Highlights
-
•
COVID-19 infection is characterized by its prominent effect on specific ethnic group.
-
•
SARS-CoV-2 cases/mortality were negatively associated with ACE1 II genotype.
-
•
The ACE1 II genotype could be a predictive marker of SARS-CoV-2 risk and severity.
Abbreviations: SARS-CoV-2, severe acute respiratory syndrome coronavirus-2; COVID-19, coronavirus disease 2019; ACE, angiotensin-converting enzyme; SARS, severe acute respiratory syndrome; MERS, Middle East respiratory syndrome; RAAS, renin angiotensin aldosterone system; I, insertion; D, deletion; bp, base pair; SNP, single nucleotide polymorphism; KPGP, Korean Personal Genome Project; FTP, File Transfer Protocol; SNV, single nucleotide variants; VQSR, Variant Quality Score Recalibration; CTSL, cathepsin L1; TMPRSS2, transmembrane serine protease 2; gnomAD, genome aggregation database; Ang, angiotensin; ARDS, acute respiratory distress syndrome; HLA, human leukocyte antigens
Keywords: COVID-19, SARS-CoV-2, ACE1 I/D polymorphism, ACE1 II genotype, Prevalence, Mortality, Ethnicity
Abstract
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) causes coronavirus disease 2019 (COVID-19). The relentless spread and pathogenicity of the virus have become a global public health emergency. One of the striking features of this pandemic is the pronounced impact on specific regions and ethnic groups. In particular, compared with East Asia, where the virus first emerged, SARS-CoV-2 has caused high rates of morbidity and mortality in Europe. This has not been experienced in past global viral infections, such as influenza, severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) and is unique to SARS-CoV-2. For this reason, we investigated the involvement of genetic factors associated with SARS-CoV-2 infection with a focus on angiotensin-converting enzyme (ACE)-related genes, because ACE2 is a receptor for SARS-CoV-2. We found that the ACE1 II genotype frequency in a population was significantly negatively correlated with the number of SARS-CoV-2 cases. Similarly, the ACE1 II genotype was negatively correlated with the number of deaths due to SARS-CoV-2 infection. These data suggest that the ACE1 II genotype may influence the prevalence and clinical outcome of COVID-19 and serve as a predictive marker for COVID-19 risk and severity.
1. Introduction
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) has spread worldwide, and coronavirus disease 2019 (COVID-19) is now a pandemic with over 7.8 million infected people and over 430,000 deaths (as of June 15, 2020) (https://coronavirus.jhu.edu/map.html). Shortly after the first reported case in Wuhan, China, the virus spread rapidly to other Asian countries. SARS-CoV-2 has also spread to Central and Northern Europe and the Americas.
Central Europe has experienced a far greater number of cases and deaths from COVID-19 than East Asia, where the disease originally occurred (Zhu et al., 2020). Such population differences are unique to COVID-19 as they were not recognized in the severe acute respiratory syndrome (SARS) and the Middle East respiratory syndrome (MERS) that emerged in 2002 and 2012, respectively.
In addition to apparent socio-behavioral differences, European and East Asian people have differences that could facilitate SARS-CoV-2 infection including virological and immunological factors. In infectious diseases, such as COVID-19, the host genetic factors thought to define resistance or susceptibility to infection are also important. The angiotensin-converting enzyme (ACE) 2, a host cell receptor for SARS-CoV-2 (Hoffmann et al., 2020), has an analog called ACE1, and these molecules, together with renin and angiotensin, constitute the renin angiotensin aldosterone system (RAAS) (Fountain and Lappin, 2019). RAAS is a general term for hormone systems related to regulation of blood pressure and extracellular volume. The human ACE1 gene on chromosome 17 has an insertion (I) or deletion (D) of a 287 base pair (bp) Alu repeat sequence in intron 16 (Rieder et al., 1999). Therefore, in the I/D polymorphism, there are three different genotypes, II, ID and DD.
European and Asian people have continued a long-term migration eastward and westward across the vast Eurasian continent since the exodus of modern human ancestors from Africa about 200,000 years ago (Stringer and Galway-Witham, 2018) and during that time have achieved genetic variation. Therefore, in the current study, we investigated the genotypic differences in RAAS-related genes, ACE1 and ACE2, to explain the ethnic difference in infection rate/mortality rates due to SARS-CoV-2 infection in European and Asian countries.
2. Materials and methods
2.1. Determination of ACE1 I/D genotypes
ACE1 insertion/deletion (I/D) genotypes were determined from high-coverage sequenced data of the phase 3 panel of the international 1000 Genomes Project (1000Genomes) and the Korean Personal Genome Project (KPGP) (doi: https://doi.org//10.1038/s41598-018-23837-x). The mapped read files in CRAM format of the phase 3 panel of the 1000 Genomes Project were retrieved from the European Nucleotide Archive. The raw read files in FASTQ format were retrieved from a File Transfer Protocol (FTP) server of KPGP. Sequence reads were aligned to GRCh38 reference sequences by BWA-mem (ver. 0.7.17-r1188) followed by the removal of duplicate reads by PicardTools (version 1.93). Single nucleotide variants (SNV) and short insertion/deletion (indel) were identified with GATK 4.1 according to the developer’s protocol. SNV and indel discovery was conducted by HaplotypeCaller for each sample. Genotypes were determined using the GenotypeGVCFs program followed by filtration by Variant Quality Score Recalibration (VQSR). The genotype of ACE1 polymorphism which is a 287 bp insertion of an ALU element at chr17:63488529 was determined by paraGRAPH v2.3 (https://doi.org/10.1186/s13059-019–1909-7).
