Abstract
The SARS-CoV-2 virus responsible for coronavirus 2019 (COVID-19) poses a significant challenge to healthcare systems worldwide. According to the World Health Organization (WHO), the outbreak of COVID-19 has been a pandemic that infected more than 25.32 million people and caused more than 848.25 thousand deaths worldwide at the time of 1st September 2020. Despite governmental initiatives aimed to contain the spread of the disease, several countries are experiencing unmanageable increases in medical equipment and larger testing capacity. The current diagnosis based on nuclear acid requires specialized instruments, time-consuming, and laborious, the low-cost and convenient technologies were still urgently needed. Both spike and nucleocapsid are key structural proteins of COVID-19 with good immunogenicity, can serve as primary targets for immunoassay. After comparative research, we certified nucleocapsid antigen-monoclonal antibody (mAbs) system was more suitable for the COVID-19 immunodetection. Subsequently, we designed a rapid test strip based on it that can be used in large-scale screening of COVID-19 in population and more suitable for some remote and special needs areas were restricted by a medical condition or for quick and large quantities of screenings.
Keywords: Covid-19, N protein, S protein, mAbs, Immunoassay strips
1. Introduction
A sudden outbreak of viral pneumonia swept across the world in early 2020, causing panic and severe economic loss, generating global concern given its rapid spread around the world and fatal progression. This global pandemic of coronavirus disease 2019 (COVID-19) was named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by the International Committee on Taxonomy of Viruses (ICTV) on February 11, 2020 (Gorbalenya et al., 2020; Yang and Wang, 2020; Su et al., 2016), and also was declared by the WHO in March 2020.
Coronaviruses are enveloped RNA viruses, belong to the Coronaviridae family distributed broadly among humans, other mammals and birds, and cause respiratory, enteric, hepatic, renal and neurologic diseases. According to the genome structure and phylogenetic analysis of coronaviruses, the Coronaviridae family can be divided into four genera: α, β, γ, and δ. The coronaviruses of the α and β genera generally infect mammals and humans, while the coronaviruses of the γ and δ genera mainly infect birds (Weiss and Leibowitz, 2011; Gorbalenya et al., 2020). There have already been two zoonotic outbreaks of β-CoVs before the COVID-19 outbreak in the last two decades: Severe acute respiratory syndrome coronavirus (SARS-CoV, 2002–2003) and Middle East respiratory syndrome coronavirus (MERS-CoV, 2012) (Zaki, 2012 Zhong et al., 2003 Zhu et al., 2019). The main symptoms of COVID-19 caused by SARS-CoV-2 are respiratory tract infection-like syndrome: fever, dry cough, upper airway congestion, runny nose, sore throat, myalgia, headache, and fatigue, a few patients may have diarrhea. Some patients may have dyspnea, and those who have a severe form of COVID-19 may rapidly progress to acute respiratory distress syndrome, coagulation dysfunction, and septic shock (Chang et al., 2020).
Clinically and commercially available COVID-19 tests currently fall into two major categories. The first category includes molecular assays for the detection of SARS-CoV-2 viral RNA using polymerase chain reaction (PCR)-based techniques or nucleic acid hybridization-related strategies. The second category includes serological and immunological assays that mainly rely on detecting antibodies produced by individuals due to exposure to the virus or on the detection of antigenic proteins in infected individuals (Carter et al., 2020). However, such methods require specialized instruments, time-consuming and laborious, the rapid, convenient, and low-cost technologies were still urgently needed to further specifically identify. For these reasons, increasing concern has been paid to the immunogold labeling technique, which has high sensitivity, simple procedure, and rapid detection in immunoassay. It is essential to select a specific antigen as the target for COVID-19 immunodetection. Both spike glycoprotein (S-protein) and nucleocapsid (N-proteins) are vital structural proteins with good immunogenicity, can serve as primary targets for immunoassay. The coronavirus spike is a trimeric single-pass membrane protein that mediates receptor binding and fusion of the viral and host cell membranes, which are the initial and critical steps in the coronavirus infection cycle. S-protein contains three segments: a large ectodomain, a single-pass transmembrane anchor, and a short intracellular tail. The ectodomain consists of a receptor-binding domain (RBD) subunit S1 and a membrane-fusion subunit S2. During virus entry, S1 binds to a receptor on the host cell surface for viral attachment, and S2 fuses the host and viral membranes, allowing viral genomes to enter host cells (Li, 2016; Wong et al., 2017). The coronavirus nucleocapsid is a structural protein with multifunctional that forms complexes with genomic RNA, interacts with the viral membrane protein during virion assembly. It plays a critical role in enhancing the efficiency of virus transcription, translation and assembly (McBride et al., 2014). N protein is extremely antigenic, and coronavirus infection causes a highly restricted, immunoglobulin G-dominated antibody response that is directed most frequently and predominantly at the nucleocapsid (Lee et al., 2008). Besides, N protein has developed mechanisms to overcome the host innate immune by produce the β-interferon, which plays an important role in viral pathogenesis response (Spiegel et al., 2005).
In this research, we have prepared monoclonal antibodies (mAbs) against S proteins and N proteins respectively. The comparative studies showed that the mAbs against N proteins were more suitable for rapid immunoassay in specificity, accuracy and stability. Subsequently, we designed a rapid test strip based on it that can be used in large-scale screening of COVID-19 in population and conducive to take the corresponding medical measures will undoubtedly save precious time for the control of the epidemic. These rapid and sensitive strips are more suitable for some remote and special needs areas restricted by a medical condition or for quick and large screenings.
