral lung deposition ranged from 17 to 20% [206]. Similarly, inner lung deposition of radiolabeled nebulized drug with average diameter of 4.6 μm (56.8% droplets had a diameter < 5 μm) was 62.8–73.3%, while 28.7–34.8% deposition was observed in the upper airways and only ~0.99% was exhaled by the patients [207]. As high as 80% lung deposition has been achieved with nebulizers that generate particles with average diameters of 3 μm [208]. Nebulizers can be operated with additional systems to filter out particles larger than the desired size, and in that way increase drug deposition in the targeted lung region [209]. Rapid lung deposition of therapeutics can be achieved even with modest lung deposition efficiencies. For instance, even with a lung deposition rate of 32%, 43.9 mg of tobramycin could be delivered in 2.5 min with a mesh nebulizer [210].
Fig. 5.
Relationship between aerosol diameter and lung deposition predicted by the Model by the International Commission on Radiological Protection. Aerosols with diameters >5 μm deposit mainly on the oropharynx, whereas particles with sizes <5 μm can travel deeper into the lung and deposit in the bronchial/conducting airways or reach the alveoli (diameter < 1 μm). Reproduced with permission of the © ERS 2020: European Respiratory Journal 37 (6) 1308–1417; DOI: https://doi.org/10.1183/09031936.00166410 Published 1 June 2011.
Jet nebulizers have been successfully used to nebulize protein therapeutics that are FDA-approved, namely Pulmozyme®. In jet nebulizers, a compressed gas, often air, passes through a liquid reservoir containing the drug, atomizing the liquid intro primary aerosols. These aerosolized particles are then pushed into a baffle. Larger droplets collide with the baffle and are forced back into the reservoir, whereas small aerosols of a specific size are carried out by the gas for inhalation. Several cycles of aerosolization are required to nebulize the therapeutic, and the repeated recirculation of the aerosols, together with the air-liquid interface, can both increase protein aggregation [211]. Aggregated proteins in turn could potentially more readily induce anti-drug antibodies (ADA), as was observed with omalizumab that was nebulized using a jet nebulizer [193]. Readers are referred to an excellent review on the mechanism of protein aggregation during nebulization [212]. Finally, jet nebulizers typically have considerable “dead volume”; residual mass can be up to 50% of the loaded drug mass [213,214]. This wasted volume would be a significant concern with an expensive, complex, biological molecule like mAb.
Ultrasonic nebulizers have been used for pulmonary delivery of a variety of therapeutics. In ultrasonic nebulizers, a piezo-electric crystal at the bottom of the medical reservoir vibrates at frequencies between 1 and 3 mHz to generate a geyser that releases aerosols [215]. Small particles are collected in the nebulization chamber, while larger particles cycle back into the liquid chamber after aggregating or colliding with baffles. Similar to jet nebulizers, droplet recirculation increases aggregation of biomolecules [212]. Importantly, ultrasonic nebulizers are generally not suitable for nebulization of heat labile biomolecules; excess energy from the ultrasonic vibration is converted into heat, which substantially increases the temperature of the liquid reservoir by as much as 20 °C [215,216] which, in turn, leads to aggregation or degradation of the therapeutic. Finally, just like jet nebulizers, ultrasonic nebulizers also have large dead volumes, as high as 30% [214].
6.3.2. Vibrating mesh nebulizers (VMN)
VMNs aerosols are formed by pushing a liquid containing the therapeutic molecule through a vibrating mesh. The mesh contains conical holes, with the broader end at the liquid interface. Droplets are generated in a predictable range of sizes, and the aerodynamic diameter of droplets can be tailored by changing the pore size of the vibrating mesh [217]. There are two types of VMN, active mesh nebulizers and passive mesh nebulizers. These devices are small, portable, and can be powered by batteries. They achieve up to twice the lung deposition of jet nebulizers and have residual volumes of less than 10% [212,214]. Recirculation of the liquid containing the therapeutic inside the machine is not necessary, which is a major concern in jet and ultrasonic nebulizers since it can lead to protein aggregation and denaturation. Indeed, VMNs nebulize mAbs with far fewer aggregates [218]. For instance, during preclinical development of ALX-0171, a trivalent nanobody for RSV, it was observed that jet nebulizers had up to 40% increase in high-molecular weight species, whereas the APIXNEB VMN only produced a 2–3% increase [219].
Given these advantages, it is not surprising that VMN have become the preferred method for nebulization of therapeutics by the pharmaceutical industry and clinicians since 2004; mesh nebulizers have been used in 60% of registered clinical trials in the USA and EU, from 2006 to 2016. Jet nebulizers have only been used in 36% of the trials in the same time frame [208]. The FDA approved the PARI eRapid® VMN for nebulized delivery of Pulmozyme® in 2015; compared to jet nebulizers, the eRapid® VMN showed equal efficacy and safety, 73% shorter nebulization times, and greater patient satisfaction [220]. We refer readers to excellent articles detailing the advantages of VMN over other nebulizers [199,208,214,221].
The integration of breath-activated or smart functions into VMN has further improved their efficiency. One such technology is Adaptive aerosol delivery (AAD®). This technology that was developed to predict the patient's own inhalation pattern to optimize the time for aerosol delivery [222]. This decreases the amount of drug wasted during exhalation, which is a problem with jet nebulizers due to their continued flow, and reduces variations in lung deposition [221]. Some of the latest VMD with AAD includes the I-neb AAD System [223], the AKITA® system [224] and Pulmonary Drug Delivery Systems [178]. These devices improve the overall deposition efficiency from ~60% with conventional VMNs to north of 70% [225].
Few inhalable monoclonal antibodies have been tested or are being tested in clinical trials [199]; however, evidence of successful antibody nebulization by VMN is available from various preclinical studies. For instance, an anti-IL13 neutralizing antibody was nebulized with eFlow® vibrating mesh nebulizer. It retained high affinity and potency following nebulization, and no aggregation or degradation was observed [226]. The antibody was tested in a model of chronic asthma in cynomolgus macaques, where it showed it can inhibit lung inflammation due to allergen exposure [227]. Nebulization and deposition of IgGs, IgAs, and IgMs into the lungs of rats and non-human primates by eFlow® mesh nebulizer was examined by Vonarburg [192]. For all their Ig formulations, >69% of their particles had a diameter smaller than 5 μm. They also observed that IgG, IgA, and IgM maintain their structural integrity, did not induce aggregation vs. unnebulized controls, and retain their activity after nebulization, as shown in Table 2 . This study demonstrated that even large molecules such as IgM (970 kDa) can be nebulized by VMN. Respaud et al effectively nebulized two mAbs, an IgG1 and an IgG4, using Omron, PARI eFlow®, and Aerogen Solo mesh nebulizers [228]. Protein formulation, such as the presence of surfactants and protein concentration, can be optimized to further reduce mAb aggregation.
Table 2.
Physical characterization of immunoglobulin aerosol generated with a vibrating mesh nebulizer. Ig formulations were analyzed by size exclusion chromatography after nebulization. Table adapted from [192].
| Formulation | Condition | High molecular weight species (%) | Monomers and Dimers (%) | Fragments (%) | Respirable Fraction <5 μm (%) |
|---|---|---|---|---|---|
| IgG 10% | Non-nebulized Nebulized |
<1 <1 |
>98 >98 |
<1 <1 |
76.87 ± 2.04 |
| IgG 5% | Non-nebulized Nebulized |
<1 <1 |
>98 >98 |
1 <1 |
69.68 ± 2.33 |
| IgA 5% | Non-nebulized Nebulized |
23 23 |
74 74 |
3 3 |
73.13 ± 2.97 |
| IgAM 5% | Non-nebulized Nebulized |
62 60 |
34 35 |
4 5 |
76.46 ± 3.24 |
Finally, studies of the nebulized delivery of a variety of biologics have generally shown them to be exceptionally safe. No serious adverse effects were observed in clinical trials testing nebulized delivery of Omalizumab [193], Pulmozyme® [229], DAS-181 [99], SNG001 (interpheron beta-1a) [230], Alpha-1 antitrypsin [231], Molgramostim [232], or the ALX-0171 nanobody (including in adults with hypersensitive airways [219]). Both inhalable DAS181 (NCT04324489) and SNG001 (NCT04385095) are currently being evaluated in phase I/II clinical trials for COVID-19. These results underscore that inhalation of human mAb using VMNs is a potentially safe and efficacious route of delivering mAbs into the lung.
7. Conclusion and perspectives
ARIs have long posed a significant public and global health challenge. While the COVID-19 pandemic has brought about sufferings on a societal and economic scale rarely seen in the past, it has also catalyzed efforts across the entire medical and scientific spectrum in a race against time to advance effective interventions. With the integration of the latest advances in mAb discovery, large scale mAb manufacturing and drug delivery technologies, we believe all the tools are in place to enable mAbs to serve as an effective therapeutic intervention for COVID-19, with particularly efficient opportunities in pulmonary delivery, harnessing the mucosal effector functions of mAbs. The lessons learned from the intensive development of mAbs for SARS-CoV-2 will likely provide an important platform for the scientific community to better develop alternative interventions to address the diverse array of others ARIs that to this day still lack effective vaccines and treatment.
Competing interests
S.K.L is founder of Mucommune, LLC and currently serves as its interim CEO. S.K.L is also founder of Inhalon Biopharma, Inc., and currently serves as its CSO, Board of Director, and Scientific Advisory Board. S.K.L has equity interests in both Mucommune and Inhalon Biopharma; S.K.L's relationships with Mucommune and Inhalon are subject to certain restrictions under University policy. The terms of these arrangements are managed by UNC-CH in accordance with its conflict of interest policies. M.M. has equity interests in Inhalon Biopharma.
Acknowledgements
This work was financially supported by the Eshelman Institute of Innovation (S.K.L.); The David and Lucile Packard Foundation (2013-39274; S.K.L); National Center for Advancing Translational Sciences (NCATS), National Institutes of Health, through Grant Award Number UL1TR002489; National Institutes of Health under grants R43AI155185 (M.M.), R44AI138728 (M.M.), R44AI141054 (M.M.), and R43AI149894-01A1 (S.K.L, M.M. and RP); and National Science Foundation (DMS-2028758; S.K.L.). This project was also supported by the North Carolina Policy Collaboratory at the University of North Carolina at Chapel Hill with funding from the North Carolina Coronavirus Relief Fund established and appropriated by the North Carolina General Assembly.The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH and other funders.