Data on the geographical variation of the I/D polymorphism of the ACE1 were also collected from published studies by Saab et al and others (Table 1 ), which include studies of 19 countries from Europe, the Middle East, South Asia and East Asia (Sweden, Denmark, United Kingdom, the Netherlands, Hungary, Belgium, Germany, France, Spain, Italy, Turkey, Lebanon, Kuwait, United Arab Emirates, India, China, South Korea, Taiwan and Japan (Saab et al., 2007). Additionally, data on ACE1 I/D genotypes from six countries (Portugal, Switzerland, Poland, Slovakia, Iran and Israel) were retrieved from the literature (Borzyszkowska et al., 2012, Freitas et al., 2008, Frishberg et al., 1998, Nikzamir et al., 2008, Siváková et al., 2009, Walder et al., 1998).
Table 1.
ACE1 II genotype frequency and the number of COVID-19 cases and deaths among European, Middle Eastern, South Asian and East Asian countries.
| Country | Study authors | Year of |
No. of |
ACE1 II |
#COVID-19 cases | case/pop |
deaths | death/pop |
|---|---|---|---|---|---|---|---|---|
| publication | subjects | frequency (%) | (n/million) | (n/million) | ||||
| Sweden | Kurland et al. | 2001 | 59 | 27 | 33,188 | 3307 | 3,992 | 398 |
| Denmark | Bladbjerg et al. | 1999 | 199 | 23 | 11,487 | 1990 | 561 | 97 |
| United Kingdom | Kehoe et al. | 1999 | 386 | 23 | 258,504 | 3828 | 36,757 | 544 |
| United Kingdom | Steeds et al. | 2001 | 507 | 22 | 258,504 | 3828 | 36,757 | 544 |
| United Kingdom | Narain et al. | 2000 | 342 | 18 | 258,504 | 3828 | 36,757 | 544 |
| Netherlands | Hosoi et al. | 1996 | 61 | 20 | 45,265 | 2648 | 5,830 | 341 |
| Hungary | Barkai et al. | 2005 | 120 | 27 | 3,741 | 386 | 482 | 50 |
| Belgium | Gu et al. | 1994 | 109 | 19 | 56,810 | 4923 | 9,237 | 800 |
| Germany | Ebert et al. | 2005 | 145 | 23 | 179,986 | 2155 | 8,261 | 99 |
| Germany | Filler et al. | 2001 | 200 | 18 | 179,986 | 2155 | 8,261 | 99 |
| France | Blanche et al. | 2001 | 560 | 18 | 182,036 | 2795 | 28,218 | 433 |
| France | Girerd et al. | 1998 | 340 | 17 | 182,036 | 2795 | 28,218 | 433 |
| Spain | Alvarez et al. | 1999 | 400 | 15 | 235,290 | 5034 | 28,678 | 614 |
| Spain | Coll et al. | 2003 | 133 | 15 | 235,290 | 5034 | 28,678 | 614 |
| Italy | Di Pasquale et al. | 2005 | 684 | 18 | 229,327 | 3787 | 32,735 | 541 |
| Italy | Panza et al. | 2002 | 252 | 13 | 229,327 | 3787 | 32,735 | 541 |
| Portugal | Freitas et al. | 2008 | 510 | 16 | 36,690 | 3569 | 1,517 | 148 |
| Switzerland | Walder et al. | 1998 | 199 | 25 | 31,117 | 3631 | 1,938 | 226 |
| Poland | Borzyszkowska et al. | 2012 | 632 | 29 | 29,392 | 774 | 1,247 | 33 |
| Slovakia | Siváková et al. | 2009 | 209 | 25 | 1,548 | 284 | 28 | 5 |
| Iran | Nikzamiret et al. | 2008 | 51 | 16 | 187,427 | 2291 | 8,837 | 108 |
| Israel | Frishberg et al. | 1998 | 216 | 10 | 19,121 | 2152 | 302 | 34 |
| Turkey | Tanriverdi et al. | 2005 | 102 | 23 | 150,388 | 1834 | 4,182 | 51 |
| Turkey | Serdaroglu et al. | 2005 | 287 | 23 | 150,388 | 1834 | 4,182 | 51 |
| Turkey | Bedir et al. | 1999 | 143 | 23 | 150,388 | 1834 | 4,182 | 51 |
| Lebanon | Saab et al. | 2007 | 570 | 7 | 1089 | 159 | 27 | 4 |
| Kuwait | Al-Eisa et al. | 2001 | 48 | 2 | 19,858 | 4800 | 145 | 35 |
| United Arab Emirates | Saeed et al. | 2005 | 130 | 6 | 27,198 | 2824 | 231 | 24 |
| India | Patil et al. | 2005 | 300 | 26 | 124,476 | 92 | 4,059 | 3 |
| China | Thomas et al. | 2001 | 119 | 33 | 84,084 | 59 | 4,638 | 3 |
| China | Ohishi et al. | 1994 | 175 | 37 | 84,084 | 59 | 4,638 | 3 |
| China | Young et al. | 1998 | 183 | 39 | 84,084 | 59 | 4,638 | 3 |
| China | Iwai et al. | 1994 | 122 | 41 | 84,084 | 59 | 4,638 | 3 |
| China | Yan et al. | 2005 | 352 | 41 | 84,084 | 59 | 4,638 | 3 |
| Korea, South | Ryu et al. | 2002 | 167 | 34 | 11,190 | 218 | 266 | 5 |
| Korea, South | Um et al. | 2003 | 613 | 37 | 11,190 | 218 | 266 | 5 |
| Taiwan | Lee et al. | 2002 | 750 | 47 | 441 | 19 | 7 | 0 |
| Japan | Katoh et al. | 2005 | 270 | 41 | 16,536 | 130 | 808 | 6 |
| Japan | Odawara et al. | 1997 | 248 | 42 | 16,536 | 130 | 808 | 6 |
| Japan | Mannami et al. | 2001 | 3657 | 43 | 16,536 | 130 | 808 | 6 |
| Japan | Maguchi et al. | 1996 | 84 | 48 | 16,536 | 130 | 808 | 6 |
| Japan | Ishigami et al. | 1995 | 87 | 51 | 16,536 | 130 | 808 | 6 |
| Japan | 1000G JPT | – | 104 | 33 | 16,536 | 130 | 808 | 6 |
| Korea, South | KPGP | – | 88 | 41 | 11,190 | 218 | 266 | 5 |
Information of ACE1 II genotype frequencies in each country were found in the references listed in the left half of the table, while those on the number of COVID-19 cases and deaths were collected from the Center for Systems Science and Engineering at Johns Hopkins University (https://coronavirus.jhu.edu/map.html) (as of May 23, 2020) and listed in the right half of the table, respectively.