2. Materials and methods
2.1. Acquisition of S protein, N protein and clinical specimens
The recombinant S-trimer proteins were purchased from Novoprotein Scientific Inc., produced by mammalian expression system and the target gene encoding Cys15-Gln1208 is expressed with a 6His tag at the C-terminus. The recombinant N proteins were purchased from Zhenkang Sci. & Tech. Inc., produced by E.coli expression system and the target gene encoding Met1-Ala419 is expressed with a 6His tag at the N-terminus. The amino acid sequences of S protein and N protein were displayed in supplementary material Fig S1 and Fig S2 (Wu et al., 2020). The 15 positive nasopharyngeal swabs are all COVID-19 mild symptoms patients with RT-PCR Ct value between 26 and 32 (see supplementary material, Table S2), which were collected by Shenzhen center for disease control and prevention.
2.2. Establish of monoclonal antibody
2.2.1. Immunization of mice
Four 4−6-week-old Balb/C mice were injected on leg intramuscular with 20 μg S proteins and N proteins respectively, which mixed with an equal volume of adjuvant(Quick antibody-Mouse 3 W)for twice with a 2-week interval. The tail blood was collected to detect the titer at 7 days after the second immunization. Mice received a final booster immunization to the spleen 3 days prior to hybridoma fusion.
2.2.2. Generation of monoclonal antibody
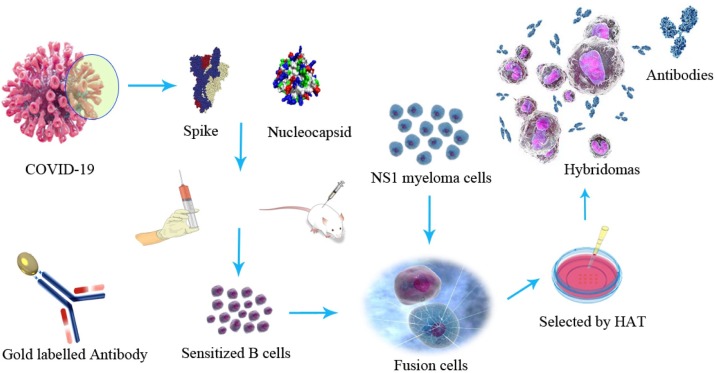
Mice were sacrificed followed ethical rules and their spleen cells harvested. Mice NS1 myeloma cells were in log-phase growth prior to fusion with spleen cells as a ratio of 9:1 by 50 % PEG 1500 (de StGroth and Scheidegger, 1980; Yokoyama et al., 2006). Hybridomas were discarded in HAT medium and the culture supernatants were screened using ELISA. When the desired clones were identified, they were expanded in flasks. 7 days later, the hybridoma suspension was harvested via centrifugation at 800 g for 10 min, followed by the collection of the supernatant and storage at −80 ◦C. And in the meantime, these hybridoma cells were transplanted in mice and the monoclonal antibody were purified from ascites produced in Balb/C mice by protein-G affinity chromatography (Fig. 1 ).
Fig. 1.
The flow chart of monoclonal antibodies production against spike and nucleocapsid.
2.3. Enzyme-linked immunosorbent assay
2.3.1. Evaluation of hybridoma secretion of monoclonal antibody
The hybridoma cells supernatant was incubated in 96-well microplates coated with nucleocapsid protein and spike protein respectively, and the bound antibody probed with a 1:4000 dilution of horseradish peroxidase (HRP) labeled goat anti-mouse immunoglobulin. After extensive washing by PBS-T, the plates were incubated with o-phenylenediamine dihydrochloride (OPD) for 10 min and the reaction was stopped by the addition of 2 M H2SO4. The absorbances at 490 nm were read using the microplate reader.
2.3.2. Development of antigen capture sandwich ELISA
The 96-well flat bottom microtiter plates were coated with 200 ng of purified IgG in 100 μL carbonate coating buffer per well, incubated at 37 ◦C for 1 h or at 4 ◦C overnight. Plates were washed three times with PBS-T and blocked with 200 μL of blocking solution (1% BSA in PBS-T) and incubated at 37 ◦C for 2 h. After the plates were rinsed with PBS-T for three times and pat dry, 100 μL of purified nucleocapsid protein or spike protein in PBS-T containing 1% BSA was added. The plate was washed three times with PBS-T, and 100 μL of mAb was added to each well, after incubated at 37 ◦C for 1 h, the plate was washed with PBS-T for three times again and 100 μL of goat anti-mice immunoglobulin HRP-conjugated antibodies were added at 1:4000 dilution and incubated at 37 ◦C for 1 h. After another extensive wash with PBS-T, 100 μL of OPD was added to each well. Plates were incubated at room temperature for 10 min, the absorbance was read at 490 nm after the reaction was stopped by 2 M H2SO4 stop solution.
2.3.3. Preparation of colloidal gold nanoparticles
Gold nanoparticles (AuNPs) with an average diameter of 40 nm were prepared by the citric acid reduction method. 10 mL of 0.1 % (w/v) chloroauric acid diluted in 90 mL of deionized water was heated to boiling point and then reduced in the presence of 3 mL of 1 % (w/v) sodium citrate. The solution was boiled for an additional 10 min, till it turned to wine-red color and then was allowed to cool down to room temperature. The colloidal gold particles were stored at 4 °C for use.