References
- 1.Cohen J. Why Flu Vaccines Don't Protect People for Long. 2020. https://www.sciencemag.org/news/2020/08/why-flu-vaccines-don-t-protect-people-long
- 2.CDC Vaccine Effectiveness: How Well Do the Flu Vaccines Work? 2020. https://www.cdc.gov/flu/vaccines-work/vaccineeffect.htm
- 3.Wright P.F., Karron R.A., Belshe R.B., Thompson J., Crowe J.E., Jr., Boyce T.G., Halburnt L.L., Reed G.W., Whitehead S.S., Anderson E.L., Wittek A.E., Casey R., Eichelberger M., Thumar B., Randolph V.B., Udem S.A., Chanock R.M., Murphy B.R. Evaluation of a live, cold-passaged, temperature-sensitive, respiratory syncytial virus vaccine candidate in infancy. J. Infect. Dis. 2000;182:1331–1342. doi: 10.1086/315859. [DOI] [PubMed] [Google Scholar]
- 4.Brandenburg A.H., Groen J., van Steensel-Moll H.A., Claas E.C., Rothbarth P.H., Neijens H.J., Osterhaus A.D. Respiratory syncytial virus specific serum antibodies in infants under six months of age: limited serological response upon infection. J. Med. Virol. 1997;52:97–104. doi: 10.1002/(sici)1096-9071(199705)52:1<97::aid-jmv16>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 5.Ray G.T., Lewis N., Klein N.P., Daley M.F., Wang S.V., Kulldorff M., Fireman B. Intraseason waning of influenza vaccine effectiveness. Clin. Infect. Dis. 2019;68:1623–1630. doi: 10.1093/cid/ciy770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis C.W., Jackson K.J.L., McCausland M.M., Darce J., Chang C., Linderman S.L., Chennareddy C., Gerkin R., Brown S.J., Wrammert J., Mehta A.K., Cheung W.C., Boyd S.D., Waller E.K., Ahmed R. Influenza vaccine–induced human bone marrow plasma cells decline within a year after vaccination. Science. 2020;370:237. doi: 10.1126/science.aaz8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knudsen L., Ochs M. The micromechanics of lung alveoli: structure and function of surfactant and tissue components. Histochem. Cell Biol. 2018;150:661–676. doi: 10.1007/s00418-018-1747-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y., Tang Z., Huang H., Li J., Wang Z., Yu Y., Zhang C., Li J., Dai H., Wang F., Cai T., Tang N. Pulmonary alveolar type I cell population consists of two distinct subtypes that differ in cell fate. Proc. Natl. Acad. Sci. 2018;115:2407. doi: 10.1073/pnas.1719474115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamaya M., Finkbeiner W.E., Chun S.Y., Widdicombe J.H. Differentiated structure and function of cultures from human tracheal epithelium. Am. J. Phys. 1992;262:L713–L724. doi: 10.1152/ajplung.1992.262.6.L713. [DOI] [PubMed] [Google Scholar]
- 10.Whitcutt M.J., Adler K.B., Wu R. A biphasic chamber system for maintaining polarity of differentiation of cultured respiratory tract epithelial cells. In Vitro Cell. Develop. Biol. 1988;24:420–428. doi: 10.1007/BF02628493. [DOI] [PubMed] [Google Scholar]
- 11.Pickles R.J. Human airway epithelial cell cultures for modeling respiratory syncytial virus infection. Curr. Top. Microbiol. Immunol. 2013;372:371–387. doi: 10.1007/978-3-642-38919-1_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cifuentes-Muñoz N., Dutch R.E., Cattaneo R. Direct cell-to-cell transmission of respiratory viruses: The fast lanes. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Momose F., Sekimoto T., Ohkura T., Jo S., Kawaguchi A., Nagata K., Morikawa Y. Apical transport of influenza A virus ribonucleoprotein requires Rab11-positive recycling endosome. PLoS One. 2011;6 doi: 10.1371/journal.pone.0021123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matlin K.S., Reggio H., Helenius A., Simons K. Infectious entry pathway of influenza virus in a canine kidney cell line. J. Cell Biol. 1981;91:601–613. doi: 10.1083/jcb.91.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson C.I., Barclay W.S., Zambon M.C., Pickles R.J. Infection of human airway epithelium by human and avian strains of influenza a virus. J. Virol. 2006;80:8060–8068. doi: 10.1128/JVI.00384-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodriguez Boulan E., Sabatini D.D. Asymmetric budding of viruses in epithelial monlayers: a model system for study of epithelial polarity. Proc. Natl. Acad. Sci. U. S. A. 1978;75:5071–5075. doi: 10.1073/pnas.75.10.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bruce E.A., Digard P., Stuart A.D. The Rab11 pathway is required for influenza A virus budding and filament formation. J. Virol. 2010;84:5848. doi: 10.1128/JVI.00307-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barman S., Adhikary L., Chakrabarti A.K., Bernas C., Kawaoka Y., Nayak D.P. Role of transmembrane domain and cytoplasmic tail amino acid sequences of influenza a virus neuraminidase in raft association and virus budding. J. Virol. 2004;78:5258–5269. doi: 10.1128/JVI.78.10.5258-5269.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts S.R., Compans R.W., Wertz G.W. Respiratory syncytial virus matures at the apical surfaces of polarized epithelial cells. J. Virol. 1995;69:2667–2673. doi: 10.1128/jvi.69.4.2667-2673.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brock S.C., Goldenring J.R., Crowe J.E., Jr. Apical recycling systems regulate directional budding of respiratory syncytial virus from polarized epithelial cells. Proc. Natl. Acad. Sci. U. S. A. 2003;100:15143–15148. doi: 10.1073/pnas.2434327100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mellow T.E., Murphy P.C., Carson J.L., Noah T.L., Zhang L., Pickles R.J. The effect of respiratory synctial virus on chemokine release by differentiated airway epithelium. Exp. Lung Res. 2004;30:43–57. doi: 10.1080/01902140490252812. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L., Peeples M.E., Boucher R.C., Collins P.L., Pickles R.J. Respiratory syncytial virus infection of human airway epithelial cells is polarized, specific to ciliated cells, and without obvious cytopathology. J. Virol. 2002;76:5654–5666. doi: 10.1128/JVI.76.11.5654-5666.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wright P.F., Ikizler M.R., Gonzales R.A., Carroll K.N., Johnson J.E., Werkhaven J.A. Growth of respiratory syncytial virus in primary epithelial cells from the human respiratory tract. J. Virol. 2005;79:8651. doi: 10.1128/JVI.79.13.8651-8654.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L., Bukreyev A., Thompson C.I., Watson B., Peeples M.E., Collins P.L., Pickles R.J. Infection of ciliated cells by human parainfluenza virus type 3 in an in vitro model of human airway epithelium. J. Virol. 2005;79:1113–1124. doi: 10.1128/JVI.79.2.1113-1124.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pyrc K., Sims A.C., Dijkman R., Jebbink M., Long C., Deming D., Donaldson E., Vabret A., Baric R., van der Hoek L., Pickles R. Culturing the unculturable: Human coronavirus HKU1 infects, replicates, and produces progeny virions in human ciliated airway epithelial cell cultures. J. Virol. 2010;84:11255. doi: 10.1128/JVI.00947-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sims A.C., Baric R.S., Yount B., Burkett S.E., Collins P.L., Pickles R.J. Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs. J. Virol. 2005;79:15511–15524. doi: 10.1128/JVI.79.24.15511-15524.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milewska A., Kula-Pacurar A., Wadas J., Suder A., Szczepanski A., Dabrowska A., Owczarek K., Ochman M., Stacel T., Rajfur Z., Labaj P., Branicki W., Pyrc K. Replication of SARS-CoV-2 in human respiratory epithelium. bioRxiv. 2020 doi: 10.1101/2020.03.20.999029. (2020.2003.2020.999029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jia H.P., Look D.C., Shi L., Hickey M., Pewe L., Netland J., Farzan M., Wohlford-Lenane C., Perlman S., McCray P.B., Jr. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J. Virol. 2005;79:14614–14621. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia H.P., Look D.C., Shi L., Hickey M., Pewe L., Netland J., Farzan M., Wohlford-Lenane C., Perlman S., McCray P.B., Jr. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J. Virol. 2005;79:14614–14621. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ortiz Bezara M.E., Thurman A., Pezzulo A.A., Leidinger M.R., Klesney-Tait J.A., Karp P.H., Tan P., Wohlford-Lenane C., McCray P.B., Meyerholz D.K. Heterogeneous expression of the SARS-Coronavirus-2 receptor ACE2 in the human respiratory tract. bioRxiv. 2020 doi: 10.1101/2020.04.22.056127. (2020.2004.2022.056127) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poritz M.A., Blaschke A.J., Byington C.L., Meyers L., Nilsson K., Jones D.E., Thatcher S.A., Robbins T., Lingenfelter B., Amiott E., Herbener A., Daly J., Dobrowolski S.F., Teng D.H.F., Ririe K.M. FilmArray, an automated nested multiplex PCR system for multi-pathogen detection: development and application to respiratory tract infection. PLoS One. 2011;6 doi: 10.1371/journal.pone.0026047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanson K.E., Couturier M.R. Multiplexed molecular diagnostics for respiratory, gastrointestinal, and central nervous system infections. Clin. Infect. Dis. 2016;63:1361–1367. doi: 10.1093/cid/ciw494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stramer S.L., Collins C., Nugent T., Wang X., Fuschino M., Heitman J.W., Law J., Krysztof D.E., Kiely N., Todd D., Vermeulen N.M., Harrington K., Kamel H., Kelvin D.J., Busch M.P., George K. St, Hewlett I.K., Linnen J.M., Norris P.J. Sensitive detection assays for influenza RNA do not reveal viremia in US blood donors. J. Infect. Dis. 2012;205:886–894. doi: 10.1093/infdis/jir863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Domachowske J.B., Rosenberg H.F. Respiratory syncytial virus infection: immune response, immunopathogenesis, and treatment. Clin. Microbiol. Rev. 1999;12:298–309. doi: 10.1128/cmr.12.2.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schuster J.E., Williams J.V. Human metapneumovirus. Pediatr. Rev. 2013;34:558–565. doi: 10.1542/pir.34-12-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang B., Zhou X., Zhu C., Feng F., Qiu Y., Feng J., Jia Q., Song Q., Zhu B., Wang J. Immune phenotyping based on neutrophil-to-lymphocyte ratio and IgG predicts disease severity and outcome for patients with COVID-19. medRxiv. 2020 doi: 10.1101/2020.03.12.20035048. (2020.2003.2012.20035048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mason R.J. Pathogenesis of COVID-19 from a cell biology perspective. Eur. Respir. J. 2020;55 doi: 10.1183/13993003.00607-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tse H., To K.K.W., Wen X., Chen H., Chan K.-H., Tsoi H.-W., Li I.W.S., Yuen K.-Y. Clinical and virological factors associated with viremia in pandemic influenza A/H1N1/2009 virus infection. PLoS One. 2011;6 doi: 10.1371/journal.pone.0022534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morbidity and Mortality Weekly Report. Vol. 52. 2003. Preliminary clinical description of severe acute respiratory syndrome, MMWR; pp. 255–256. [PubMed] [Google Scholar]