2.2. Single nucleotide polymorphism (SNP) and the allele frequency of the ACE2, CTSL and TMPRSS2
We obtained the allele frequency of the genes, ACE2, cathepsin L (CTSL) and transmembrane serine protease 2 (TMPRSS2) likely to be involved in the infection/pathogenicity of SARS-CoV-2 (Hoffmann et al., 2020; Ou et al., 2020), from the Genome Aggregation Database (gnomAD) (https://gnomad.broadinstitute.org/). gnomAD summarizes the variant information found in the whole genome sequences of about 140,000 people.
2.3. COVID-19 cases in each country
The number of COVID-19 cases and related fatalities were collected from the Center for Systems Science and Engineering at Johns Hopkins University, as of May 23, 2020 (https://coronavirus.jhu.edu/map.html). The population of each country was obtained from the United Nations Population Division to calculate the number of cases and deaths per 1 million people.
ACE1 II genotype frequencies and the number of COVID-19 cases and deaths among European, Middle Eastern, South Asian and East Asian countries are summarized in Table 1.
3. Results
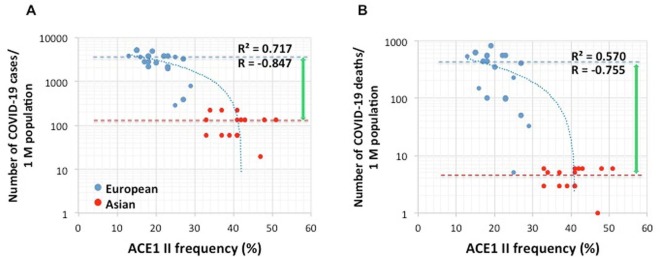
First, we investigated the correlation between ACE1 I/D genotype and SARS-CoV-2 morbidity and mortality in Europe and Asia. Both the number of patients infected with SARS-CoV-2 (Fig. 1 A) and the number of deaths from COVID-19 (Fig. 1B) were negatively correlated with the ACE1 II genotype frequency, and the negative correlations were strong (R= − 0.847 and − 0.755, respectively). This suggests that the ACE1 II genotype may affect the prevalence and clinical outcome of COVID-19. The European population has a lower ACE1 II genotype frequency and a higher prevalence of and mortality due to COVID-19 than the Asian population. The ACE1 II genotype frequency increases according to an eastward trend from European to Asian countries (Saab et al., 2007). A comparison of the median values obtained for cases/population and deaths/population of the European and Asian populations shows that the difference between deaths/population is greater than that between cases/population. This finding indicates that the European population has, in addition to a higher incidence of SARS-CoV-2 infection and associated disease, a higher probability of dying from the disease.
Fig. 1.
Correlation between COVID-19 prevalence and ACE1 II allele frequency (%) in Europe and Asia: (A) the number of COVID-19 cases/ million populations, R2 = 0.717, R=- 0.847, (B) the number of COVID-19-related deaths/ million population, R2 = 0.570, R=- 0.755. The dashed lines indicate the median values.
A similar analysis was performed, adding data points from countries in the Middle East and India collected from the literature (Table 1), and the same pattern as described above was observed (Fig. 2 ). Though weaker than the correlation observed without the Middle Eastern populations, the number of infected individuals still showed a fairly strong correlation with the ACE1 II genotype (R = − 0.732) (Fig. 2A). Again, the comparison of the median values indicated that the European and Middle Eastern populations have a higher probability of acquiring SARS-CoV-2 infection compared with the Asian population. The correlation between the ACE1 II genotype and the number of deaths was also weaker than that observed without Middle Eastern population but still demonstrated a moderate negative correlation (R = −0.452) (Fig. 2B).
Fig. 2.
Correlation between COVID-19 prevalence and ACE1 II allele frequency (%) in Europe, the Middle East, India and East Asia: (A) the number of COVID-19 cases/ million populations, R2 = 0.536, R=- 0.732, (B) the number of COVID-19-related deaths/ million population, R2 = 0.204, R=- 0.452. The dashed lines indicate the median values.
Other genes involved in the pathogenicity of SARS-CoV-2 such as ACE2, CTSL and TMPRSS2 were also studied; however, no significant correlation with COVID-19 prevalence or mortality was observed so far.
4. Discussion
Our results suggest that the ACE1 I/D polymorphism may be one of the genetic markers for SARS-CoV-2 infectivity and pathogenicity. In addition to medical issues, a polymorphism in the ACE1 , which is located in chromosome 17, is anthropologically very interesting. The geographical eastward migration of humans seems to coincide with a gradual increase in the frequency of the ACE1 II genotype. Based on the Mantel test results, there is a significant correlation between geographical distance and genetic distance between populations (r = 0.478984, P < 0.0001) (Saab et al., 2007). It is therefore noteworthy that an increased frequency of the II genotype in the ACE1 was inversely correlated with susceptibility to SARS-CoV-2 infection and consequent mortality. Therefore, mortality was not simply proportional to the number of infected/diseased cases, but appeared to be affected independently and additionally to the infected/diseased state. Under physiological conditions, the signal from angiotensin (Ang) II generated by ACE1 is related to pathological conditions such as vasoconstriction, inflammation and fibrosis, and ACE2-derived peptide, Ang 1–7, reverses the action of Ang II via the Mas receptor (Xiao et al., 2020). Therefore, the balance between ACE1 and ACE2 is considered to be very important for maintaining homeostasis in the body (Chappel and Ferrario, 2006, Gemmati et al., 2020).