2.3.4. Preparation of colloidal gold antibodies conjugates
The optimal ratio between colloidal gold and antibodies required for conjugation was determined before. The pH of the solution was adjusted to 7.5 with 0.1 M potassium carbonate. 1 mL of colloidal gold solution was mixed with 8 μg purified mAb PBS solution and stirred for 60 min at room temperature. Subsequently, 100 μL of 10 % (w/v) BSA was added with gentle stirring for 60 min to block unreacted sites of gold particles. The solution was incubated for 60 min at room temperature and centrifuged at 12,000 rpm for 20 min at 4 °C. The supernatant was discarded, and the pellet was washed with PBS and was finally resuspended in 0.5 mL PBS buffered saline (0.02 M, pH 7.4) containing 1 % (w/v) BSA. The conjugate was stored at 4 °C for use.
2.3.5. Development of colloidal gold immunochromatographic assay test strips
The sample pad was treated with sample pad buffer containing 0.02 % (v/v) Tween-20, 0.5 % (w/v) BSA, 0.05 % (w/v) SDS and completely air-dried. The colloidal-gold-conjugated solution of the mAbs raised against the S proteins and N proteins were applied to the conjugate pad and dried at 37 °C for 2 h. The capture antibodies against the antigen were immobilized at the test line, and goat anti-mouse IgG was applied at the control line, parallel to the test line over the nitrocellulose membrane. The membrane was incubated at 37 °C for 2 h for complete drying. The absorption pad used had no treatment.
2.3.6. Practicability of the colloidal gold immunochromatographic assay test strips in COVID-19 samples and other pathogens
To evaluate the practicability of the colloidal gold immunochromatographic assay test strip in the real COVID-19 positive samples, we collected 15 COVID-19 infected mild symptoms positive swab samples from Shenzhen center for disease control and prevention (CDC) and 42 random healthy negative nasopharyngeal swabs and oropharyngeal swabs as control check. Specifically, 50 μL virus lysis solution (0.5 % NP40, 0.2 % tween 20 and 0.2 % LNL in 0.02 M pH 7.4 PBS) was added into 50 μL swab storage solution (or pathogen solution) incubate for 1 min after mix well. 60 μL of sample mixture were added on to the sample window of the test strip. The strip was then placed flat to allow the solution to migrate up the membrane, the results were evaluated after 10 min.
2.3.7. Confirming the sensitivity of colloidal gold immunochromatographic test strip assay
To determine the sensitivity of the strip, the assay was carried out by applying samples including 10 pg/mL, 102 pg/mL, 103 pg/mL, 104 pg/mL, 105 pg/mL, and 106 pg/mL of the appropriate test N protein solution and S protein solution to the bottom of the device. More specifically, 60 μL of a standard solution or sample were added on to the sample pad of the test strip, the results will be shown on the T line area in 10 min.
3. Results
3.1. Production of mAb-secreting cell lines and screening of the matched mAbs
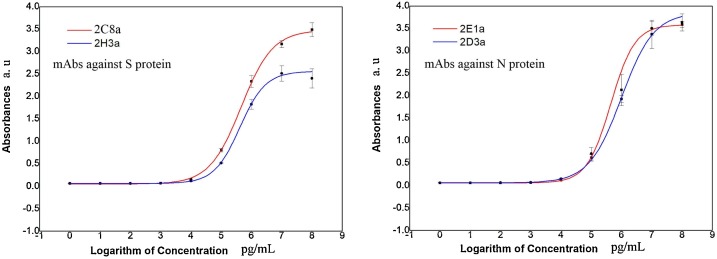
The fusion of spleen cells from immunized Balb/C mice with NS1 myeloma cells produced several hybridomas clones secreting mAbs against N proteins and S proteins. We have selected four groups of positive clones of hybridomas with superior and mAbs secreting ability by indirect ELISA (Fig. 2 .), The chosen clones showing at least a 2.1 fold higher absorbance than that of the negative control (P/N ≥ 2.1). We acquired hybridomas marked as 2E1a and 2D3a producing mAbs against N proteins, 2C1a and 2H3a producing mAbs against S proteins. Meanwhile, the mAbs secreted by these cells were screened from the numerous mAb-secreting cell lines to confirm whether they matched. These hybridomas cell lines yielding a relatively high antibody titer and were selected to mAbs for further analyses and experiments.
Fig. 2.
Indirect ELISA identification of mAb titer produced by selected hybridoma cells. All 10-fold serial dilutions values were taken as common logarithm as the abscissa.