- 40.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. Jama. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang X., Yu Y., Xu J., Shu H., Xia J.A., Liu H., Wu Y., Zhang L., Yu Z., Fang M., Yu T., Wang Y., Pan S., Zou X., Yuan S., Shang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Backer J.A., Klinkenberg D., Wallinga J. Incubation period of 2019 novel coronavirus (2019-nCoV) infections among travellers from Wuhan, China, 20–28 January 2020. Eur. Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.5.2000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McBride J.M., Lim J.J., Burgess T., Deng R., Derby M.A., Maia M., Horn P., Siddiqui O., Sheinson D., Chen-Harris H., Newton E.M., Fillos D., Nazzal D., Rosenberger C.M., Ohlson M.B., Lambkin-Williams R., Fathi H., Harris J.M., Tavel J.A. Phase 2 randomized trial of the safety and efficacy of MHAA4549A, a broadly neutralizing monoclonal antibody, in a human influenza a virus challenge model. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.01154-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goodman M.R., Link D.W., Brown W.R., Nakane P.K. Ultrastructural evidence of transport of secretory IgA across bronchial epithelium. Am. Rev. Respir. Dis. 1981;123:115–119. doi: 10.1164/arrd.1981.123.1.115. [DOI] [PubMed] [Google Scholar]
- 46.Mostov K.E., Blobel G. A transmembrane precursor of secretory component. The receptor for transcellular transport of polymeric immunoglobulins. J. Biol. Chem. 1982;257:11816–11821. [PubMed] [Google Scholar]
- 47.Newkirk M.M., Klein M.H., Katz A., Fisher M.M., Underdown B.J. Estimation of polymeric IgA in human serum: an assay based on binding of radiolabeled human secretory component with applications in the study of IgA nephropathy, IgA monoclonal gammopathy, and liver disease. J. Immunol. 1983;130:1176–1181. [PubMed] [Google Scholar]
- 48.Stockley R.A., Afford S.C., Burnett D. Assessment of 7S and 11S immunoglobulin A in sputum. Am. Rev. Respir. Dis. 1980;122:959–964. doi: 10.1164/arrd.1980.122.6.959. [DOI] [PubMed] [Google Scholar]
- 49.Igarashi Y., Skoner D.P., Doyle W.J., White M.V., Fireman P., Kaliner M.A. Analysis of nasal secretions during experimental rhinovirus upper respiratory infections. J. Allergy Clin. Immunol. 1993;92:722–731. doi: 10.1016/0091-6749(93)90016-9. [DOI] [PubMed] [Google Scholar]
- 50.Wang Y.-Y., Harit D., Subramani D.B., Arora H., Kumar P.A., Lai Samuel K. Influenza-binding antibodies immobilise influenza viruses in fresh human airway mucus. Eur. Respir. J. 2017;49 doi: 10.1183/13993003.01709-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deuschl H., Johansson S.G. Immunoglobulins in tracheo-bronchial secretion with special reference to IgE. Clin. Exp. Immunol. 1974;16:401–412. [PMC free article] [PubMed] [Google Scholar]
- 52.Nahm D.H., Kim H.Y., Park H.S. Elevation of specific immunoglobulin A antibodies to both allergen and bacterial antigen in induced sputum from asthmatics. Eur. Respir. J. 1998;12:540–545. doi: 10.1183/09031936.98.12030540. [DOI] [PubMed] [Google Scholar]
- 53.Reynolds H.Y. Immunoglobulin G and Its function in the human respiratory tract*. Mayo Clin. Proc. 1988;63:161–174. doi: 10.1016/s0025-6196(12)64949-0. [DOI] [PubMed] [Google Scholar]
- 54.Burnett D. Immunoglobulins in the lung. Thorax. 1986;41:337–344. doi: 10.1136/thx.41.5.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Delacroix D.L., Dive C., Rambaud J.C., Vaerman J.P. IgA subclasses in various secretions and in serum. Immunology. 1982;47:383–385. [PMC free article] [PubMed] [Google Scholar]
- 56.Soutar C.A. Distribution of plasma cells and other cells containing immunoglobulin in the respiratory tract in chronic bronchitis. Thorax. 1977;32:387–396. doi: 10.1136/thx.32.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nijhuis-Heddes J.M., Lindeman J., Otto A.J., Snieders M.W., Kievit-Tyson P.A., Dijkman J.H. Distribution of immunoglobulin-containing cells in the bronchial mucosa of patients with chronic respiratory disease. Eur. J. Respir. Dis. 1982;63:249–256. [PubMed] [Google Scholar]
- 58.Hill S.L., Mitchell J.L., Burnett D., Stockley R.A. IgG subclasses in the serum and sputum from patients with bronchiectasis. Thorax. 1998;53:463–468. doi: 10.1136/thx.53.6.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burton D.R., Poignard P., Stanfield R.L., Wilson I.A. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science. 2012;337:183–186. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murin C.D., Wilson I.A., Ward A.B. Antibody responses to viral infections: a structural perspective across three different enveloped viruses. Nat. Microbiol. 2019;4:734–747. doi: 10.1038/s41564-019-0392-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Plotkin S.A. Vaccines: correlates of vaccine-induced immunity. Clin. Infect. Dis. 2008;47:401–409. doi: 10.1086/589862. [DOI] [PubMed] [Google Scholar]
- 62.Pelegrin M., Naranjo-Gomez M., Piechaczyk M. Antiviral monoclonal antibodies: can they be more than simple neutralizing agents? Trends Microbiol. 2015;23:653–665. doi: 10.1016/j.tim.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dunkelberger J.R., Song W.-C. Complement and its role in innate and adaptive immune responses. Cell Res. 2010;20:34–50. doi: 10.1038/cr.2009.139. [DOI] [PubMed] [Google Scholar]
- 64.Blue C.E., Spiller O.B., Blackbourn D.J. The relevance of complement to virus biology. Virology. 2004;319:176–184. doi: 10.1016/j.virol.2003.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Binley J.M., Clas B., Gettie A., Vesanen M., Montefiori D.C., Sawyer L., Booth J., Lewis M., Marx P.A., Bonhoeffer S., Moore J.P. Passive infusion of immune serum into simian immunodeficiency virus-infected rhesus macaques undergoing a rapid disease course has minimal effect on plasma viremia. Virology. 2000;270:237–249. doi: 10.1006/viro.2000.0254. [DOI] [PubMed] [Google Scholar]
- 66.Garagiola D.M., Huard T.K., LoBuglio A.F. Comparison of monocyte and alveolar macrophage antibody-dependent cellular cytotoxicity and Fc-receptor activity. Cell. Immunol. 1981;64:359–370. doi: 10.1016/0008-8749(81)90487-1. [DOI] [PubMed] [Google Scholar]
- 67.Naegel G.P., Young K.R., Reynolds H.Y. Receptors for human IgG subclasses on human alveolar macrophages. Am. Rev. Respir. Dis. 1984;129:413–418. doi: 10.1164/arrd.1984.129.3.413. [DOI] [PubMed] [Google Scholar]
- 68.Lin P.M., Wright J.R. AJP: Lung Cellular and Molecular Physiology. Vol. 291. 2006. Surfactant protein A binds to IgG and enhances phagocytosis of IgG-opsonized erythrocytes; pp. L1199–L1206. [DOI] [PubMed] [Google Scholar]
- 69.Wang Y.-Y., Kannan A., Nunn K.L., Murphy M.A., Subramani D.B., Moench T., Cone R., Lai S.K. IgG in cervicovaginal mucus traps HSV and prevents vaginal Herpes infections. Mucosal Immunol. 2014;7:1036–1044. doi: 10.1038/mi.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Henry C.E., Wang Y.Y., Yang Q., Hoang T., Chattopadhyay S., Hoen T., Ensign L.M., Nunn K.L., Schroeder H., McCallen J., Moench T., Cone R., Roffler S.R., Lai S.K. Anti-PEG antibodies alter the mobility and biodistribution of densely PEGylated nanoparticles in mucus. Acta Biomater. 2016;43:61–70. doi: 10.1016/j.actbio.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang B., Schaefer A., Wang Y.-Y., McCallen J., Lee P., Newby J.M., Arora H., Kumar P.A., Zeitlin L., Whaley K.J., McKinley S.A., Fischer W.A., II, Harit D., Lai S.K. ZMapp reinforces the airway mucosal barrier against ebola virus. J. Infect. Dis. 2018;218:901–910. doi: 10.1093/infdis/jiy230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Newby J., Schiller J.L., Wessler T., Edelstein J., Forest M.G., Lai S.K. A blueprint for robust crosslinking of mobile species in biogels with weakly adhesive molecular anchors. Nat. Commun. 2017;8 doi: 10.1038/s41467-017-00739-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schroeder H.A., Newby J., Schaefer A., Subramani B., Tubbs A., Gregory Forest M., Miao E., Lai S.K. LPS-binding IgG arrests actively motile Salmonella Typhimurium in gastrointestinal mucus. Mucosal Immunol. 2020;13:814–823. doi: 10.1038/s41385-020-0267-9. [DOI] [PubMed] [Google Scholar]