It has been reported that serum levels of ACE1 are significantly higher in those with the DD genotype compared with those with either the ID or II genotypes (Rigat et al., 1990). Moreover, because ACE2 is a receptor for SARS-COV-2 (Hoffmann et al., 2020), viral infection may lead to the suppression of ACE2 function and cause ACE1/ACE2 imbalance responsible for RAAS over-activation and pulmonary shut-down (Gemmati et al., 2020). This can further reduce the effects of ACE2, which counteract the pathophysiological effects of Ang II produced by ACE1, and may worsen the pathology. In patients with the D allele, especially those with the DD genotype, the risk of morbidity and mortality from acute respiratory distress syndrome (ARDS) (Marshall et al., 2002, Adamzik et al., 2007) and certain heart, lung and inflammatory conditions (Gard, 2010) is reported to be higher. The ACE1/ACE2 imbalance predicts that COVID-19 patients with the D allele of ACE1, especially the DD genotype will have a higher severity of disease as seen in SARS patients with an ACE1 DD genotype (Itoyama et al., 2004). However, one report found that the ACE1 I/D polymorphism is not directly related to susceptibility to SARS-CoV infection nor the development of SARS (Chan et al., 2005).
ACE1 I/D polymorphism is also thought to be associated with the cough reflex (Morimoto et al., 2002). It is known that aspiration due to a decrease in cough with aging is the main cause of pneumonia in the elderly. Morimoto et al. reported that the ACE1 D allele contributes to the risk of pneumonia in elderly Japanese individuals (Morimoto et al., 2002). They propose a model in which the high expression of ACE1 in the ACE1 DD genotype reduces substance P and increases the risk of pneumonia due to a reduced cough reflex. Additionally, a Chinese group conducted a meta-analysis of 12 studies on pneumonia and ACE1 including studies from Japan and the Netherlands, and concluded that the ACE1 I/D polymorphism and pneumonia risk are significantly associated (Nie et al., 2014). These studies, which were conducted during the pre-COVID-19 era, are in relative agreement with the observation of many fewer cases and deaths due to the COVID-19 in Asian populations with a high frequency of ACE1 II genotypes.
We discovered that a gene (ACE1) and its genotype (I/D) seem to provide a plausible explanation for why some ethnic groups, especially Europeans populations, have been more heavily affected by SARS-CoV-2 than Asians populations. We believe that this is a very unique and important issue in infectious diseases. Such an ethnic difference was never experienced in the past, even in the influenza pandemic of 1918 that caused high rates of mortality or with SARS and MERS, which in many respects are thought to be very similar to COVID-19. In this analysis, we have shown for the first time that the prevalence of the D allele in the ACE1 gene is integrally involved in susceptibility to SARS-CoV-2 infection and the exacerbation of COVID-19 symptoms such as pneumonia. The association of the I/D polymorphism in the ACE1 with susceptibility and outcome of ARDS has been reported (Marshall et al., 2002). This finding is closely linked to the current results observed in patients infected with SARS-CoV-2.
Middle Eastern populations, especially, those from Lebanon, are believed to be ancestral with regard to the ACE polymorphism and have a relatively low frequency of the insertion allele (Saab et al., 2007). In our analysis of data from the literature, which included Middle Eastern countries, the association between SARS-CoV-2 infections/mortalities and ACE1 I/D genotype was clearly reduced (Fig. 2). Indeed, Delanghe et al. recently showed a correlation between increasing D alleles and decreasing COVID-19 morbidity/mortality from an analysis of 33 countries in Europe, North Africa and the Middle East (Delanghe et al., 2020). This is contrary to our observation. In North Africa and the Middle East, the onset of SARS-CoV-2 was clearly delayed compared with Asia and Europe. However, it is reported that COVID-19 now poses a formidable threat to fragile countries in the Middle East (https://coronavirus.jhu.edu/map.html). This suggests that more complex factors are involved in the Middle East. This view may be further supported from our observations that people in Europe and the Middle East have a higher probability of acquiring SARS-CoV-2 infection compared with the Asian people, whereas the correlation between ACE1 II genotype and COVID-19 mortality was weakened when data from the Middle East was added (Fig. 1, Fig. 2). Will the Middle East continue to have the unique situation of high numbers of infected people and low deaths? Therefore, the relationship between SARS-CoV-2 and genetic factors in the Middle East is an important focus for future investigation.
In the Americas, although the onset of infection was slightly delayed compared with Europe, the number of cases of SARS-CoV-2 infection (at the time of this communication) is rapidly increasing. The situation in the United States is the most serious with the number of people infected or deceased now as large as nearly 30% of the infected/deceased worldwide. Similarly, the number of new cases is rapidly increasing in Latin America and the Caribbean, including Brazil, Peru, Chile and Mexico. Populations in these countries were generally formed by immigrants from around the world in addition to native Americans originally coming from Asia. In this sense, analytical research focusing on ethnic differences is important. The advantage of research in these countries is that it makes it possible to investigate the racial differences that COVID-19 infection causes in very similar environments. It is anticipated that, in the near future, the accumulated data from the United States and other countries will provide a more accurate understanding of the role of ACE1 I/D polymorphism in SARS-CoV-2 infection and COVID-19-related morbidity/mortality.
Regarding the genetic predisposition of COVID-19, other genes may be involved, and a combination of multiple genes may affect the severity of infection. Although SNPs occur with high frequency in ACE2, CTSL and TMPRSS2, and over 1% of differences among populations are present in these genes, the significance of these SNPs in SARS-CoV-2 infection/pathogenicity is not clear. Human leukocyte antigens (HLA) gene polymorphisms are associated with various diseases such as autoimmune diseases and infectious diseases. Because HLA is a protein of the immune system responsible for antigen presentation, HLA has been attracting attention in relation to disease susceptibility. However, there is no correlation between the global distribution of HLA alleles frequency and allele ability to bind SARS-CoV-2 peptides (Barquera et al., 2020, Nguyen et al., 2020). Furthermore, there is as yet no report of HLA involvement in SARS-CoV-2 infection and COVID-19 pathology.