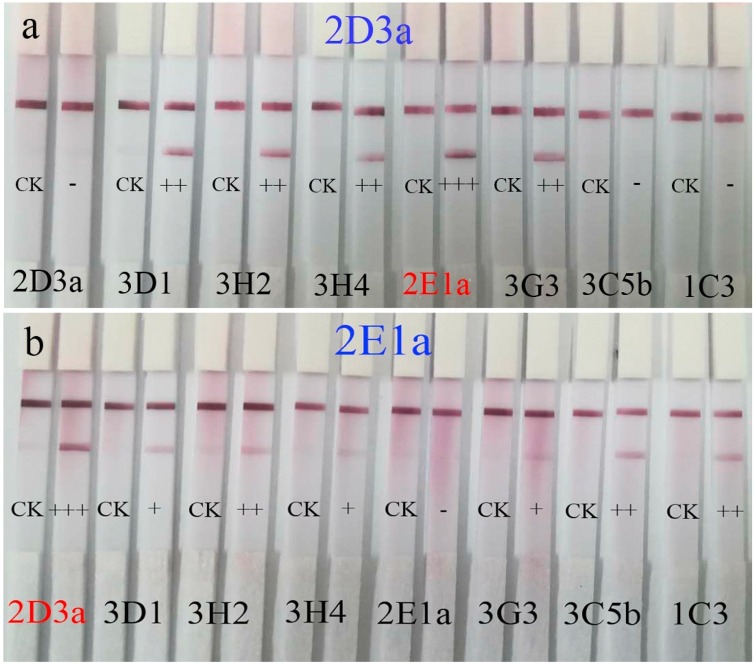
The screening of matched mAbs against N proteins by test strips was shown in Table 1 . and Fig. 3 . The 2D3a-2E1a couple showed the darkest color in reciprocal crosses labeling experiments. By contrast, the screening of matched mAbs against S proteins, without satisfactory results as the reciprocal crosses labeling experiments were out of accord (details see Supplementary information Fig S3 and Table S1).
Table 1.
The screening of matched mAbs against N protein by the checkerboard method.
| Gold labblled antibodies | Coated antibodies |
|||||||
|---|---|---|---|---|---|---|---|---|
| 2D3a | 3D1 | 3H2 | 3H4 | 2E1a | 3G3 | 3C5b | 1C3 | |
| 2D3a | – | ++ | ++ | ++ | +++ | ++ | – | – |
| 3D1 | ++ | – | + | + | + | + | – | – |
| 3H2 | ++ | + | – | + | ++ | + | – | + |
| 3H4 | ++ | – | – | + | + | + | – | – |
| 2E1a | +++ | – | ++ | ++ | – | + | + | ++ |
| 3G3 | ++ | – | ++ | + | + | + | + | ++ |
| 3C5b | – | – | ++ | ++ | ++ | ++ | – | – |
| 1C3 | – | – | ++ | ++ | ++ | ++ | – | – |
Fig. 3.
The results of the screening of matched mAbs against N proteins by test strips. The 2D3a mAbs were immobilized at the test line, and the 2D3a, 3D1, 3H2, 3H4, 2E1a, 3G3, 3C5b and 1C3 mAbs were labeled by colloidal gold nanoparticles respectively to construct a sandwich immunoassay structure (Fig. 3a). The reciprocal crosses labeling experiments (the 2E1a mAbs were immobilized at the test line, and the 2D3a, 3D1, 3H2, 3H4, 2E1a, 3G3, 3C5b and 1C3 mAbs were labeled by colloidal gold nanoparticles Fig. 3b) showed high consistency which suggests 2D3a-2E1a couple mAbs could perfectly bind with opposite epitopes of N protein without steric hindrance.
3.2. Minimal detection limit by sandwich ELISA
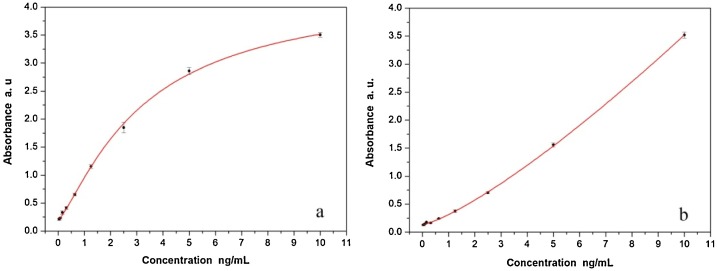
In this research, 0.1 ng/mL of N proteins (102 pg/mL dilution) and 1.25 ng/mL of S proteins were considered positive according to the cut-off threshold calculation formula P/N ≥ 2.1(Fig. 4 .), which means the rapid immunoassay based on these monoclonal antibodies could identify accurately and reliably to as little as 0.1 ng/mL target N protein antigens and 1.25 ng/mL of S protein antigens respectively in upper respiratory track secretion and throat swab samples. The sensitivity based on the N protein mAbs assay is almost 10 fold higher than S protein mAbs based detection. In contrast, the mAbs against N protein we prepared are more suitable for the development of a rapid immunoassay system. In order to further quantitative study of nucleocapsid, the concentration of N proteins and S proteins and the corresponding absorbance were well fitted by the nonlinear equation; the variables satisfy the Logistic function model, the confident function expression is as follow: the associated parameters were shown in Table 2 .
Fig. 4.
The minimal detection limit of N proteins and S proteins by the capture sandwich ELISA. (a) Absorption at different amounts of N proteins samples concentration using 2-fold serial dilutions, (b) Absorption at different amounts of S protein samples concentration using 2-fold serial dilutions.
Table 2.