- 74.Schroeder H.A., Nunn K.L., Schaefer A., Henry C.E., Lam F., Pauly M.H., Whaley K.J., Zeitlin L., Humphrys M.S., Ravel J., Lai S.K. Herpes simplex virus-binding IgG traps HSV in human cervicovaginal mucus across the menstrual cycle and diverse vaginal microbial composition. Mucosal Immunol. 2018;11:1477–1486. doi: 10.1038/s41385-018-0054-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Olmsted S.S., Padgett J.L., Yudin A.I., Whaley K.J., Moench T.R., Cone R.A. Diffusion of macromolecules and virus-like particles in human cervical mucus. Biophys. J. 2001;81:1930–1937. doi: 10.1016/S0006-3495(01)75844-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Saltzman W.M., Radomsky M.L., Whaley K.J., Cone R.A. Antibody diffusion in human cervical mucus. Biophys. J. 1994;66:508–515. doi: 10.1016/s0006-3495(94)80802-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang Y.-Y., Harit D., Subramani D.B., Arora H., Kumar P.A., Lai S.K. Influenza-binding antibodies immobilise influenza viruses in fresh human airway mucus. Eur. Respir. J. 2017;49 doi: 10.1183/13993003.01709-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tortorici M.A., Veesler D. Structural insights into coronavirus entry. Adv. Virus Res. 2019;105:93–116. doi: 10.1016/bs.aivir.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Walls A.C., Tortorici M.A., Snijder J., Xiong X., Bosch B.J., Rey F.A., Veesler D. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc. Natl. Acad. Sci. U. S. A. 2017;114:11157–11162. doi: 10.1073/pnas.1708727114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Neuman B.W., Adair B.D., Yoshioka C., Quispe J.D., Orca G., Kuhn P., Milligan R.A., Yeager M., Buchmeier M.J. Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. J. Virol. 2006;80:7918–7928. doi: 10.1128/JVI.00645-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (New York, N.Y.) 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., Wang Q., Zhou H., Yan J., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. (e899) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. (e278) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. (e286) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cai Y., Zhang J., Xiao T., Peng H., Sterling S.M., Walsh R.M., Rawson S., Rits-Volloch S., Chen B. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;1592 doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Benton D.J., Wrobel A.G., Xu P., Roustan C., Martin S.R., Rosenthal P.B., Skehel J.J., Gamblin S.J. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature. 2020 doi: 10.1038/s41586-020-2772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wrobel A.G., Benton D.J., Xu P., Roustan C., Martin S.R., Rosenthal P.B., Skehel J.J., Gamblin S.J. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat. Struct. Mol. Biol. 2020;27:763–767. doi: 10.1038/s41594-020-0468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 91.Song W., Gui M., Wang X., Xiang Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14:1–19. doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Walls A.C., Xiong X., Park Y.J., Tortorici M.A., Snijder J., Quispe J., Cameroni E., Gopal R., Dai M., Lanzavecchia A., Zambon M., Rey F.A., Corti D., Veesler D. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell. 2019;176:1026–1039. doi: 10.1016/j.cell.2018.12.028. (e1015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gui M., Song W., Zhou H., Xu J., Chen S., Xiang Y., Wang X. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27:119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kirchdoerfer R.N., Wang N., Pallesen J., Wrapp D., Turner H.L., Cottrell C.A., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci. Rep. 2018;8:1–11. doi: 10.1038/s41598-018-34171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Roy S., Jaiswar A., Sarkar R. Dynamic asymmetry exposes 2019-nCoV prefusion spike. J. Phys. Chem. Lett. 2020;11:7021–7027. doi: 10.1021/acs.jpclett.0c01431. [DOI] [PubMed] [Google Scholar]
- 96.Yuan M., Wu N.C., Zhu X., Lee C.C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y., Ying T. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microb. Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., Peter A., Guarino B., Spreafico R., Cameroni E., Case J.B., Chen R.E., Havenar-Daughton C., Snell G., Telenti A., Virgin H.W., Lanzavecchia A., Diamond M.S., Fink K., Veesler D., Corti D. Structural and functional analysis of a potent sarbecovirus neutralizing antibody. bioRxiv. 2020;45 (S-102) [Google Scholar]
- 99.Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., Hägglöf T., Oliveira T.Y., Viant C., Hurley A., Hoffmann H.H., Millard K.G., Kost R.G., Cipolla M., Gordon K., Bianchini F., Chen S.T., Ramos V., Patel R., Dizon J., Shimeliovich I., Mendoza P., Hartweger H., Nogueira L., Pack M., Horowitz J., Schmidt F., Weisblum Y., Michailidis E., Ashbrook A.W., Waltari E., Pak J.E., Huey-Tubman K.E., Koranda N., Hoffman P.R., West A.P., Rice C.M., Hatziioannou T., Bjorkman P.J., Bieniasz P.D., Caskey M., Nussenzweig M.C. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S., Sutton R.E., Suryadevara N., Martinez D.R., Williamson L.E., Chen E.C., Jones T., Day S., Myers L., Hassan A.O., Kafai N.M., Winkler E.S., Fox J.M., Shrihari S., Mueller B.K., Meiler J., Chandrashekar A., Mercado N.B., Steinhardt J.J., Ren K., Loo Y.M., Kallewaard N.L., McCune B.T., Keeler S.P., Holtzman M.J., Barouch D.H., Gralinski L.E., Baric R.S., Thackray L.B., Diamond M.S., Carnahan R.H., Crowe J.E. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Andreano E., Nicastri E., Paciello I., Pileri P., Manganaro N., Piccini G., Manenti A., Pantano E., Kabanova A., Troisi M., Vacca F., Cardamone D., Santi C.D., Agrati C., Capobianchi M.R., Castilletti C., Emiliozzi A., Fabbiani M., Montagnani F., Montomoli E., Sala C., Ippolito G., Rappuoli R. Identification of neutralizing human monoclonal antibodies from Italian Covid-19 convalescent patients. bioRxiv. 2020 doi: 10.1101/2020.05.05.078154. (1=29–21=29) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., Bentlage A.E.H., van Haaren M.M., Guerra D., Burger J.A., Schermer E.E., Verheul K.D., van der Velde N., van der Kooi A., van Schooten J., van Breemen M.J., Bijl T.P.L., Sliepen K., Aartse A., Derking R., Bontjer I., Kootstra N.A., Wiersinga W.J., Vidarsson G., Haagmans B.L., Ward A.B., de Bree G.J., Sanders R.W., van Gils M.J. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., Chai X., He R., Li X., Lv Q., Zhu H., Deng W., Xu Y., Wang Y., Qiao L., Tan Y., Song L., Wang G., Du X., Gao N., Liu J., Xiao J., Su X.-D., Du Z., Feng Y., Qin C., Qin C., Jin R., Xie X.S. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell. 2020;182:73–84. doi: 10.1016/j.cell.2020.05.025. (e16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen X., Li R., Pan Z., Qian C., Yang Y., You R., Zhao J., Liu P., Gao L., Li Z., Huang Q., Xu L., Tang J., Tian Q., Yao W., Hu L., Yan X., Zhou X., Wu Y., Deng K., Zhang Z., Qian Z., Chen Y., Ye L. Human monoclonal antibodies block the binding of SARS-CoV-2 spike protein to angiotensin converting enzyme 2 receptor. Cell. Mol. Immunol. 2020;17:647–649. doi: 10.1038/s41423-020-0426-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., Chung K.M., Hermann A., Ullman E., Cruz J., Rafique A., Huang T., Fairhurst J., Libertiny C., Malbec M., Lee W.-Y., Welsh R., Farr G., Pennington S., Deshpande D., Cheng J., Watty A., Bouffard P., Babb R., Levenkova N., Chen C., Zhang B., Hernandez A. Romero, Saotome K., Zhou Y., Franklin M., Sivapalasingam S., Lye D.C., Weston S., Logue J., Haupt R., Frieman M., Chen G., Olson W., Murphy A.J., Stahl N., Yancopoulos G.D., Kyratsous C.A. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kreer C., Zehner M., Weber T., Ercanoglu M.S., Gieselmann L., Rohde C., Halwe S., Korenkov M., Schommers P., Vanshylla K., Di Cristanziano V., Janicki H., Brinker R., Ashurov A., Krähling V., Kupke A., Cohen-Dvashi H., Koch M., Eckert J.M., Lederer S., Pfeifer N., Wolf T., Vehreschild M.J.G.T., Wendtner C., Diskin R., Gruell H., Becker S., Klein F. Longitudinal isolation of potent near-germline SARS-CoV-2-neutralizing antibodies from COVID-19 patients. Cell. 2020;182:843–854. doi: 10.1016/j.cell.2020.06.044. (e812) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.-T., Limbo O., Smith C., Song G., Woehl J., Yang L., Abbott R.K., Callaghan S., Garcia E., Hurtado J., Parren M., Peng L., Ramirez S., Ricketts J., Ricciardi M.J., Rawlings S.A., Wu N.C., Yuan M., Smith D.M., Nemazee D., Teijaro J.R., Voss J.E., Wilson I.A., Andrabi R., Briney B., Landais E., Sok D., Jardine J.G., Burton D.R. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;963 doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Seydoux E., Homad L.J., MacCamy A.J., Parks K.R., Hurlburt N.K., Jennewein M.F., Akins N.R., Stuart A.B., Wan Y.-H., Feng J., Nelson R.E., Singh S., Cohen K.W., McElrath M.J., Englund J.A., Chu H.Y., Pancera M., McGuire A.T., Stamatatos L. Characterization of neutralizing antibodies from a SARS-CoV-2 infected individual. bioRxiv. 2020 doi: 10.1101/2020.05.12.091298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lv Z., Deng Y.-Q., Ye Q., Cao L., Sun C.-Y., Fan C., Huang W., Sun S., Sun Y., Zhu L., Chen Q., Wang N., Nie J., Cui Z., Zhu D., Shaw N., Li X.-F., Li Q., Xie L., Wang Y., Rao Z., Qin C.-F., Wang X. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science. 2020;5881 doi: 10.1126/science.abc5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang C., Li W., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A.D.M.E., van Kuppeveld F.J.M., Haagmans B.L., Grosveld F., Bosch B.J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:1–6. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wu F., Wang A., Liu M., Wang Q., Chen J., Xia S., Ling Y., Zhang Y., Xun J., Lu L., Jiang S., Lu H., Wen Y., Huang J. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. medRxiv. 2020 doi: 10.1101/2020.03.30.20047365. [DOI] [Google Scholar]