5. Conclusions
There was a strong negative correlation between the number of SARS-CoV-2 cases and the number of deaths due to viral infection, which decreased with increasing ACE1 II genotype frequency. This suggests that the ACE1 I/D genotype may be involved in various pathological conditions caused by SARS-CoV-2 infection such as pneumonia, disseminated intravascular coagulation and thrombosis, ischemic stroke, renal injury and immune response such as cytokine storm. Therefore, an urgent task is to assess the clinical outcome of SARS-CoV-2 infection in DD, ID, and II carriers and study the exact role of ACE1. Further studies on COVID-19 and ACE1 polymorphisms are expected to promote the prediction of high-risk groups and the treatment of COVID-19 patients.
CRediT authorship contribution statement
Naoki Yamamoto: Writing - original draft, Investigation, Supervision. Yasuo Ariumi: Writing - original draft, Investigation. Nao Nishida: Formal analysis, Software. Rain Yamamoto: Formal analysis, Software. Georg Bauer: Investigation, Conceptualization. Takashi Gojobori: Investigation, Conceptualization. Kunitada Shimotohno: Investigation, Conceptualization, Supervision. Masashi Mizokami: Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
References
- Adamzik M., Frey U., Sixt S., Knemeyer L., Beiderlinden M., Peters J., Siffert W. ACE I/D but not AGT (-6)A/G polymorphism is a risk factor for mortality in ARDS. Eur. Respir. J. 2007;29:482–488. doi: 10.1183/09031936.00046106. [DOI] [PubMed] [Google Scholar]
- Al-Eisa A., Haider M.Z., Srivastva B.S. Angiotensin converting enzyme gene insertion/deletion polymorphism in idiopathic nephrotic syndrome in Kuwaiti Arab children. Scand. J. Urol. Nephrol. 2001;35:239–242. doi: 10.1080/003655901750292033. [DOI] [PubMed] [Google Scholar]
- Alvarez R., Alvarez V., Lahoz C.H., Martínez C., Peña J., Sánchez J.M., Guisasola L.M., Salas-Puig J., Morís G., Vidal J.A., Ribacoba R., Menes B.B., Uría D., Coto E. Angiotensin converting enzyme and endothelial nitric oxide synthase DNA polymorphisms and late onset Alzheimer’s disease. J. Neurol. Neurosurg. Psychiatry. 1999;67:733–736. doi: 10.1136/jnnp.67.6.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkai L., Soos A., Vamosi I. Association of angiotensin-converting enzyme DD genotype with 24-h blood pressure abnormalities in normoalbuminuric children and adolescents with type 1 diabetes. 2005. Diabet. Med. 2005;22:1054–1059. doi: 10.1111/j.1464-5491.2005.01601.x. [DOI] [PubMed] [Google Scholar]
- Barquera R., Collen E., Di D., Buhler S., Teixeira J., Llamas B., Nunes J.M., Sanchez-Mazas A. Binding affinities of 438 HLA proteins to complete proteomed of seven pandemic viruses and distributions of strongest and weakest HLA peptide binders in populations worldwide. HLA, in press. 2020 doi: 10.1111/tan.13956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedir A., Arik N., Adam B., Kilinc K., Gumus T., Guner E. Angiotensin converting enzyme gene polymorphism and activity in Turkish patients with essential hypertension. Ame. J. Hypertens. 1999;12:1038–1043. doi: 10.1016/S0895-7061(99)00096-5. [DOI] [PubMed] [Google Scholar]
- Bladbjerg, E.M., Andersen-Ranberg, K., de Maat, M.P., Kristensen, S.R., Jeune, B., Gram, J., Jespersen. 1999. Longevity is independent of common variations in genes associated with cardiovascular risk. Thromb. Haemost., 82, 1100-1105 https://doi.org/10.1055/s-0037-1614336. [PubMed]
- Blanche H., Cabanne L., Sahbatou M., Thomas G. A study of French centenarians: are ACE and APOE associated with longevity? C. R. Acad. Sci. III. 2001;324:129–135. doi: 10.1016/s0764-4469(00)01274-9. [DOI] [PubMed] [Google Scholar]
- Borzyszkowska J., Stanislawska-Sachadyn A., Wirtwein M., Sobiczewski W., Ciecwierz D., Targonski R., Gruchala M., Rynkiewicz A., Limon J. Angiotensin converting enzyme gene polymorphism is associated with severity of coronary artery disease in men with high total cholesterol levels. J. Appl. Genet. 2012;53:175–182. doi: 10.1007/s13353-012-0083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.C., Tang N.L.S., Hui D.S.C., Chung G.T.Y., Wu A.K.L., Chim S.S.C., Chiu R.W.K., Lee N., Choi K.W., Sung Y.M., Chan P.K.S., Tong Y.K., Lai S.T., Yu W.C., Tsang O., Lo Y.M.D. Absence of association between angiotensin converting enzyme polymorphism and development of adult respiratory distress syndrome in patients with severe acute respiratory syndrome: a case control study. BMC Infect. Dis. 2005;5:26. doi: 10.1186/1471-2334-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappel M.C., Ferrario C.M. ACE and ACE2: Their Role to Balance the Expression of Angiotensin II and angiotensin-(1–7) Kidney Int. 2006;70(1):8–10. doi: 10.1038/sj.ki.5000321. [DOI] [PubMed] [Google Scholar]