The sandwich ELISA parameters and function of fitting nonlinear equation between concentration and absorbance of N proteins and S proteins.
| Ag | A1 | A2 | x0 | b | Function | R2 |
|---|---|---|---|---|---|---|
| N | 0.202 | 4.317 | 3.249 | 1.26 | 0.9992 | |
| S | 0.133 | 5438.08 | 3386.38 | 1.27 | 0.9965 |
3.3. Detection by immunochromatographic assay test strip
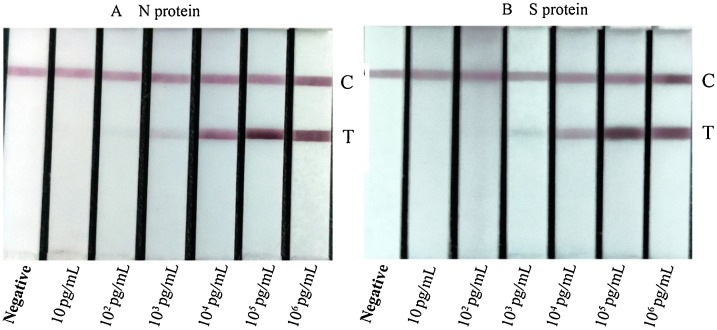
The sensitivity of the test strip was determined by testing the N protein and S protein standard samples. These standard samples were thereafter examined and judged with the naked eye, the differential color intensity between limit concentration and negative control is easy to tell instantly by healthy color vision. The test line’s color intensity decreased significantly with the decreasing sample concentration (Fig. 5 .). This differential color intensity of the test line represented the concentration of samples that are particularly noticeable at the range of 102 pg/mL-105 pg/mL for N proteins and 103 pg/mL-105 pg/mL for S proteins. The test limit concentrations of N proteins using the strips are 100 pg/mL(0.1 ng/mL), and the limit concentrations of S proteins are 1000 pg/mL(1 ng/mL).
Fig. 5.
Detection of N proteins and S proteins using colloidal gold immunochromatographic assay test strips at various dilutions (C: control line, T: Test line).
3.4. Specificity of test strips
In order to identify the specificity, the prepared test strips were taken to detect 20 different pathogens, which including common coronaviruses in the population (HCoV-229E, HCoV-OC43, HCoV-NL63, and HCoV-HKU1, provided by Shenzhen Center for Disease Control and Prevention), influenza A (H1N1, H3N2, H5N1, H7N9, and H9N2), influenza B (Victoria strain and Yamagata strain), Paramyxoviridae (measles virus and mumps virus), rubella virus, varicella zoster virus, and pyogenic bacteria (staphylococcus aureus and pseudomonas aeruginosa), SARS-CoV N protein, MARS-CoV N protein and chicken IBV. As display in Table 3 , HCoV-OC43, HCoV-NL63, and HCoV-HKU1 yielded positive results by S protein test strips compared with negative results by N protein test strips; SARS-CoV yielded positive results by N strips because SARS-CoV and COVID-19 are highly similar (Fig S6). The other pathogen tests were all negative. By contrast, the test strips based on N protein were higher sensitivity and specificity than S protein test strips, suitable for developing colloidal gold immunochromatographic assay test strips and clinical sample detection.
Table 3.
Cross reaction between several common coronaviruses and pathogens on N protein & S protein test strips. (Ct, cycle threshold; TCID50, median tissue culture infective dose; CFU, colony formation unit; EID50, egg infectious dose).
| Samples (Viruses and bacteria) |
Ct/titer/CFU/ug | N- strips results | S- strips results | |
|---|---|---|---|---|
| HCoV-229E | Ct 27 | – | – | |
| HCoV-OC43 | Ct 25 | – | + | |
| HCoV-NL63 | Ct 23 | – | + | |
| HCoV-HKU1 | Ct 25 | – | + | |
| influenza A H1N1 | 1.22 × 104 TCID50/mL | – | – | |
| influenza A H3N2 | 1.25 × 104 TCID50/mL | – | – | |
| influenza A H5N1 | 5.25 × 104 TCID50/mL | – | – | |
| influenza A H7N9 | 1.00 × 104 TCID50/mL | – | – | |
| influenza A H9N2 | 1.25 × 104 TCID50/mL | – | – | |
| influenza B Victoria strain | 1.00 × 104 TCID50/mL | – | – | |
| influenza B Yamagata strain | 1.00 × 104TCID50/mL | – | – | |
| Measles virus | 1 × 104 TCID50/mL | – | – | |
| Mumps virus | 1 × 104 TCID50/mL | – | – | |
| Rubella virus | 1 × 104 TCID50/mL | – | – | |
| Varicella zoster virus | 1 × 104 TCID50/mL | – | – | |
| Staphylococcus aureus | 2.3 × 107 CFU/mL | – | – | |
| Pseudomonas aeruginosa | 1.8 × 107 CFU/mL | – | – | |
| SARS-CoV N protein | 2 μg/mL | + | – | |
| MERS-CoV N protein | 2 μg/mL | – | – | |
| Chicken IBV | 1 × 104 EID50/mL | – | – | |
3.5. Detection of clinical samples and compared with RT-PCR
Fifteen identified positive nasopharyngeal swab samples from the COVID-19 infected who underwent routine clinical tests by real-time reverse transcription-polymerase chain reaction (RT-PCR) were prepared using the virus solution. 42 negative nasopharyngeal swabs and 42 negative oropharyngeal swabs were from healthy people as control (Fig S4), the all samples were mixed well with virus split solution at a volume ratio of 1:1 after incubation for 1 min at room temperature, followed by adding 60 μL mix samples solution into the sample window of the test strip, the test results were evaluated after 10 min. Results can be determined as follows: the positive result is judged by the appearance of two red lines in the control and test region. The negative result is judged by the appearance of only a single line in the control region. If no line is present in the test strip or only one line appears in the test region, the test is invalid. The contrast between test strips and RT-PCR in terms of detection time, device, procedure and detection rate are showing in Table 4 .
Table 4.