- 112.Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., Gao G., Hu X., Zhang Y., Tong Z., Huang W., Liu W.J., Wu G., Zhang B., Wang L., Qi J., Feng H., Wang F.S., Wang Q., Gao G.F., Yuan Z., Yan J. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 113.Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., Tang X., Yu J., Lan J., Yuan J., Wang H., Zhao J., Zhang S., Wang Y., Shi X., Liu L., Zhao J., Wang X., Zhang Z., Zhang L. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 114.Wec A.Z., Wrapp D., Herbert A.S., Maurer D.P., Haslwanter D., Sakharkar M., Jangra R.K., Dieterle M. Eugenia, Lilov A., Huang D., Tse L.V., Johnson N.V., Hsieh C.L., Wang N., Nett J.H., Champney E., Burnina I., Brown M., Lin S., Sinclair M., Johnson C., Pudi S., Bortz R., Wirchnianski A.S., Laudermilch E., Florez C., Fels J. Maximilian, O'Brien C.M., Graham B.S., Nemazee D., Burton D.R., Baric R.S., Voss J.E., Chandran K., Dye J.M., McLellan J.S., Walker L.M. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Byrnes J.R., Zhou X.X., Lui I., Elledge S.K., Glasgow J.E., Lim S.A., Loudermilk R.P., Chiu C.Y., Wang T.T., Wilson M.R., Leung K.K., Wells J.A. Competitive SARS-CoV-2 serology reveals most antibodies targeting the spike receptor-binding domain compete for ACE2 binding. mSphere. 2020;5:1–11. doi: 10.1128/mSphere.00802-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Soo-Young L., Cheolmin K., Dong-Kyun R., Jihun L., Young-Il K., Ji-Min S., Yeon-Gil K., Jae-Hee J., Minsoo K., Jong-In K., Pankyeom K., Soo B. Jin, Yeong S. Eun, Seob L. Min, Su K. Man, Hanmi N., Geun-Soo P., Sang P. Jae, Dain S., Yongjin A., No L. Jeong, Ki-Sung K., Joo-Yeon L., Hansaem L., Jeong-Sun Y., Kyung-Chang K., Soon K. Sung, Hye-Min W., Jun-Won K., Man-Seong P., Kwang-Min Y., Se-Mi K., Eun-Ha K., Su-Jin P., Tae J. Seong, Ho Y. Chi, Youngjo S., Hun G. Se, Hanseul O., Bon-Sang K., Joo H. Jung, Choong-Min R., Beom P. Wan, Myoung-don O., Ki C. Young. A novel neutralizing antibody targeting receptor binding domain of SARS-CoV-2. Nat. Res. Forum. 2020 doi: 10.21203/rs.3.rs-59639/v1. [DOI] [Google Scholar]
- 117.Luchsinger L.L., Ransegnola B., Jin D., Muecksch F., Weisblum Y., Bao W., George P.J., Rodriguez M., Tricoche N., Schmidt F., Gao C., Jawahar S., Pal M., Schnall E., Zhang H., Strauss D., Yazdanbakhsh K., Hillyer C.D., Bieniasz P.D., Hatziioannou T. Serological assays estimate highly variable SARS-CoV-2 neutralizing antibody activity in recovered COVID19 patients. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.02005-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Davis C.W., Jackson K.J.L., McElroy A.K., Halfmann P., Huang J., Chennareddy C., Piper A.E., Leung Y., Albariño C.G., Crozier I., Ellebedy A.H., Sidney J., Sette A., Yu T., Nielsen S.C.A., Goff A.J., Spiropoulou C.F., Saphire E.O., Cavet G., Kawaoka Y., Mehta A.K., Glass P.J., Boyd S.D., Ahmed R. Longitudinal analysis of the human B cell response to ebola virus infection. Cell. 2019;177:1566–1582. doi: 10.1016/j.cell.2019.04.036. (e1517) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wec A.Z., Haslwanter D., Abdiche Y.N., Shehata L., Pedreño-Lopez N., Moyer C.L., Bornholdt Z.A., Lilov A., Nett J.H., Jangra R.K., Brown M., Watkins D.I., Ahlm C., Forsell M.N., Rey F.A., Barba-Spaeth G., Chandran K., Walker L.M. Longitudinal dynamics of the human B cell response to the yellow fever 17D vaccine. Proc. Natl. Acad. Sci. U. S. A. 2020;117:6675–6685. doi: 10.1073/pnas.1921388117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pascal K.E., Coleman C.M., Mujica A.O., Kamat V., Badithe A., Fairhurst J., Hunt C., Strein J., Berrebi A., Sisk J.M., Matthews K.L., Babb R., Chen G., Lai K.-M.V., Huang T.T., Olson W., Yancopoulos G.D., Stahl N., Frieman M.B., Kyratsous C.A. Pre- and postexposure efficacy of fully human antibodies against Spike protein in a novel humanized mouse model of MERS-CoV infection. Proc. Natl. Acad. Sci. 2015;112:8738. doi: 10.1073/pnas.1510830112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen W.C., Murawsky C.M. Strategies for generating diverse antibody repertoires using transgenic animals expressing human antibodies. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Muyldermans S. Nanobodies: Natural single-domain antibodies. Annu. Rev. Biochem. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 123.English H., Hong J., Ho M. Ancient species offers contemporary therapeutics: an update on shark VNAR single domain antibody sequences, phage libraries and potential clinical applications. Antibody Ther. 2020;3:1–9. doi: 10.1093/abt/tbaa001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jovčevska I., Muyldermans S. The therapeutic potential of nanobodies. BioDrugs. 2020;34:11–26. doi: 10.1007/s40259-019-00392-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Scully M., Cataland S.R., Peyvandi F., Coppo P., Knöl P., Kremer Hovinga J.A., Metjian A., De La Rubia J., Pavenski K., Callewaert F., Biswas D., De Winter H., Zeldin R.K. Caplacizumab treatment for acquired thrombotic thrombocytopenic purpura. N. Engl. J. Med. 2019;380:335–346. doi: 10.1056/NEJMoa1806311. [DOI] [PubMed] [Google Scholar]
- 126.Larios Mora A., Detalle L., Gallup J.M., Van Geelen A., Stohr T., Duprez L., Ackermann M.R. Delivery of ALX-0171 by inhalation greatly reduces respiratory syncytial virus disease in newborn lambs. mAbs. 2018;10:778–795. doi: 10.1080/19420862.2018.1470727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chi X., Liu X., Wang C., Zhang X., Li X., Hou J., Ren L., Jin Q., Wang J., Yang W. Humanized single domain antibodies neutralize SARS-CoV-2 by targeting the spike receptor binding domain. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-18387-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gai J., Ma L., Li G., Zhu M., Qiao P., Li X., Zhang H., Zhang Y., Chen Y., Gong R., Wan Y. A potent neutralizing nanobody against SARS-CoV-2 with inhaled delivery potential. bioRxiv. 2020 doi: 10.1101/2020.08.09.242867. (2020.2008.2009.242867–242020.242808.242809.242867) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Huo J., Le Bas A., Ruza R.R., Duyvesteyn H.M.E., Mikolajek H., Malinauskas T., Tan T.K., Rijal P., Dumoux M., Ward P.N., Ren J., Zhou D., Harrison P.J., Weckener M., Clare D.K., Vogirala V.K., Radecke J., Moynié L., Zhao Y., Gilbert-Jaramillo J., Knight M.L., Tree J.A., Buttigieg K.R., Coombes N., Elmore M.J., Carroll M.W., Carrique L., Shah P.N.M., James W., Townsend A.R., Stuart D.I., Owens R.J., Naismith J.H. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct. Mol. Biol. 2020;27:846–854. doi: 10.1038/s41594-020-0469-6. [DOI] [PubMed] [Google Scholar]
- 130.Schoof M., Faust B., Saunders R.A., Sangwan S., Hoppe N., Boone M., Bache Billesbølle C., Zimanyi M., Deshpande I., Liang J., Anand A.A., Dobzinski N., Shoshana Zha B., Barsi-Rhyne B., Belyy V., Barile-Hill A.W., Gupta S., Leon K., White K.M., Nock S., Liu Y. An ultra-high affinity synthetic nanobody blocks SARS-CoV-2 infection by locking Spike into an inactive conformation. bioRxiv. 2020 doi: 10.1101/2020.08.08.238469. (2020.2008.2008.238469–232020.238408.238408.238469) [DOI] [Google Scholar]
- 131.Wrapp D., De Vlieger D., Corbett K.S., Torres G.M., Wang N., Van Breedam W., Roose K., van Schie L., Hoffmann M., Pöhlmann S., Graham B.S., Callewaert N., Schepens B., Saelens X., McLellan J.S. Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell. 2020:1–12. doi: 10.1016/j.cell.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Xiang Y., Nambulli S., Xiao Z., Liu H., Sang Z., Duprex W.P., Schneidman-Duhovny D., Zhang C., Shi Y. Versatile, multivalent nanobody cocktails efficiently neutralize SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.08.24.264333. (2020.2008.2024.264333–262020.264308.264324.264333) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Linsky T.W., Vergara R., Codina N., On J.W. Nels, Walker M.J., Su W., Hsiang T.-Y., Esser-Nobis K., Yu K., Hou Y.J., Priya T., Mitsumoto M., Pong A., Lau U.Y., Mason M.L., Chen J., Chen A., Berrocal T., Peng H., Clairmont N.S., Castellanos J., Lin Y.-R., Josephson-Day A., Baric R., Walkey C.D., Swanson R., Jr M.G., Blancas-Mejia L.M., Yen H.-L., Silva D.-A. De novo design of ACE2 protein decoys to neutralize SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.08.03.231340. (2020.2008.2003.231340–232020.231308.231303.231340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cao L., Goreshnik I., Coventry B., Case J.B., Miller L., Kozodoy L., Chen R.E., Carter L., Walls A.C., Park Y.-J., Strauch E.-M., Stewart L., Diamond M.S., Veesler D., Baker D. De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science. 2020;21 doi: 10.1126/science.abd9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bracken C.J., Lim S.A., Solomon P., Rettko N.J., Nguyen D.P., Zha B.S., Schaefer K., Byrnes J.R., Zhou J., Lui I., Liu J., Pance K., Zhou X.X., Leung K.K., Wells J.A. Bi-paratopic and multivalent human VH domains neutralize SARS-CoV-2 by targeting distinct epitopes within the ACE2 binding interface of Spike. bioRxiv. 2020 doi: 10.1101/2020.08.08.242511. (2020.2008.2008.242511–242020.242508.242508.242511) [DOI] [Google Scholar]
- 136.Sun Z., Chen C., Li W., Martinez D.R., Drelich A., Baek D.S., Liu X., Mellors J.W., Tseng C.T., Baric R.S., Dimitrov D.S. Potent neutralization of SARS-CoV-2 by human antibody heavy-chain variable domains isolated from a large library with a new stable scaffold. mAbs. 2020;12 doi: 10.1080/19420862.2020.1778435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Y. Weisblum, F. Schmidt, F. Zhang, J. DaSilva, D. Poston, J.C.C. Lorenzi, F. Muecksch, M. Rutkowska, H.-H. Hoffmann, E. Michailidis, C. Gaebler, M. Agudelo, A. Cho, Z. Wang, A. Gazumyan, M. Cipolla, L. Luchsinger, C.D. Hillyer, M. Caskey, D.F. Robbiani, C.M. Rice, M.C. Nussenzweig, T. Hatziioannou, P.D. Bieniasz, Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants, bioRxiv, 53 (2020) 1689–1699. [DOI] [PMC free article] [PubMed]