- Coll E., Campos B., González-Núñez D., Botey A., Poch E. Association between the A1166C polymorphism of the angiotensin II receptor type 1 and progression of chronic renal insufficiency. J. Nephrol. 2003;16:357–364. [PubMed] [Google Scholar]
- Delanghe J.R., Speeckaert M.M., Buyzere M.L. COVID-19 infections are also affected by human ACE1 D/I polymorphism. Clin. Chem. Lab. Med. 2020;58:1125–1126. doi: 10.1515/cclm-2020-0425. [DOI] [PubMed] [Google Scholar]
- Di Pasquale P., Cannizzaro S., Scalzo S., Maringhini G., Pipitone F., Fasullo S., Giubilato A., Ganci F., Vitale G., Sarullo F.M., Paterna S. Cardiovascular effects of I/D angiotensin-converting enzyme gene polymorphism in healthy subjects. Findings after follow-up of six years. Acta Cardiol. 2005;60:427–435. doi: 10.2143/AC.60.4.2004993. [DOI] [PubMed] [Google Scholar]
- Ebert M.P.A., Lendeckel U., Westphal S., Dierkes J., Glas J., Folwaczny C., Roessner A., Stolte M., Malfertheiner P., Röcken C. The angiotensin I-converting enzyme gene insertion/deletion polymorphism is linked to early gastric cancer. Cancer Epidemiol. Biomarkers Prev. 2005;14:2987–2989. doi: 10.1158/1055-9965.EPI-05-0411. [DOI] [PubMed] [Google Scholar]
- Filler G., Yang F., Martin A., Stolpe J., Neumayer H.H., Hocher B. Renin angiotensin system gene polymorphisms in pediatric renal transplant recipients. Pediatr. Transplant. 2001;5:166–173. doi: 10.1034/j.1399-3046.2001.00053.x. [DOI] [PubMed] [Google Scholar]
- Fountain, J. H., Lappin, S.L (5 May 2019). “Physiology, Renin-Angiotensin System”. NCBI. NIH. Retrieved 9 May2019. [PubMed]
- Freitas, A.I., Mendonça, I., Brión, M. Sequeira, M.M., Reis, R.P., Carracedo, A., Brehm, A. 2008. RAS gene polymorphisms, classical risk factors and the advent of coronary artery disease in the Portuguese population. BMC Cardiovasc. Disord., 8 (2008) 15 http://www.biomedcentral.com/1471-2261/8/15. [DOI] [PMC free article] [PubMed]
- Frishberg Y., Becker-Cohen R., Halle D., Feigin E., Eisenstein B., Halevy R., Lotan D., Juabeh I., Ish-Shalom N., Magen D., Shvil Y., Sinai-Treiman L., Drukker A. Kidney Int. 1998;54:1843–1849. doi: 10.1046/j.1523-1755.1998.00218.x. [DOI] [PubMed] [Google Scholar]
- Gard, P.R. 2010. Implications of the angiotensin converting enzyme gene insertion/deletion polymorphism in health and disease: a snapshot review. Int. J. Mol. Epidemiol.Genet., 1, 145-157 https://www.ijmeg.org/ijmeg102003. [PMC free article] [PubMed]
- Gemmati D., Bramanti B., Serino M.L., Secchiero P., Zauli G., Tisato V. COVID-19 and individual genetic susceptibility/receptivity: Role of ACE1/ACE2 genes, immunity, inflammation and coagulation. Might the double X-chromosome in females be protective against SARS-CoV-2 compared to the single X-chromosome in male? Int. J. Mol. Sci. 2020;21:3474. doi: 10.3390/ijms21103474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girerd, X., Hanon, O., Mourad, J.J., Boutouyrie, P., Laurent, S., Jeunemaitre. 1998. Lack of association between renin-angiotensin system, gene polymorphisms, and wall thickness of the radial and carotid arteries. Hypertension, 32, 579-583 https://doi.org/10.1161/01.HYP.32.3.579. [DOI] [PubMed]
- Gu X.X., Spaepen M., Guo C., Fagard R., Amery A., Lijnen P., Cassiman J.J. Lack of association between the I/D polymorphism of the angiotensin-converting enzyme gene and essential hypertension in a Belgian population. J. Hum. Hypertens. 1994;8:683–685. [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrier T., Erichsen S., Schiergens T.S., Herrier G., Wu N.-H., Nitsche A., Müller M.A., Drostern C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven proteaseinhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoi M., Nishizawa Y., Kogawa K., Kawagishi T., Konishi T., Maekawa K., Emoto M., Fukumoto S., Shioi A., Shoji T., Inaba M., Okuno Y., Morii H. Angiotensin-converting enzyme gene polymorphism is associated with carotid arterial wall thickness in non-insulin-dependent diabetic patients. Circulation. 1996;94:704–707. doi: 10.1161/01.CIR.94.4.704. [DOI] [PubMed] [Google Scholar]
- Ishigami T., Iwamoto T., Tamura K., Yamaguchi S., Iwasawa K., Uchino K., Umemura S., Ishii M. Angiotensin I converting enzyme (ACE) gene polymorphism and essential hypertension in Japan. Ethnic difference of ACE genotype. Ame. J. Hypertens. 1995;8:95–97. doi: 10.1016/0895-7061(94)00184-D. [DOI] [PubMed] [Google Scholar]
- Itoyama S., Keicho N., Quy T., Phi C.P., Long H.T., Ha L.D., Ban V.V., Ohashi J., Hijikata M., Matsushita I., Kawana A., Yanai H., Kirikae T., Kuratsuji T., Sasazuki T. ACE1 polymorphism and progression of SARS. Biochem. Biophys. Res. Commun. 2004;323:1124–1129. doi: 10.1016/j.bbrc.2004.08.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai N., Ohmichi N., Nakamura Y., Kinoshita M. DD genotype of the angiotensin-converting enzyme gene is a risk factor for left ventitricular hypertrophy. Circulation. 1994;90:2622–2628. doi: 10.1161/01.CIR.90.6.2622. [DOI] [PubMed] [Google Scholar]