The comparison of characteristics between test strips and RT-PCR.
| Contrastive items | RT-PCR | N-Test strips |
|---|---|---|
| Positive samples (total 15) | P = 15 | P = 14 |
| Negative nasopharyngeal swabs (total 42) | N = 42 | N = 42 |
| Negative oropharyngeal swabs (total 42) |
N = 42 | N = 42 |
| Detection time | 4 h | 11 min |
| Device requirement | + | – |
| Operation procedure | Complicated | 1 step |
| Detection rate | 100 % | 93.33 % |
4. Discussion
Globally, COVID-19 still has the potential to overburden health systems, although China has made remarkable achievements in fighting against it. This novel coronavirus is highly homologous to the known SARS-CoV-2 coronavirus in respiratory specimens or blood samples and can be used as a diagnostic. Various methodologies based on nuclear acid have been introduced to discriminate COVID-9 in clinical diagnosis and investigate suspected cases (Kong et al., 2020), but the current diagnosis mainly depends on a quantitative reverse transcriptase-polymerase chain reaction to detect the SARS-CoV-2 nucleic acids (Yan et al., 2020; Bordi et al., 2020). These valid technologies can make detection more efficient. However, such methods require specialized instruments, time-consuming, and laborious, the rapid, convenient, and low-cost technologies were still urgently needed to further identify. In this study, we first developed the colloidal gold immunochromatographic assay test strips based on nucleocapsid and spike protein of COVID-19 for rapid and sensitive immunodiagnosis in vitro. In this research, we found that N protein and its mAbs have better specificity and lower cross-reaction to suit for test strip preparation compared with the S protein Ag-mAb system.
COVID-19 spike protein is essential for viral infection and nucleocapsid protein is important for viral RNA transcription. As mentioned above, the coronavirus spike is responsible for binding the cellular receptor, angiotensin-converting enzyme 2 (ACE2). It contains three segments: a large ectodomain, a single-pass transmembrane anchor, and a short intracellular tail. The ectodomain consists of a receptor-binding subunit S1 and a membrane-fusion subunit S2. Previous studies revealed that the spike is a clove-shaped trimer with three S1 heads and a trimeric S2 stalk (Kirchdoerfer et al., 2016; Walls et al., 2016; Li et al., 2003; Wang et al., 2020). Upon receptor binding, conformational changes lead to exposure of a second cleavage site (S2') allowing the fusion of viral and cell membranes. Interestingly, most reports indicate that SARS-CoV-2 S binds to human ACE2 with higher affinity than the SARS-CoV S protein, which may impact viral infectivity for SARS-CoV-2 (Wrapp et al., 2020). The S proteins of SARS-CoV-2 and SARS-CoV are highly homologous, and the structural basis of the interactions between ACE2 and the RBD of SARS-CoV-2 and SARS-CoV, located in the C-terminal portion of S1 (CTD). Previous research suggests that the two S1 domains, the N-terminal portion and C-terminal portion, from different coronavirus genera, have related structural topologies. The sequences, structures and membrane-fusion mechanisms of the S2 subunits are conserved among different coronavirus genera; the S1 subunits of different coronavirus genera, on the contrary, share the same evolutionary origin but have undergone extensive divergent evolution. For instance, S2 of SARS-CoV and SARS-CoV-2 sequence similarity than the respective S1 subunits (∼90 %), and importantly, S2 of SARS-CoV-2 might contain analogously neutralizing epitopes (Li, 2012; Zheng et al., 2006; Xu et al., 2004; Karamloo and König, 2020). It seems to be the conceivable reason for the relatively significant cross-reaction among HCoV−OC43, HCoV-NL63 and HCoV-HKU1 based on S mAbs. Also, previous researches showing the antibodies against the nucleocapsid protein are longer lived and occur in greater abundance in β-CoVs represented by SARS infected patients than antibodies against other viral components such as the spike, membrane and envelope proteins suggest that to some extent, the antibodies against N protein are more stable. Despite, both spike and nucleocapsid are key structural proteins with good immunogenicity, can serve as primary targets for immunoassay theoretically, the N protein Ag-mAb system is more specific and suitable for immunoassay as the sequence conservation of nucleocapsid proteins within the genus is low (Ou et al., 2020; Chen et al., 2004; Kim et al., 2004), it in accordance with our experimental results. Significantly, because SARS-CoV and COVID-19(SARS-CoV-2) are strikingly similar and belong to β genus of Coronaviridae (Fig S6), these immunostrips, of course, also can be used to detect SARS-CoV, and the limit of detection is 1 ng/mL (Fig S5. b).
Besides, it is necessary to screen the matched mAbs to build sandwich immunochromatographic assay test strips. The “matched mAbs” mean the mAbs, which could recognize the epitopes of antigen molecular in the opposite position to avoid the steric hindrance. The reciprocal cross labeling experiments showed high consistency, which suggests the matched mAbs could correctly bind with opposite epitopes of N protein without steric hindrance. In contrast, the reciprocal cross labeling experiments of mAbs against S protein presented unsatisfactory results. Despite, the 2C8a-2H3a couple mAbs showed the darkest color on test strips when 2C8a as capture antibodies and 2H3a as labeled antibodies, the experiment in reverse showed negative (see Supplementary information). It may further suggest that mAbs against N proteins are more suitable for developing immunochromatographic assay test strips.
According to the WHO, the outbreak of COVID-19 has been a pandemic that infected more than 25.32 million people and caused more than 848.25 thousand deaths worldwide at the time of 1st September 2020. Under the global pandemic of coronavirus 2019, RT-PCR in molecular diagnostic labs has played an essential role in clinical diagnosis and the investigation of suspected cases of COVID-19. However, these conventional approaches are laborious, require specialized instruments, and need more than 4 h to detect COVID-19 in clinical samples, for these obvious reasons, not to satisfy a large number of suspected patients, asymptomatic patients, and close contact. In this research, we prepared mAbs against S proteins and N proteins; after detected 20 different respiratory pathogens, we confirmed N protein mAbs has its characteristic signs on the diagnosis of COVID-19 rapid detection and is an excellent method to be selected. The colloidal gold immunochromatographic assay test strips based on N protein mAbs can detect as little as 0.1 ng/mL nucleocapsid antigens, the sensitivity is 10 times high than S protein based (Fig. 5.), the detection limit was consisted with sandwich ELISA results but with much less detection time and much simpler process. It was also worth mentioning that compared with RT-PCR detection for 15 positive nasopharyngeal swab samples, N protein strips can quickly detect 14 positive in 15 within 11 min with a 93.33 % accuracy rate (Table 4.). These rapid test strips can be used in large-scale screening of infected persons and conducive to take the corresponding medical measures will undoubtedly save precious time for the control of COVID-19.
5. Conclusion
The test strip based on N protein mAbs can rapidly discern the COVID-19 in 11 min from the visible signal by the naked eye and exhibited good specificity, accuracy, and stability than S protein mAbs strips. These rapid and sensitive strips are more suitable for some remote and special needs areas restricted by a medical condition or for quick and large screenings.
Ethics declarations
Balb/C mice were kept in a pathogen-free environment and ad-lib fed. The procedures for care and use of animals were approved by the Ethics Committee of the International Graduate School at Shenzhen, Tsinghua University)and all applicable institutional and governmental regulations concerning the ethical use of animals were followed.
Contributions
Dan Liu drafted the manuscript, analyzed data and helped in performing experiments. Feng Wu performed and optimized experiments and helped in analyzing data. Yu Cen, Lei Ye, Xueying Shi, and Yulan Huang helped in performing experiments and preparing monoclonal antibodies. Lan Ma conceptualized, devised and supervised experiments and edited manuscript draft, Shisong Fang performed COVID-19 positive samples detection and contributed to the experimental design and analyses.
Author statement
We have made substantial contributions to the conception or design of the acquisition, analysis, or interpretation of data for the work; and we have drafted the work or revised it critically for important intellectual content; and we have approved the final version to be published; and we agree to be accountable for all aspect of the work in ensuring that questions related to the accuracy or integrity of any part of work are appropriately investigated and resolved. Dan Liu drafted the manuscript, analyzed data and helped in performing experiments. Feng Wu performed and optimized experiments and helped in analyzing data. Yu Cen, Lei Ye, Xueying Shi, and Yulan Huang helped in performing experiments and preparing monoclonal antibodies. Lan Ma conceptualized, devised and supervised experiments and edited manuscript draft, Shisong Fang performed COVID-19 positive samples detection and contributed to the experimental design and analyses.
Declaration of Competing Interest
The authors declare no competing interests.
Acknowledgements
This work was supported by National Key R & D Plan in China (2016YFD0501103), Shenzhen Strategic Emerging Industry Development Special Funds (JCYJ20170816143646446), Shenzhen Science and Technology Research and Development Funds (JCYJ20200109143018683).
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.molimm.2021.01.005.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Yang P., Wang X. COVID-19: a new challenge for human beings. Cell. Mol. Immunol. 2020;17:555–557. doi: 10.1038/s41423-020-0407-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S.R., Leibowitz J.L. Vol. 81. Academic Press; 2011. pp. 85–164. (ADV VIRUS RES). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong N.S., Zheng B.J., Li Y.M., Poon L.L.M., Xie Z.H., Chan K.H., Li P.H., Tan S.Y., Chang Q., Xie J.P., Liu X.Q., Xu J., Li D.X., Yuen K.Y., Peiris J.S.M., Guan Y. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. LANCET. 2003;362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China. New England J. Med. Surg. Collat. Branches Sci. 2019;382(2020):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D., Lin M., Wei L., Xie L., Zhu G., Dela Cruz C.S., Sharma L. Epidemiologic and clinical characteristics of novel coronavirus infections involving 13 patients outside Wuhan. China. JAMA. 2020;323:1092–1093. doi: 10.1001/jama.2020.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter L., Garner L., Smoot J., Li Y., Zhou Q., Saveson C., Sasso J., Gregg A., Soares D., Beskid T., Jervey S., Liu C. Assay techniques and test development for COVID-19 diagnosis. ACS CENTRAL SCI. 2020;6:591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016;3 doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A.H.M., Tomlinson A.C.A., Zhou D., Satkunarajah M., Chen K., Sharon C., Desforges M., Talbot P.J., Rini J.M. Receptor-binding loops in alphacoronavirus adaptation and evolution. Nat. Commun. 