- 138.Moore M.J., Dorfman T., Li W., Wong S.K., Li Y., Kuhn J.H., Coderre J., Vasilieva N., Han Z., Greenough T.C., Farzan M., Choe H. Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus spike protein efficiently infect cells expressing angiotensin-converting enzyme 2. J. Virol. 2004;78:10628–10635. doi: 10.1128/JVI.78.19.10628-10635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Iwanaga N., Cooper L., Rong L., Beddingfield B., Crabtree J., Tripp R.A., Kolls J.K. Novel ACE2-IgG1 fusions with improved activity against SARS-CoV2. bioRxiv. 2020 doi: 10.1101/2020.06.15.152157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chan K.K., Dorosky D., Sharma P., Abbasi S.A., Dye J.M., Kranz D.M., Herbert A.S., Procko E. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science. 2020;0870 doi: 10.1126/science.abc0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Glasgow A., Glasgow J., Limonta D., Solomon P., Lui I., Zhang Y., Nix M.A., Rettko N.J., Lim S.A., Zha S., Yamin R., Kao K., Rosenberg O.S., Ravetch J.V., Wiita A.P., Leung K.K., Zhou X.X., Hobman T.C., Kortemme T., Wells J.A. Engineered ACE2 receptor traps potently neutralize SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.07.31.231746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Higuchi Y., Suzuki T., Arimori T., Ikemura N., Kirita Y. High affinity modified ACE2 receptors prevent SARS-CoV-2 infection. bioRxiv. 2020 doi: 10.1101/2020.09.16.299891. [DOI] [Google Scholar]
- 143.Xiao A.T., Lu J., Zhang J., Johnson R.I. A trimeric human angiotensin-converting enzyme 2 as an anti- SARS-CoV-2 agent in vitro. bioRxiv. 2020 doi: 10.1101/2020.09.18.301952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Guo L., Bi W., Wang X., Xu W., Yan R., Zhang Y., Zhao K., Li Y., Zhang M., Bao X., Cai X., Li Y., Qu D., Jiang S., Xie Y., Zhou Q., Lu L., Dang B. Engineered trimeric ACE2 binds and locks “Three-up” spike protein to potently inhibit SARS-CoVs and mutants. bioRxiv. 2020;1 doi: 10.1038/s41422-020-00438-w. (2020.2008.2031.274704–272020.274708.274731.274704) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Miao X., Luo Y., Huang X., Lee S.M.Y., Yuan Z., Tang Y., Chen L., Wang C., Wu F., Xu Y., Jiang W., Gao W., Song X., Yan Y., Pang T., Chen C., Zou Y., Fu W., Wan L., Gilbert-Jaramillo J., Knight M., Tan T.K., Rijal P., Townsend A., Sun J., Liu X., James W., Tsun A., Xu Y. A novel biparatopic hybrid antibody-ACE2 fusion that blocks SARS-CoV-2 infection: implications for therapy. mAbs. 2020;12 doi: 10.1080/19420862.2020.1804241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Johansen M.D., Irving A., Montagutelli X., Tate M.D., Rudloff I., Nold M.F., Hansbro N.G., Kim R.Y., Donovan C., Liu G., Faiz A., Short K.R., Lyons J.G., McCaughan G.W., Gorrell M.D., Cole A., Moreno C., Couteur D., Hesselson D., Triccas J., Neely G.G., Gamble J.R., Simpson S.J., Saunders B.M., Oliver B.G., Britton W.J., Wark P.A., Nold-Petry C.A., Hansbro P.M. Animal and translational models of SARS-CoV-2 infection and COVID-19. Mucosal Immunol. 2020 doi: 10.1038/s41385-020-00340-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Muñoz-Fontela C., Dowling W.E., Funnell S.G.P., Gsell P.-S., Balta X.R., Albrecht R.A., Andersen H., Baric R.S., Carroll M.W., Cavaleri M., Qin C., Crozier I., Dallmeier K., de Waal L., de Wit E., Delang L., Dohm E., Duprex W.P., Falzarano D., Finch C.L., Frieman M.B., Graham B.S., Gralinski L., Guilfoyle K., Haagmans B.L., Hamilton G.A., Hartman A.L., Herfst S., Kaptein S.J.F., Klimstra W., Knezevic I., Krause P.R., Kuhn J.H., Le Grand R., Lewis M., Liu W.-C., Maisonnasse P., McElroy A.K., Munster V., Oreshkova N., Rasmussen A.L., Rocha-Pereira J., Rockx B., Rodríguez E., Rogers T., Salguero F.J., Schotsaert M., Stittelaar K., Thibaut H. Jan, Tseng C.-T., Vergara-Alert J., Beer M., Brasel T., Chan J.F.W., García-Sastre A., Neyts J., Perlman S., Reed D.S., Richt J.A., Roy C.J., Segalés J., Vasan S.S., Henao-Restrepo A.M., Barouch D.H. Animal models for COVID-19. Nature. 2020 doi: 10.1038/s41586-020-2787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.E.L.A. Company Lilly Announces Proof of Concept Data for Neutralizing Antibody LY-CoV555 in the COVID-19 Outpatient Setting. 2020. https://investor.lilly.com/news-releases/news-release-details/lilly-announces-proof-concept-data-neutralizing-antibody-ly
- 149.Chen P., Nirula A., Heller B., Gottlieb R.L., Boscia J., Morris J., Huhn G., Cardona J., Mocherla B., Stosor V., Shawa I., Adams A.C., Van Naarden J., Custer K.L., Shen L., Durante M., Oakley G., Schade A.E., Sabo J., Patel D.R., Klekotka P., Skovronsky D.M. SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Regeneron Regeneron's Regn-Cov2 Antibody Cocktail Reduced Viral Levels And Improved Symptoms In Non-Hospitalized Covid-19 Patients. https://investor.regeneron.com/news-releases/news-release-details/regenerons-regn-cov2-antibody-cocktail-reduced-viral-levels-and
- 151.T.A. Society COVID-19 Biologics Tracker. https://www.antibodysociety.org/covid-19-biologics-tracker/
- 152.Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., Wei Y., Atwal G.S., Murphy A.J., Stahl N., Yancopoulos G.D., Kyratsous C.A. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;1018 doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rondy M., El Omeiri N., Thompson M.G., Levêque A., Moren A., Sullivan S.G. Effectiveness of influenza vaccines in preventing severe influenza illness among adults: A systematic review and meta-analysis of test-negative design case-control studies. J. Inf. Secur. 2017;75:381–394. doi: 10.1016/j.jinf.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mulholland K. Global burden of acute respiratory infections in children: implications for interventions. Pediatr. Pulmonol. 2003;36:469–474. doi: 10.1002/ppul.10344. [DOI] [PubMed] [Google Scholar]
- 155.Ali S.O., Takas T., Nyborg A., Shoemaker K., Kallewaard N.L., Chiong R., Dubovsky F., Mallory R.M. Evaluation of MEDI8852, an anti-influenza a monoclonal antibody, in treating acute uncomplicated influenza. Antimicrob. Agents Chemother. 2018;62 doi: 10.1128/AAC.00694-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kallewaard N.L., Corti D., Collins P.J., Neu U., McAuliffe J.M., Benjamin E., Wachter-Rosati L., Palmer-Hill F.J., Yuan A.Q., Walker P.A., Vorlaender M.K., Bianchi S., Guarino B., De Marco A., Vanzetta F., Agatic G., Foglierini M., Pinna D., Fernandez-Rodriguez B., Fruehwirth A., Silacci C., Ogrodowicz R.W., Martin S.R., Sallusto F., Suzich J.A., Lanzavecchia A., Zhu Q., Gamblin S.J., Skehel J.J. Structure and function analysis of an antibody recognizing all influenza A subtypes. Cell. 2016;166:596–608. doi: 10.1016/j.cell.2016.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Nakamura G., Chai N., Park S., Chiang N., Lin Z., Chiu H., Fong R., Yan D., Kim J., Zhang J., Lee Wyne P., Estevez A., Coons M., Xu M., Lupardus P., Balazs M., Swem Lee R. An in vivo human-plasmablast enrichment technique allows rapid identification of therapeutic Influenza A antibodies. Cell Host Microbe. 2013;14:93–103. doi: 10.1016/j.chom.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 158.Tharakaraman K., Subramanian V., Cain D., Sasisekharan V., Sasisekharan R. Broadly neutralizing influenza hemagglutinin stem-specific antibody CR8020 targets residues that are prone to escape due to host selection pressure. Cell Host Microbe. 2014;15:644–651. doi: 10.1016/j.chom.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ekiert D.C., Bhabha G., Elsliger M.A., Friesen R.H., Jongeneelen M., Throsby M., Goudsmit J., Wilson I.A. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Throsby M., van den Brink E., Jongeneelen M., Poon L.L., Alard P., Cornelissen L., Bakker A., Cox F., van Deventer E., Guan Y., Cinatl J., ter Meulen J., Lasters I., Carsetti R., Peiris M., de Kruif J., Goudsmit J. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One. 2008;3 doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hershberger E., Sloan S., Narayan K., Hay C.A., Smith P., Engler F., Jeeninga R., Smits S., Trevejo J., Shriver Z., Oldach D. Safety and efficacy of monoclonal antibody VIS410 in adults with uncomplicated influenza A infection: Results from a randomized, double-blind, phase-2, placebo-controlled study. EBioMedicine. 2019;40:574–582. doi: 10.1016/j.ebiom.2018.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Baranovich T., Jones J.C., Russier M., Vogel P., Szretter K.J., Sloan S.E., Seiler P., Trevejo J.M., Webby R.J., Govorkova E.A. The hemagglutinin stem-binding monoclonal antibody VIS410 controls influenza virus-induced acute respiratory distress syndrome. Antimicrob. Agents Chemother. 2016;60:2118. doi: 10.1128/AAC.02457-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wu H., Pfarr D.S., Johnson S., Brewah Y.A., Woods R.M., Patel N.K., White W.I., Young J.F., Kiener P.A. Development of motavizumab, an ultra-potent antibody for the prevention of respiratory syncytial virus infection in the upper and lower respiratory tract. J. Mol. Biol. 2007;368:652–665. doi: 10.1016/j.jmb.2007.02.024. [DOI] [PubMed] [Google Scholar]
- 164.Wegzyn C., Toh L.K., Notario G., Biguenet S., Unnebrink K., Park C., Makari D., Norton M. Safety and effectiveness of palivizumab in children at high risk of serious disease due to respiratory syncytial virus infection: a systematic review. Infect. Dis. Ther. 2014;3:133–158. doi: 10.1007/s40121-014-0046-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Tang A., Chen Z., Cox K.S., Su H.-P., Callahan C., Fridman A., Zhang L., Patel S.B., Cejas P.J., Swoyer R., Touch S., Citron M.P., Govindarajan D., Luo B., Eddins M., Reid J.C., Soisson S.M., Galli J., Wang D., Wen Z., Heidecker G.J., Casimiro D.R., DiStefano D.J., Vora K.A. A potent broadly neutralizing human RSV antibody targets conserved site IV of the fusion glycoprotein. Nat. Commun. 2019;10:4153. doi: 10.1038/s41467-019-12137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hart T.K., Cook R.M., Zia-Amirhosseini P., Minthorn E., Sellers T.S., Maleeff B.E., Eustis S., Schwartz L.W., Tsui P., Appelbaum E.R., Martin E.C., Bugelski P.J., Herzyk D.J. Preclinical efficacy and safety of mepolizumab (SB-240563), a humanized monoclonal antibody to IL-5, in cynomolgus monkeys. J. Allergy Clin. Immunol. 2001;108:250–257. doi: 10.1067/mai.2001.116576. [DOI] [PubMed] [Google Scholar]