- Katoh T., Suzuki H., Sakuma Y., Watanabe T. Relationship of PAI-I 4G/5G polymorphism and IgA nephropathy. Nephrology. 2005;10:A434. doi: 10.1111/j.1440-1797.2005.00517.x. [DOI] [Google Scholar]
- Kehoe P.G., Russ C., Mcllory S., Williams H., Holmans P., Holmes C., Liolitsa D., Vahidassr D., Powell J., McGleenon B., Liddell M., Plomin R., Dynan K., Williams N., Neal J., Cairns N.J., Wilcock G., Passmore P., Lovestone S., Williams J., Owen M.J. Variation in DCP1, encoding ACE, is associated with susceptibility to Alzheimer Disease. Nat. Genet. 1999;21:71–72. doi: 10.1038/5009. [DOI] [PubMed] [Google Scholar]
- Kurland, L. Melhus, H., Karlsson, J., Kahan, T., Malmqvist, K., Ohman, K.P., Nyström, F., Hägg, A., Lind, L., Swedish Irbesartan Left Ventricular Hypertrophy Investigation versus Atenolol (SILVHIA) Trial. 2001. Angiotensin converting enzyme gene polymorphism predicts blood pressure response to angiotensin II receptor type 1 antagonist treatment in hypertensive patients. J. Hypertens., 19, 1783-1787 https://doi.org/10.1097/00004872-200110000-00012. [DOI] [PubMed]
- Lee Y.-J., Tsai J.C. ACE gene insertion/deletion polymorphism associated with 1998 World Health Organization definition of metabolic syndrome in Chinese type 2 diabetic patients. Diabetes Care. 2002;25:1002–1008. doi: 10.2337/diacare.25.6.1002. [DOI] [PubMed] [Google Scholar]
- Maguchi M., Kohara K., Okura T., Li S., Takezaki M., Nishida W., Hiwada K. Angiotensin-converting enzyme gene polymorphism in essential hypertensive patients in Japanese population. Angiology. 1996;47:643–648. doi: 10.1177/000331979604700702. [DOI] [PubMed] [Google Scholar]
- Mannami T., Katsuya T., Baba S., Inamoto N., Ishikawa K., Higaki J., Ogihara T., Ogata J. Low potentiality of angiotensin-converting enzyme gene insertion/deletion polymorphism as a useful predictive marker for carotid atherogenesis in a large general population of a Japanese city: The Suita Study. Stroke. 2001;32:1250–1256. doi: 10.1161/01.STR.32.6.1250. [DOI] [PubMed] [Google Scholar]
- Marshall R.P., Webb S., Bellingan G.J., Montgomery H.E., Chaudhari B., McAnulty R.J., Humphries S.E., Hill M.R., Laurent G.J. Angiotensin converting enzyme insertion/deletion polymorphism is associated with susceptibility and outcome in acute respiratory distress syndrome. Ame. J. Respir. Crit. Care Med. 2002;166:646–650. doi: 10.1164/rccm.2108086. [DOI] [PubMed] [Google Scholar]
- Morimoto S., Okaishi K., Onishi M., Katsuya T., Yang J., Okuro M., Sakurai S., Onishi T., Ogihara T. Deletion allele of the angiotensin- converting enzyme gene as a risk factor for pneumonia in elderly patients. Ame. J. Med. 2002;112:89–94. doi: 10.1016/S0002-9343(01)01071-3. [DOI] [PubMed] [Google Scholar]
- Narain Y., Murphy A.Y.T., Brayne C., Easton D., Evans J.G., Xuereb J., Cairns N., Esiri M.M., Furlong A., Rubinsztein D.C. The ACE gene and Alzheimer’s disease susceptibility. J. Med. Genet. 2000;37:695–697. doi: 10.1136/jmg.37.9.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, A., David, J.K., Maden, S.K., Wood, M.A., Weeder, B.R., Nellore, A., Thompson, R.F. 2020. Human leukocyte antigen susceptibility map for SARS-CoV-2. J. Virol., in press. https://doi.org/10.1128/JVI.00510-20. [DOI] [PMC free article] [PubMed]
- Nie W., Zang Y., Chen J., Chen J., Liu T., Xiao L., Xiu Q. Angiotensin-converting enzyme I/D polymorphism is associated with pneumonia risk: a meta-analysis. J. Renin Angiotensin Aldosterone Syst. 2014;15:585–592. doi: 10.1177/1470320313507622. [DOI] [PubMed] [Google Scholar]
- Nikzamir, A., Nakhjavani, M., Golmohamadi, T., Dibai, L. 2008. Association of angiotensin-converting enzyme gene insetion/deletion polymorphism with metabolic syndrome in Iranians with type 2 diabetes Mellitus. Arch. Iranian Med., 11, 3-9 http://www.ams.ac.ir/AIM/NEWPUB/08/11/1/004.pdf. [PubMed]
- Odawara M., Matsunuma A., Yamashita K. Mistyping frequency of the angiotensin-converting enzyme gene polymorphism and an improved method for its avoidance. Hum. Genet. 1997;100:163–166. doi: 10.1007/s004390050484. [DOI] [PubMed] [Google Scholar]
- Ohishi M., Rakugi H., Ogihara T. Association between a deletion polymorphism of the angiotensin-converting-enzyme gene and left ventricular hypertrophy. N. Engl. J. Med. 1994;331:1097–1098. doi: 10.1056/NEJM199410203311616. [DOI] [PubMed] [Google Scholar]
- Panza F., Solfrizzi V., D’Introno A., Capurso C., Colaiccco A.M., Argentieri G., Capurso A. Lack of association between Ace polymorphism and Alzheimer’s disease in Southern Italy. Arch. Gerontol. Geriatr. Suppl. 2002;8:239–245. doi: 10.1016/s0167-4943(02)00140-1. [DOI] [PubMed] [Google Scholar]
- Patil S., Gulati S., Khan F., Tripathi M., Ahmed M., Agrawal S. Angiotensin converting enzyme gene polymorphism in Indian children with steroid sensitive nephrotic syndrome. Indian J. Med. Sci. 2005;59:431–435. doi: 10.4103/0019-5359.17049. [DOI] [PubMed] [Google Scholar]