2017;8:1735. doi: 10.1038/s41467-017-01706-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride R., van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. VIRUSES. 2014;1999-4915(6):2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.K., Lee Bh Fau - Dutta N.K., Dutta Nk Fau - Seok S.-H., Seok Sh Fau - Baek M.-W., Baek Mw Fau - Lee H.-Y., Lee Hy Fau - Kim D.-J., Kim Dj Fau - Na Y.-R., Na Yr Fau - Noh K.-J., Noh Kj Fau - Park S.-H., Park Sh Fau - Kariwa H., Kariwa H Fau - Nakauchi M., Nakauchi M Fau - Mai L.Q., Mai le Q Fau - Heo S.-J., Heo Sj Fau - Park J.-H., Park J.H. Detection of antibodies against SARS-Coronavirus using recombinant truncated nucleocapsid proteins by ELISA . J MICROBIOL BIOTECHN. 2008;18:1717–1721. [PubMed] [Google Scholar]
- Spiegel M., Pichlmair A., Martínez-Sobrido L., Cros J., García-Sastre A., Haller O., Weber F. Inhibition of Beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J. Virol. 2005;79:2079–2086. doi: 10.1128/JVI.79.4.2079-2086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Fan, Zhao Su, Yu Bin, Chen Yan-Mei, Wang Wen, Song Zhi-Gang, Hu Yi, Tao Zhao-Wu, Tian Jun-Hua, Pei Yuan-Yuan, Yuan Ming-Li, Zhang Yu-Ling, Dai Fa-Hui, Liu Yi, Wang Qi-Min, Zheng Jiao-Jiao, Xu Lin, Holmes Edward C., Zhang Yong-Zhen. A new coronavirus associated with human respiratory disease in China. NAT. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de StGroth S.F., Scheidegger D. Production of monoclonal antibodies: strategy and tactics. J. Immunol. Methods. 1980;35:1–21. doi: 10.1016/0022-1759(80)90146-5. [DOI] [PubMed] [Google Scholar]
- Yokoyama W.M., Christensen M., Santos G.D., Miller D. Production of monocl-onal antibodies. CURR PROT IMMUNOL. 2006;74 doi: 10.1002/0471142735.im0205s74. 2.5.1-2.5.25. [DOI] [PubMed] [Google Scholar]
- Kong W.-H., Li Y., Peng M.-W., Kong D.-G., Yang X.-B., Wang L., Liu M.-Q. SARS-CoV-2 detection in patients with influenza-like illness. Nat. Microbiol. 2020;5:675–678. doi: 10.1038/s41564-020-0713-1. [DOI] [PubMed] [Google Scholar]
- Yan C., Cui J., Huang L., Du B., Chen L., Xue G., Li S., Zhang W., Zhao L., Sun Y. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. CLIN MICROBIOL INFEC. 2020;26:773–779. doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordi L., Piralla A., Lalle E., Giardina F., Colavita F., Tallarita M., Sberna G., Novazzi F., Meschi S., Castilletti C., Brisci A., Minnucci G., Tettamanzi V., Baldanti F., Capobianchi M.R. Rapid and sensitive detection of SARS-CoV-2 RNA using the SimplexaTM COVID-19 direct assay. J. Clin. Virol. 2020 doi: 10.1016/j.jcv.2020.104416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Cottrell C.A., Wang N., Pallesen J., Yassine H.M., Turner H.L., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Pre-fusion structure of a human coronavirus spike protein. NATURE. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Tortorici M.A., Bosch B.J., Frenz B., Rottier P.J.M., DiMaio F., Rey F.A., Veesler D. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. NATURE. 2016;531:114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. NATURE. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.-Y., Wang Q., Zhou H., Yan J., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. CELL. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. SCIENCE. 2020;367:1260. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Evidence for a common evolutionary origin of coronavirus spike protein receptor-binding subunits. J. Virol. 2012;86:2856–2858. doi: 10.1128/JVI.06882-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moh Zaki Ali, van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a Novel Coronavirus from a Man with Pneumonia in Saudi Arabia. New England J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. In press. [DOI] [PubMed] [Google Scholar]
- Zheng Q., Deng Y., Liu J., van der Hoek L., Berkhout B., Lu M. Core structure of S2 from the human coronavirus NL63 spike glycoprotein. BIOCHEMISTRY. 2006;45:15205–15215. doi: 10.1021/bi061686w. [DOI] [PubMed] [Google Scholar]
- Xu Y., Lou Z., Liu Y., Pang H., Tien P., Gao G.F., Rao Z. Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core. J. Biol. Chem. 2004;279:49414–49419. doi: 10.1074/jbc.M408782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.L.M., Samborskiy D.V., Sidorov I.A., Sola I. Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. NAT MICROBIOL. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamloo F., König R. SARS-CoV-2 immunogenicity at the crossroads. ALLERGY. 2020;75:1822–1824. doi: 10.1111/all.14360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. NAT CONMMUN. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Pei D., Jiang L., Song Y., Wang J., Wang H., Zhou D., Zhai J., Du Z., Li B., Qiu M., Han Y., Guo Z., Yang R. Antigenicity analysis of different regions of the severe acute respiratory syndrome coronavirus nucleocapsid protein. Clin. Chem. 2004;50:988–995. doi: 10.1373/clinchem.2004.031096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Lee J.H., Hung C.-F., Peng S., Roden R., Wang M.-C., Viscidi R., Tsai Y.-C., He L., Chen P.-J., Boyd D.A.K., Wu T.C. Generation and characterization of DNA vaccines targeting the nucleocapsid protein of severe acute respiratory syndrome coronavirus. J. Virol. 2004;78:4638–4645. doi: 10.1128/JVI.78.9.4638-4645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.