- 167.Dall'Acqua W.F., Kiener P.A., Wu H. Properties of human IgG1s engineered for enhanced binding to the neonatal Fc receptor (FcRn) J. Biol. Chem. 2006;281:23514–23524. doi: 10.1074/jbc.M604292200. [DOI] [PubMed] [Google Scholar]
- 168.T.I.-R.S. Group Palivizumab, a humanized respiratory syncytial virus monoclonal antibody, reduces hospitalization from respiratory syncytial virus infection in high-risk infants. Pediatrics. 1998;102:531. [PubMed] [Google Scholar]
- 169.Carbonell-Estrany X., Simões E.A., Dagan R., Hall C.B., Harris B., Hultquist M., Connor E.M., Losonsky G.A. Motavizumab for prophylaxis of respiratory syncytial virus in high-risk children: a noninferiority trial. Pediatrics. 2010;125 doi: 10.1542/peds.2008-1036. (e35-51) [DOI] [PubMed] [Google Scholar]
- 170.Pamela Griffin M., Yuan Y., Takas T., DeVincenzo J., Domachowske J.B., Simoes E.A., Khan A., Esser M.T., Dubovsky F., Villafana T.L. 901. MEDI8897 Prevents serious RSV disease in healthy preterm infants. Open Forum Infect Dis. 2019;6 (S27-S27) [Google Scholar]
- 171.Roche J.A., Roche R. A hypothesized role for dysregulated bradykinin signaling in COVID-19 respiratory complications. FASEB J. 2020;34:7265–7269. doi: 10.1096/fj.202000967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Herold T., Jurinovic V., Arnreich C., Lipworth B.J., Hellmuth J.C., von Bergwelt-Baildon M., Klein M., Weinberger T. Elevated levels of IL-6 and CRP predict the need for mechanical ventilation in COVID-19. J. Allergy Clin. Immunol. 2020;146:128–136. doi: 10.1016/j.jaci.2020.05.008. (e124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lipworth B., Chan R., Lipworth S., RuiWen Kuo C. Weathering the cytokine storm in susceptible patients with severe SARS-CoV-2 infection. J Allergy Clin Immunol Pract. 2020;8:1798–1801. doi: 10.1016/j.jaip.2020.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.He G., Massarella J., Ward P. Clinical pharmacokinetics of the prodrug oseltamivir and its active metabolite Ro 64–0802. Clin. Pharmacokinet. 1999;37:471–484. doi: 10.2165/00003088-199937060-00003. [DOI] [PubMed] [Google Scholar]
- 175.Respaud R., Vecellio L., Diot P., Heuzé-Vourc’h N. Nebulization as a delivery method for mAbs in respiratory diseases. Expert. Opin. Drug Deliv. 2015;12:1027–1039. doi: 10.1517/17425247.2015.999039. [DOI] [PubMed] [Google Scholar]
- 176.Guilleminault L., Azzopardi N., Arnoult C., Sobilo J., Hervé V., Montharu J., Guillon A., Andres C., Herault O., Le Pape A., Diot P., Lemarié E., Paintaud G., Gouilleux-Gruart V., Heuzé-Vourc'h N. Fate of inhaled monoclonal antibodies after the deposition of aerosolized particles in the respiratory system. J. Control. Release. 2014;196:344–354. doi: 10.1016/j.jconrel.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 177.Ramilo O., Lagos R., Sáez-Llorens X., Suzich J., Wang C.K., Jensen K.M., Harris B.S., Losonsky G.A., Griffin M.P. Motavizumab treatment of infants hospitalized with respiratory syncytial virus infection does not decrease viral load or severity of illness. Pediatr. Infect. Dis. J. 2014;33:703–709. doi: 10.1097/INF.0000000000000240. [DOI] [PubMed] [Google Scholar]
- 178.Dhand R., Sohal H. Pulmonary Drug Delivery System for inhalation therapy in mechanically ventilated patients. Expert Rev. Med. Dev. 2008;5:9–18. doi: 10.1586/17434440.5.1.9. [DOI] [PubMed] [Google Scholar]
- 179.Carbonell-Estrany X., Simões E.A.F., Dagan R., Hall C.B., Harris B., Hultquist M., Connor E.M., Losonsky G.A. Motavizumab for prophylaxis of respiratory syncytial virus in high-risk children: a noninferiority trial. Pediatrics. 2010;125 doi: 10.1542/peds.2008-1036. [DOI] [PubMed] [Google Scholar]
- 180.Zhu Q., Lu B., McTamney P., Palaszynski S., Diallo S., Ren K., Ulbrandt N.D., Kallewaard N., Wang W., Fernandes F., Wong S., Svabek C., Moldt B., Esser M.T., Jing H., Suzich J.A. Prevalence and significance of substitutions in the fusion protein of respiratory syncytial virus resulting in neutralization escape from antibody MEDI8897. J. Infect. Dis. 2018;218:572–580. doi: 10.1093/infdis/jiy189. [DOI] [PubMed] [Google Scholar]
- 181.Zhu Q., McLellan J.S., Kallewaard N.L., Ulbrandt N.D., Palaszynski S., Zhang J., Moldt B., Khan A., Svabek C., McAuliffe J.M., Wrapp D., Patel N.K., Cook K.E., Richter B.W.M., Ryan P.C., Yuan A.Q., Suzich J.A. A highly potent extended half-life antibody as a potential RSV vaccine surrogate for all infants. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aaj1928. [DOI] [PubMed] [Google Scholar]
- 182.Oswald W.B., Geisbert T.W., Davis K.J., Geisbert J.B., Sullivan N.J., Jahrling P.B., Parren P.W., Burton D.R. Neutralizing antibody fails to impact the course of Ebola virus infection in monkeys. PLoS Pathog. 2007;3 doi: 10.1371/journal.ppat.0030009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kim K.J., Fandy T.E., Lee V.H., Ann D.K., Borok Z., Crandall E.D. Net absorption of IgG via FcRn-mediated transcytosis across rat alveolar epithelial cell monolayers. Am. J. Phys. Lung Cell. Mol. Phys. 2004;287:L616–L622. doi: 10.1152/ajplung.00121.2004. [DOI] [PubMed] [Google Scholar]
- 184.Tzotzos S.J., Fischer B., Fischer H., Zeitlinger M. Incidence of ARDS and outcomes in hospitalized patients with COVID-19: a global literature survey. Crit. Care. 2020;24 doi: 10.1186/s13054-020-03240-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ragab D., Salah Eldin H., Taeimah M., Khattab R., Salem R. The COVID-19 cytokine storm; what we know so far. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zeng Q.L., Yu Z.J., Gou J.J., Li G.M., Ma S.H., Zhang G.F., Xu J.H., Lin W.B., Cui G.L., Zhang M.M., Li C., Wang Z.S., Zhang Z.H., Liu Z.S. Effect of convalescent plasma therapy on viral shedding and survival in patients with coronavirus disease 2019. J. Infect. Dis. 2020;222:38–43. doi: 10.1093/infdis/jiaa228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Spagnolo P., Balestro E., Aliberti S., Cocconcelli E., Biondini D., Casa G.D., Sverzellati N., Maher T.M. Pulmonary fibrosis secondary to COVID-19: a call to arms? Lancet Respir. Med. 2020;8:750–752. doi: 10.1016/S2213-2600(20)30222-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zuo W., Zhao X., Chen Y.-G. 2009. SARS Coronavirus and Lung Fibrosis, Molecular Biology of the SARS-Coronavirus; pp. 247–258. [DOI] [Google Scholar]
- 189.Lilly E. Lilly's Neutralizing Antibody Bamlanivimab (LY-CoV555) Receives FDA Emergency Use Authorization for the Treatment of Recently Diagnosed COVID-19. 2020. https://investor.lilly.com/news-releases/news-release-details/lillys-neutralizing-antibody-bamlanivimab-ly-cov555-receives-fda
- 190.Regeneron Regeneron's REGEN-COV2 is First Antibody Cocktail for COVID-19 to Receive FDA Emergency Use Authorization. 2020. https://www.prnewswire.com/news-releases/regenerons-regen-cov2-is-first-antibody-cocktail-for-covid-19-to-receive-fda-emergency-use-authorization-301178464.html
- 191.Guillon A., Pardessus J., Lhommet P., Parent C., Respaud R., Marchand D., Montharu J., De Monte M., Janiak P., Boixel C., Audat H., Huille S., Guillot E., Heuze-Vourc’h N. Exploring the fate of inhaled monoclonal antibody in the lung parenchyma by microdialysis. mAbs. 2019;11:297–304. doi: 10.1080/19420862.2018.1556081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Vonarburg C., Loetscher M., Spycher M.O., Kropf A., Illi M., Salmon S., Roberts S., Steinfuehrer K., Campbell I., Koernig S., Bain J., Edler M., Baumann U., Miescher S., Metzger D.W., Schaub A., Käsermann F., Zuercher A.W. Topical application of nebulized human IgG, IgA and IgAM in the lungs of rats and non-human primates. Respir. Res. 2019;20:99. doi: 10.1186/s12931-019-1057-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Fahy J.V., Cockcroft D.W., Boulet L.P., Wong H.H., Deschesnes F., Davis E.E., Ruppel J., Su J.Q., Adelman D.C. Effect of aerosolized anti-IgE (E25) on airway responses to inhaled allergen in asthmatic subjects. Am. J. Respir. Crit. Care Med. 1999;160:1023–1027. doi: 10.1164/ajrccm.160.3.9810012. [DOI] [PubMed] [Google Scholar]
- 194.Leyva-Grado V.H., Tan G.S., Leon P.E., Yondola M., Palese P. Direct administration in the respiratory tract improves efficacy of broadly neutralizing anti-influenza virus monoclonal antibodies. Antimicrob. Agents Chemother. 2015;59:4162. doi: 10.1128/AAC.00290-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Prince G.A., Hemming V.G., Horswood R.L., Baron P.A., Chanock R.M. Effectiveness of topically administered neutralizing antibodies in experimental immunotherapy of respiratory syncytial virus infection in cotton rats. J. Virol. 1987;61:1851–1854. doi: 10.1128/jvi.61.6.1851-1854.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Weltzin R., Traina-Dorge V., Soike K., Zhang J.Y., Mack P., Soman G., Drabik G., Monath T.P. Intranasal monoclonal IgA antibody to respiratory syncytial virus protects rhesus monkeys against upper and lower respiratory tract infection. J. Infect. Dis. 1996;174:256–261. doi: 10.1093/infdis/174.2.256. [DOI] [PubMed] [Google Scholar]
- 197.Burgess G., Boyce M., Jones M., Larsson L., Main M.J., Morgan F., Phillips P., Scrimgeour A., Strimenopoulou F., Vajjah P., Zamacona M., Palframan R. Randomized study of the safety and pharmacodynamics of inhaled interleukin-13 monoclonal antibody fragment VR942. EBioMedicine. 2018;35:67–75. doi: 10.1016/j.ebiom.2018.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Novartis A Randomized, Subject and Investigator-Blinded, Placebo-Controlled, Parallel-Design, Broncho-Provocation Study to Evaluate the Safety, Tolerability, Pharmacokinetics and Pharmacodynamics of Multiple Doses of Inhaled CSJ117 in Adult Subjects with Mild Atopic Asthma. 2020. https://www.novctrd.com/CtrdWeb/searchbystudyid.nov#CCSJ117X2201
- 199.Bodier-Montagutelli E., Mayor A., Vecellio L., Respaud R., Heuzé-Vourc'h N. Designing inhaled protein therapeutics for topical lung delivery: what are the next steps? Expert Opin. Drug Deliv. 2018;15:729–736. doi: 10.1080/17425247.2018.1503251. [DOI] [PubMed] [Google Scholar]
- 200.Lip Kwok P.C., Chan H.-K. In: Peptide and Protein Delivery. Van Der Walle C., editor. Academic Press; Boston: 2011. Chapter 2 - Pulmonary delivery of peptides and proteins; pp. 23–46. [Google Scholar]
- 201.Kane C., O'Neil K., Conk M., Picha K. Inhalation delivery of protein therapeutics. Inflamm. Allergy Drug Targets. 2013;12:81–87. doi: 10.2174/1871528111312020002. [DOI] [PubMed] [Google Scholar]
- 202.Carvalho T.C., Peters J.I., Williams R.O. Influence of particle size on regional lung deposition – What evidence is there? Int. J. Pharm. 2011;406:1–10. doi: 10.1016/j.ijpharm.2010.12.040. [DOI] [PubMed] [Google Scholar]
- 203.Borghardt J.M., Kloft C., Sharma A. Inhaled therapy in respiratory disease: the complex interplay of pulmonary kinetic processes. Can. Respir. J. 2018;2018 doi: 10.1155/2018/2732017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Costa A., Pinheiro M., Magalhães J., Ribeiro R., Seabra V., Reis S., Sarmento B. The formulation of nanomedicines for treating tuberculosis. Adv. Drug Deliv. Rev. 2016;102:102–115. doi: 10.1016/j.addr.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 205.Laube B.L., Janssens H.M., de Jongh F.H.C., Devadason S.G., Dhand R., Diot P., Everard M.L., Horvath I., Navalesi P., Voshaar T., Chrystyn H. What the pulmonary specialist should know about the new inhalation therapies. Eur. Respir. J. 2011;37:1308. doi: 10.1183/09031936.00166410. [DOI] [PubMed] [Google Scholar]
- 206.Behr J., Zimmermann G., Baumgartner R., Leuchte H., Neurohr C., Brand P., Herpich C., Sommerer K., Seitz J., Menges G., Tillmanns S., Keller M. Lung deposition of a liposomal cyclosporine a inhalation solution in patients after lung transplantation. J. Aerosol. Med. Pulmon. Drug Deliv. 2009;22:121–130. doi: 10.1089/jamp.2008.0714. [DOI] [PubMed] [Google Scholar]
- 207.Nikander K., Prince I., Coughlin S., Warren S., Taylor G. Mode of breathing—tidal or slow and deep—through the I-neb Adaptive Aerosol Delivery (AAD) system affects lung deposition of 99mTc-DTPA. J. Aerosol. Med. Pulmon. Drug Deliv. 2010;23:S-37–S-43. doi: 10.1089/jamp.2009.0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Pritchard J.N., Hatley R.H., Denyer J., Hollen D.V. Mesh nebulizers have become the first choice for new nebulized pharmaceutical drug developments. Ther. Deliv. 2018;9:121–136. doi: 10.4155/tde-2017-0102. [DOI] [PubMed] [Google Scholar]
- 209.Longest W., Spence B., Hindle M. Devices for improved delivery of nebulized pharmaceutical aerosols to the lungs. J. Aerosol. Med. Pulmon. Drug Deliv. 2019;32:317–339. doi: 10.1089/jamp.2018.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Coates A.L., Green M., Leung K., Chan J., Ribeiro N., Louca E., Ratjen F., Charron M., Tservistas M., Keller M. Rapid pulmonary delivery of inhaled tobramycin for Pseudomonas infection in cystic fibrosis: a pilot project. Pediatr. Pulmonol. 2008;43:753–759. doi: 10.1002/ppul.20850. [DOI] [PubMed] [Google Scholar]
- 211.Fängmark I., Carpin J.C. Protein nebulization. J. Aerosol Sci. 1996;27:S231–S232. [Google Scholar]
- 212.Hertel S.P., Winter G., Friess W. Protein stability in pulmonary drug delivery via nebulization. Adv. Drug Deliv. Rev. 2015;93:79–94. doi: 10.1016/j.addr.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 213.Clay M., Newman S., Pavia D., Lennard-Jones T. Assessment of jet nebulisers for lung aerosol therapy. Lancet. 1983;322:592–594. doi: 10.1016/s0140-6736(83)90679-7. [DOI] [PubMed] [Google Scholar]
- 214.McCarthy S.D., González H.E., Higgins B.D. Future trends in nebulized therapies for pulmonary disease. J. Pers. Med. 2020;10 doi: 10.3390/jpm10020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Taylor K.M.G., McCallion O.N.M. Ultrasonic nebulisers for pulmonary drug delivery. Int. J. Pharm. 1997;153:93–104. [Google Scholar]
- 216.Steckel H., Eskandar F. Factors affecting aerosol performance during nebulization with jet and ultrasonic nebulizers. Eur. J. Pharm. Sci. 2003;19:443–455. doi: 10.1016/s0928-0987(03)00148-9. [DOI] [PubMed] [Google Scholar]
- 217.Bohr A., Beck-Broichsitter M. Generation of tailored aerosols for inhalative drug delivery employing recent vibrating-mesh nebulizer systems. Ther. Deliv. 2015;6:621–636. doi: 10.4155/tde.15.18. [DOI] [PubMed] [Google Scholar]
- 218.Maillet A., Congy-Jolivet N., Le Guellec S., Vecellio L., Hamard S., Courty Y., Courtois A., Gauthier F., Diot P., Thibault G., Lemarié E., Heuzé-Vourc’h N. Aerodynamical, immunological and pharmacological properties of the anticancer antibody cetuximab following nebulization. Pharm. Res. 2008;25:1318–1326. doi: 10.1007/s11095-007-9481-3. [DOI] [PubMed] [Google Scholar]
- 219.Van Heeke G., Allosery K., De Brabandere V., De Smedt T., Detalle L., de Fougerolles A. Nanobodies® as inhaled biotherapeutics for lung diseases. Pharmacol. Ther. 2017;169:47–56. doi: 10.1016/j.pharmthera.2016.06.012. [DOI] [PubMed] [Google Scholar]
- 220.Sawicki G.S., Chou W., Raimundo K., Trzaskoma B., Konstan M.W. Randomized trial of efficacy and safety of dornase alfa delivered by eRapid nebulizer in cystic fibrosis patients. J. Cyst. Fibros. 2015;14:777–783. doi: 10.1016/j.jcf.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 221.Arzu A. Jet, ultrasonic, and mesh nebulizers: an evaluation of nebulizers for better clinical outcomes. Eurasian J. Pulmonol. 2014;16:1–7. [Google Scholar]
- 222.Denyer J., Nikander K., Smith N.J. Adaptive aerosol delivery (AAD®) technology. Expert. Opin. Drug Deliv. 2004;1:165–176. doi: 10.1517/17425247.1.1.165. [DOI] [PubMed] [Google Scholar]
- 223.Denyer J., Dyche T. The Adaptive Aerosol Delivery (AAD) technology: Past, present, and future. J. Aerosol. Med. Pulmon. Drug Deliv. 2010;23(Suppl. 1):S1–S10. doi: 10.1089/jamp.2009.0791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Geller D.E., Kesser K.C. The I-neb Adaptive Aerosol Delivery System enhances delivery of alpha1-antitrypsin with controlled inhalation. J. Aerosol. Med. Pulmon. Drug Deliv. 2010;23(Suppl. 1):S55–S59. doi: 10.1089/jamp.2009.0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Nikander K., Prince I., Coughlin S., Warren S., Taylor G. Mode of breathing-tidal or slow and deep-through the I-neb Adaptive Aerosol Delivery (AAD) system affects lung deposition of (99m)Tc-DTPA. J. Aerosol Med. Pulm. Drug Deliv. 2010;23(Suppl. 1):S37–S43. doi: 10.1089/jamp.2009.0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Lightwood D., O'Dowd V., Carrington B., Veverka V., Carr M.D., Tservistas M., Henry A.J., Smith B., Tyson K., Lamour S., Sarkar K., Turner A., Lawson A.D., Bourne T., Gozzard N., Palframan R. The discovery, engineering and characterisation of a highly potent anti-human IL-13 Fab fragment designed for administration by inhalation. J. Mol. Biol. 2013;425:577–593. doi: 10.1016/j.jmb.2012.11.036. [DOI] [PubMed] [Google Scholar]
- 227.Lightwood D., Tservistas M., Zehentleitner M., Sarkar K., Turner A., Bracher M., Smith B., Lamour S., Bourne T., Shaw S., Gozzard N., Palframan R.T. Efficacy of an inhaled IL-13 antibody fragment in a model of chronic asthma. Am. J. Respir. Crit. Care Med. 2018;198:610–619. doi: 10.1164/rccm.201712-2382OC. [DOI] [PubMed] [Google Scholar]
- 228.Respaud R., Marchand D., Parent C., Pelat T., Thullier P., Tournamille J.-F., Viaud-Massuard M.-C., Diot P., Si-Tahar M., Vecellio L., Heuzé-Vourc’h N. Effect of formulation on the stability and aerosol performance of a nebulized antibody. mAbs. 2014;6:1347–1355. doi: 10.4161/mabs.29938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Sala V., Murabito A., Ghigo A. Inhaled biologicals for the treatment of cystic fibrosis. Recent Pat. Inflamm. Allergy Drug Discov. 2019;13:19–26. doi: 10.2174/1872213X12666181012101444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Djukanović R., Harrison T., Johnston S.L., Gabbay F., Wark P., Thomson N.C., Niven R., Singh D., Reddel H.K., Davies D.E., Marsden R., Boxall C., Dudley S., Plagnol V., Holgate S.T., Monk P., I.S. Group The effect of inhaled IFN-β on worsening of asthma symptoms caused by viral infections. A randomized trial. Am. J. Respir. Crit. Care Med. 2014;190:145–154. doi: 10.1164/rccm.201312-2235OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Stolk J., Tov N., Chapman K.R., Fernandez P., MacNee W., Hopkinson N.S., Piitulainen E., Seersholm N., Vogelmeier C.F., Bals R., McElvaney G., Stockley R.A. Efficacy and safety of inhaled alpha-1-antitrypsin in patients with severe alpha-1-antitrypsin deficiency and frequent exacerbations of Chronic Obstructive Pulmonary Disease. Eur. Respir. J. 2019 doi: 10.1183/13993003.00673-2019. [DOI] [PubMed] [Google Scholar]
- 232.Trapnell B.C., Inoue Y., Bonella F., Morgan C., Jouneau S., Bendstrup E., Campo I., Papiris S.A., Yamaguchi E., Cetinkaya E., Ilkovich M.M., Kramer M.R., Veltkamp M., Kreuter M., Baba T., Ganslandt C., Tarnow I., Waterer G., Jouhikainen T. Inhaled molgramostim therapy in autoimmune pulmonary alveolar proteinosis. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa1913590. [DOI] [PMC free article] [PubMed] [Google Scholar]