- Rieder M.J., Taylor S.L., Clark A.G., Nickerson D.A. Sequence variation in the human angiotensin converting enzyme. Nat. Genet. 1999;22:59–62. doi: 10.1038/8760. [DOI] [PubMed] [Google Scholar]
- Rigat B., Hubert C., Alhenc-Gelas F., Cambien F., Corvol P., Soubrier F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J. Clin. Invest. 1990;86:1343–1346. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu S.K., Cho E.Y., Park H.Y., Im E.K., Jang Y.S., Shin G.J., Shim W.H., Cho S.Y. Renin-angiotensin-aldosterone system (RAAS) gene polymorphism as a risk factor of coronary in-stent restenosis. Yonsei Med. J. 2002;43:461–472. doi: 10.3349/ymj.2002.43.4.461. [DOI] [PubMed] [Google Scholar]
- Saab Y.B., Gard P.R., Overall A.D.J. The geographic distribution of the ACE II genotype: a novel finding. Genet. Res. Camb. 2007;89:259–267. doi: 10.1017/S0016672307009019. [DOI] [PubMed] [Google Scholar]
- Saeed M., Saleheen D., Siddiqui S., Khan A., Butt Z.A., Frossard P.M. Association of angiotensin converting enzyme gene polymorphisms with left ventricular hypertrophy. Hypertens. Res. 2005;28:345–349. doi: 10.1291/hypres.28.345. [DOI] [PubMed] [Google Scholar]
- Serdaroglu E., Mir S., Berdeli A., Aksu N., Bak M. ACE gene insertion/deletion polymorphism in childhood idiopathic nephrotic syndrome. Pediatr. Nephrol. 2005;20:1738–1743. doi: 10.1007/s00467-005-2010-x. [DOI] [PubMed] [Google Scholar]
- Siváková D., Lajdová A., Basistová Z., Cvícelová M., Blazícek P. ACE insertion/deletion polymorphism and its relationship to the components of metabolic syndrome in elderly Slovaks. Anthropol. Anz. 2009;67:1–11. doi: 10.1127/0003-5548/2009/0001. [DOI] [PubMed] [Google Scholar]
- Steeds R.P., Wardle A., Smith P.D., Martin D., Channer K.S., Samani N.J. Analysis of the postulated interaction between the angiotensin II sub-type 12 receptor gene A1166C polymorphism and the insertion/deletion polymorphism of the angiotensin converting enzyme gene on risk of myocardial infarction. Atherosclerosis. 2001;154:123–128. doi: 10.1016/s0021-9150(00)00438-X. [DOI] [PubMed] [Google Scholar]
- Stringer C., Galway-Witham J. When did modern humans leave Africa? Science. 2018;359:389–390. doi: 10.1126/science.aas8954. [DOI] [PubMed] [Google Scholar]
- Tanriverdi H., Evrengul H., Tanriverdi S., Turgut S., Akdag B., Kaftan H.A., Semiz E. Improved endothelium dependent vasodilation in endurance athletes and its relation with ACE I/D polymorphism. Cir. J. 2005;69:1105–1110. doi: 10.1253/circj.69.1105. [DOI] [PubMed] [Google Scholar]
- Thomas G., Tomlinson B., Chan J.C., Sanderson J.E., Cockram C.S., Critchley J.A. Renin-angiotensin system gene polymorphisms, blood pressure, dyslipidemia, and diabetes in Hong Kong Chinese: a significant association of the ACE insertion/deletion polymorphism with type 2 diabetes. Diabetes Care. 2001;24:356–361. doi: 10.2337/diacare.24.2.356. [DOI] [PubMed] [Google Scholar]
- Um J., Mun K.S., An N.H., Kim P.G., Kim S.D., Song Y.S., Lee K.N., Lee K.M., Wi D.H., You Y.O., Kim H.M. Polymorphism of angiotensin-converting enzyme gene and BMI in obese Korean women. Clin. Chim. Acta. 2003;328:173–178. doi: 10.1016/S0009-8981(02)00428-X. [DOI] [PubMed] [Google Scholar]
- Walder, B., Spanaus, K.S., Weinreich, T., Sawicki, P.T., Widmer, U. 1998. Genetic heterogeneity in the renin-angiotensin system and the risk of diabetic nephropathy: Association with the angiotensinogen gene, but not with the ACE gene. J. Clin. Bas. Cardiol., 1, 55-58 https://www.kup.at/kup/pdf/19.pdf.
- Xiao L., Sakagami H., Miwa N. ACE2: The key molecule for understanding the pathophysiology of severe and critical conditions of COVID-19: Demon or angel? Viruses. 2020;12(5):491. doi: 10.3390/v12050491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C., Zhan J., Feng W. Gene polymorphisms of angiotensin II type 1 receptor and angiotensin-converting enzyme in two ethnic groups living in Zhejiang province, China. J. Renin Angiotensin Aldosterone Syst. 2005;6:132–137. doi: 10.3317/jraas.2005.019. [DOI] [PubMed] [Google Scholar]
- Young R.P., Chan J.C., Critchley J.A., Poon E., Nicholls G., Cockram C.S. Angiotensinogen T235 and ACE insertion/deletion polymorphisms associated with albuminuria in Chinese type 2 diabetic patients. Diabetes Care. 1998;21:431–437. doi: 10.2337/diacare.21.3.431. [DOI] [PubMed] [Google Scholar]
- Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., Zhao, X., Huang, B., Shi, W., Lu, R., Niu, P., Zhan, F., Ma, X., Wang, D., Xu, W., Wu, G., Gao, G.F., Tan, W., China Novel Coronavirus Investigating and Research Team. 2020. A novel coronavirus from patients with pneumonia in China, 2019. New Engl. J. Med., 382, 727-733 https://www.nejm.org/doi/10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed]