Abstract
Objectives
The aim was to determine the antibody response against SARS-CoV-2 spike protein and nucleoprotein using four automated immunoassays and three ELISAs for the detection of total Ig antibodies (Roche) or IgG (Abbott, Diasorin, Snibe, Euroimmun, Mikrogen) in COVID-19 patients.
Methods
Sensitivity and dynamic trend to seropositivity were evaluated in 233 samples from 114 patients with moderate, severe or critical COVID-19 confirmed with PCR on nasopharyngeal swab. Specificity was evaluated in 113 samples collected before January 2020, including 24 samples from patients with non-SARS coronavirus infection.
Results
Sensitivity for all assays was 100% (95% confidence interval 83.7–100) 3 weeks after onset of symptoms. Specificity varied between 94.7% (88.7–97.8) and 100% (96.1–100). Calculated at the cut-offs that corresponded to a specificity of 95% and 97.5%, Roche had the highest sensitivity (85.0% (79.8–89.0) and 81.1% (76.6–85.7), p < 0.05 except vs. Abbott). Seroconversion occurred on average 2 days earlier for Roche total Ig anti-N and the three IgG anti-N assays (Abbott, Mikrogen, Euroimmun) than for the two IgG anti-S assays (Diasorin, Euroimmun) (≥50% seroconversion day 9–10 vs. day 11–12 and p < 0.05 for percent seropositive patients day 9–10 to 17–18). There was no significant difference in the IgG antibody time to seroconversion between critical and non-critical patients.
Discussion
Seroconversion occurred within 3 weeks after onset of symptoms with all assays and on average 2 days earlier for assays detecting IgG or total Ig anti-N than for IgG anti-S. The specificity of assays detecting anti-N was comparable to anti-S and excellent in a challenging control population.
Keywords: Coronavirus, COVID-19, Diagnosis, ELISA, Immunoassay, Nucleocapsid protein, SARS-CoV-2, Sensitivity and specificity, Seroconversion, Spike glycoprotein
Introduction
In hospitalized COVID-19 patients, seroconversion for IgG is typically detected between 5 and 14 days after symptom onset. Similar to SARS-CoV-1 [1], the time to seropositivity for IgM and IgA does not appear to be significantly shorter in most studies [[2], [3], [4], [5], [6], [7], [8], [9]]. There is still debate as to which antibodies should be measured. Serological tests typically detect antibodies against spike protein (S) and/or nucleoprotein (N) since these are the most immunogenic proteins of SARS-CoV-2 [8]. The S protein, consisting of a S2 and a S1 subunit with a receptor binding domain (RBD), is present on the envelope and is used by the virus to connect to the human cells using the ACE-2 receptor. Since anti-spike protein antibodies have been shown to possess neutralizing effects in vitro, it has been suggested that detection of antibodies against spike protein could provide a better indication of an effective immune response [10,11]. Detection of antibodies against nucleoprotein, on the other hand, has been suggested to decrease the time to seroconversion in human coronavirus (HCoV) infections including SARS-CoV-1 [12]. For SARS-CoV-2, this has not been clearly established and several authors found a similar time to seropositivity for anti-N and anti-S using home-made ELISAs [6,[13], [14], [15]]. IgG anti-SARS-CoV-2 antibody levels have been reported to correlate with disease severity [3,14,16], although this has not been confirmed by other studies [6].
Many of the studies reported in the literature have been performed using home-made or research-use only enzyme linked immunosorbent assays (ELISA's) [6,13,15,17,18]. At the end of March 2020, the first ELISA, the Euroimmun IgA and IgG ELISA, received CE marking. The first automated CE-marked assay, the Maglumi assay from the Chinese company Snibe, has been evaluated and adopted by a number of Italian laboratories for the detection of antibodies against SARS-CoV-2 [4]. Since end of April 2020, several other automated immunoassays received CE marking and FDA emergency use authorization. Diasorin and Abbott released assays for the detection of IgG anti-SARS-CoV-2 antibodies for the Liason and Architect platforms, respectively, while Roche released an assay for total anti-SARS-CoV-2 immunoglobulins (total Ig) for the Cobas platform. In May 2020 the ELISA from Mikrogen received its CE mark. These different assays detect antibodies against spike protein, nucleoprotein or both (N/S). There are currently no studies comparing the antibody response against these different proteins except studies using home-made ELISAs [6,13,15,17,18].
The aim of this study was to determine the antibody response against SARS-CoV-2 spike protein and nucleoprotein using four automated immunoassays and three ELISAs for the detection of total Ig antibodies (Roche) or IgG (Abbott, Diasorin, Snibe, Euroimmun, Mikrogen) in COVID-19 patients.
Materials and methods
Patient selection
The specificity was evaluated using selected serum samples from 113 patients collected before January 2020 as negative controls. These included (a) a disease control group of 49 consecutive patients with a respiratory infection who had a PCR test for respiratory pathogens in the period September to November 2019. The serum samples were collected day 1 to day 40 after the PCR test. (b) In addition, we tested 24 samples from patients with a confirmed non-SARS-CoV-2 coronavirus infection collected 12–42 days after the positive PCR. (c) Forty samples of patients with antibodies against other pathogens (e.g. cytomegalovirus, Epstein–Barr virus, human immunodeficiency virus) from routine serology testing (Table S1). All samples were stored at –20°C until use.
To assess the sensitivity and dynamic trend to seropositivity in PCR-positive COVID-19 patients, we used a total of 233 samples of 114 patients who were positive for SARS-CoV-2 with RT-PCR on nasopharyngeal swabs (UTM, Copan, Italy) and diagnosed with COVID-19. The number of samples used per patient ranged from one to six (please see supplementary material). Immunocompromised patients (e.g. acute leukaemia, treatment with azathioprine) were excluded. RT-PCR was performed using an in-house method complying with the World Health Organization (WHO) guidelines [19].
The date of onset of symptoms, clinical classification (moderate, severe or critical [3]) and basic demographic information (male/female, age) were recorded for each COVID-19 patient. The group consisted of 81 male and 33 female patients with a median age of 66.5 (range 23–90) years. The median time between onset of symptoms and admission to the hospital was 7 days (83.3% of patients were admitted the day of the first positive PCR result). Thirty-six (31.6%) patients were classified as critical if mechanical ventilation was required or in case of fatality [3].
Data collection and analysis
This retrospective study was performed at the University Hospitals Leuven and approved by the local ethics committee (protocol number S63897). Some of the data for the Euoimmun IgG anti-S assay were included in a previous study [20].
We evaluated the diagnostic performance of 4 automated assays from Roche, Abbott, Diasorin, and Snibe (Maglumi), two Euroimmun ELISAs and an ELISA from Mikrogen. The two assays from Euroimmun detect antibodies against S1 (Euro S1) and nucleoprotein (Euro NCP), respectively. All assays are CE in vitro diagnostics (IVD) marked and all assays except Maglumi and Mikrogen received emergency use authorization from the FDA. Please see supplementary material for more detailed information about the assays and the analysers (Table S2) and data analysis. To calculate performance characteristics, equivocal results were treated as ‘positive’.
Results
Specificity of the different assays
The specificity (95% confidence interval) varied between 96.5% (91.0–98.9) (Maglumi) and 100% (96.1–100) (Roche) for the automated assays, and between 94.7% (88.7–97.8) (Euro NCP) and 96.5% (91.0–98.9) (Euro S1 and Mikrogen) for the ELISAs (Table 1 ). There were no false-positive results with any of the assays for the 24 patients with a non-SARS coronavirus infection. Two samples were false positive with three different assays. A sample from October 2019 from a patient who had acute respiratory distress syndrome (PCR+: Entero-/Rhinovirus, Pneumocystis jirovecii, Streptococcus pneumoniae) was positive with Maglumi, Euro NCP and Mikrogen, while a sample from early November 2019 from a patient who had a necrotizing pneumonia (PCR+: entero-/rhinovirus, herpes simplex virus 1, S. pneumoniae) was false positive with Euro S1, Euro NCP (both equivocal results) and Mikrogen. One sample with IgM and IgG anti-CMV antibodies was false positive with two assays: Euro NCP and Maglumi.
Table 1.
Overall diagnostic performance of the different assays
| N | Roche Ig anti-N |
Abbott IgG anti-N | Euro NCP IgG anti-N | Mikrogen IgG anti-N |
Maglumi IgG anti-N/S | Diasorin IgG anti-S | Euro S1 IgG anti-S | |
|---|---|---|---|---|---|---|---|---|
| Sensitivity (95% CI) | 223 | 71.8% (65.5–77.3) |
70.9% (64.6–76.4) |
73.1%d (66.9–78.5) |
70.4% (64.1–76.0) |
68.6% (31.8–47.1) |
63.2% (56.7–69.3) |
64.6% (58.1–70.6) |
| D0–6 | 43 | 32.6% (20.4–47.6) |
27.9% (16.6–42.8) |
30.2% (18.5–45.2) |
30.2% (18.5–45.2) |
25.6% (14.8–40.4) |
14.0% (6.2–27.6) |
18.6% (9.5–32.9) |
| D7–13 | 98 | 69.4% (59.7–77.7) |
67.3% (57.5–75.9) |
71.4% (61.8–79.5) |
67.3% (57.5–75.9) |
64.3% (54.4–73.1) |
58.2% (48.3–67.5) |
60.2% (50.3–69.3) |
| D14–17 | 42 | 92.9% (80.3–98.2) |
95.2% (83.3–99.5) |
95.2% (83.3–99.5) |
90.5% (77.4–96.8) |
92.9% (80.3–98.2) |
90.5% (77.4–96.8) |
88.1% (74.5–95.3) |
| D18–21 | 16 | 93.8% (69.7–100) |
100% (77.3–100) |
100% (77.3–100) |
100% (77.3–100) |
100% (77.3–100) |
100% (77.3–100) |
100% (77.3–100) |
| D22–27 | 13 | 100% (73.4–100) |
100% (73.4–100) |
100% (73.4–100) |
100% (73.4–100) |
100% (73.4–100) |
100% (73.4–100) |
100% (73.4–100) |
| D28–37 | 11 | 100% (70.0–100) |
100% (70.0–100) |
100% (70.0–100) |
100% (70.0–100) |
100% (70.0–100) |
100% (70.0–100) |
100% (70.0–100) |
| Specificity (95% CI) | 113 | 100% (96.1–100) |
99.1% (94.7–100) |
94.7% (88.7–97.8) |
96.5% (91.0–98.9) |
96.5% (91.0–98.9) |
99.1% (94.7–100) |
96.5% (91.0–98.9) |
| Other coronavirus | 24 | 100% (83.7–100) |
100% (83.7–100) |
100% (83.7–100) |
100% (83.7–100) |
100% (83.7–100) |
100% (83.7–100) |
100% (83.7–100) |
| Respiratory infection | 49 | 100% (91.3–100) |
100% (91.3–100) |
93.9% (82.9–98.5) |
93.9% (82.9–98.5) |
98.0% (88.3–100) |
100% (91.3–100) |
93.9% (82.9–98.5) |
| Antiviral antibodies | 40 | 100% (89.6–100) |
97.5% (86.0–100) |
92.5% (79.4–98.1) |
97.5% (86.0–100) |
92.5% (79.4–98.1) |
97.5% (86.0–100) |
97.5% (86.0–100) |
| LR+ | +∞ | 80.1 | 13.8 | 19.9 | 19.4 | 74.1 | 18.2 | |
| ROC curve (area) | All | 0.950 | 0.907 | 0.928 | 0.864 | 0.871 | 0.865 | 0.909 |
| Cut-off (Manufacturer)a | 1.0 | 1.4 | 0.8/1.1 | 20/24 | 1.0 | 12/15 | 0.8/1.1 | |
| Sensitivity (cut-off) if | ||||||||
| Specificity 95.0% | 233 | 85.0% (0.13)b (79.8–89.0) |
78.1% (0.25) (72.4–83.0) |
73.8% (1.13) (67.8–79.0) |
74.2% (16.4) (68.3–79.5) |
71.7% (0.42) (65.6–77.0) |
67.4% (7.25) (61.1–73.1) |
69.5% (0.47) (63.3–75.1) |
| Specificity 97.5% | 233 | 81.1% (0.17)b (76.6–85.7) |
75.1% (0.59)c (69.2–80.2) |
69.1% (0.77) (62.9–74.7) |
70.8% (24.0) (64.7–76.3) |
67.0% (1.23) (60.7–72.7) |
67.0% (8.11) (60.7–72.7) |
62.7% (0.89) (56.3–68.6) |
For all calculations, equivocal results were treated as positive. 95% CI, 95% confidence interval; LR+, positive likelihood ratio; ROC, receiver operator curve.
If defined by the manufacturer, the upper and lower limit of the equivocal zone are listed.
p < 0.05 vs all except Abbott.
p < 0.05 vs Euro S1.
p < 0.05 vs Diasorin.
Sensitivity and overall diagnostic performance
None of the patients became seronegative after the first positive result for any of the assays. The overall sensitivity varied between 63.2% (56.7–69.3) (Diasorin) and 73.1% (66.9–78.5) (Euro NCP) (Table 1). In a limited number of samples (n = 24) obtained >21 days after the onset of symptoms, anti-SARS-CoV-2 antibodies could be detected with all seven assays (Table 1). To account for the fact that a lower cut-off increases sensitivity at the cost of a lower specificity, we calculated the positive likelihood ratio (LR+), performed receiver operating characteristic (ROC) analysis, and calculated the sensitivity at a cut-off corresponding to a specificity of 95% and 97.5% (Table 1). The assays of Roche, Abbott and Diasorin had a LR+ ≥ 74, while the likelihood ratios of the other assays varied between 13.8 and 19.9 (Table 1).
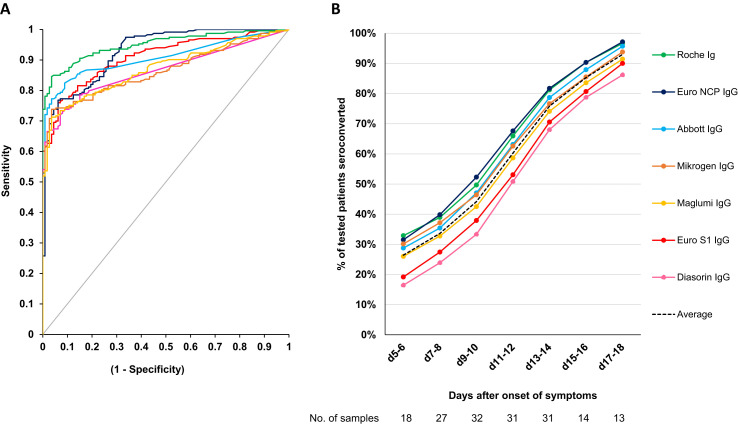
The Roche assay had the highest area under the ROC curve (0.95, p < 0.05 vs. all except Euro NCP) (Fig. 1 A). The Euro NCP and Euro S1 assays had a higher AUC than Abbott (p = NS) although the Abbott assay had a better performance in the clinically relevant area with a specificity of ≥90%. This can be explained by the fact that the ROC curves of the Euro NCP and Euro S1 assays cross the Abbott curve in the area where specificity is <75%. This artefact is the reason we did not include the statistical results of the AUC comparison in Table 1. When the sensitivity was calculated in our study cohort at a cut-off corresponding to a specificity of 95% and 97.5%, the assay of Roche had the highest sensitivity followed by Abbott (p < 0.05 for Roche vs. all except Abbott for both cut-offs, Table 1).
Fig. 1.
Diagnostic performance of the different assays. (A) ROC curve (samples used to calculate sensitivity and specificity, n = 346). (B) Dynamic trend to seropositivity in 222 samples from 106 patients with COVID-19. Of note, the average time to seroconversion lags behind the true time of seroconversion by a couple of days since patients were not tested daily and a patient is only considered to have seroconverted after the first positive result.
The agreement between the different assays is shown in Table 2 . The agreement between the 3 IgG anti-N assays varied between 93.3% (89.1–96.0) and 96.9% (93.5–98.6). The agreement between the 2 IgG anti-S assays was significantly lower than between Abott and Euro NCP (91.5% (87.0–94.5), p < 0.05).
Table 2.
Percentage agreement between the different assays in COVID-19 patients (223 samples for sensitivity) (95% confidence interval)
| Abbott | Euro NCP | Maglumi | Mikrogen | Diasorin | Euro S1 | |
|---|---|---|---|---|---|---|
| Roche | 94.6% (90.7–97.0) | 95.1% (91.3–97.3) | 89.7% (84.9–93.1) | 91.5% (87.0–94.5) | 84.3% (78.9–88.5) | 90.1% (85.4–93.4) |
| Abbott | 96.9% (93.5–98.6) | 90.6% (86.0–93.8) | 93.3% (89.1–96.0) | 85.2% (79.9–89.3) | 90.1% (85.4–93.4) | |
| Euro NCP | 87.4% (82.4–91.2) | 95.5% (91.8–97.7) | 84.8% (79.4–88.9) | 88.8% (83.9–92.3) | ||
| Maglumi | 96.4% (93.0–98.2) | 85.7% (80.4–89.7) | 87.9% (82.9–91.6) | |||
| Mikrogen | 83.9% (78.4–88.1) | 87.9% (82.9–91.6) | ||||
| Diasorin | 91.5% (87.0–94.5) |
Dynamic trend to seropositivity with the different assays
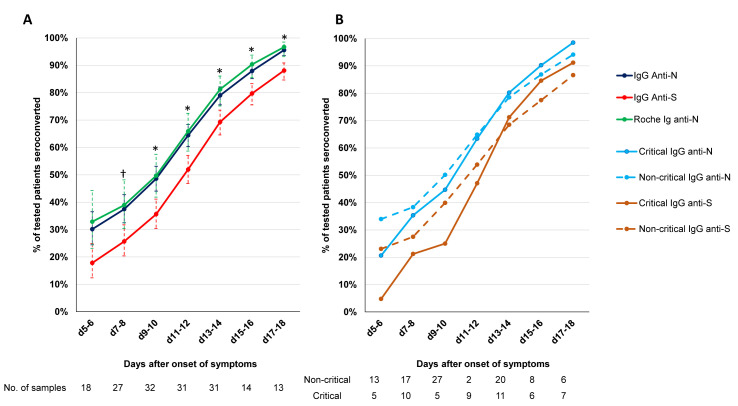
The dynamic trend to seropositivity of all the assays is shown in Fig. 1B. Seroconversion occurred significantly faster with the Roche total Ig anti-N assay and three IgG anti-N assays (Abbott, Mikrogen, Euro NCP) than with the two anti-S assays (Diasorin, Euro S1) (Fig. 2 A, ≥50% seroconversion day 9–10 vs. day 11–12 and p < 0.05 for % seropositive patients day 9–10 to day 17–18). The dynamic trend to seropositivity with the assay that detects anti-N and anti-S (Maglumi) was in between the trend for anti-N and anti-S assays. We did not observe any difference in time to seroconversion for IgG between critical and non-critical patients for the IgG anti-N and IgG anti-S assays (Fig. 2B).
Fig. 2.
Antibody response to SARS-CoV-2 N-antigen and S-antigen. (A) Dynamic trend to seropositivity for Roche total Ig, the 3 IgG anti-N assays (Abbott, Mikrogen, Euro NCP) and IgG anti-S assays (Diasorin, Euro S1). †p <0.05 for IgG anti-S1 assays vs. IgG anti-N assays. ∗p < 0.05 for anti-S assays vs. IgG anti-N assays and Roche total Ig anti-N. (B) Dynamic trend to seropositivity for IgG anti-N and IgG anti-S assays in critical and non-critical patients.
Seropositivity at the time of admission and 1 week after admission
The median time to presentation after onset of symptoms was 7 days, both for critical and non-critical patients. The percentage of patients with IgG antibodies at the time of admission varied between 21.1% (14.4–29.7) and 36.8% (28.3–46.3) and was comparable for critical and non-critical (moderate and severe) patients (Table 3 ). The Roche assay was the only assay with a difference of more than 10% between critical and non-critical, but this difference was not significant.
Table 3.
Presence of anti-SARS-CoV-2 at time of admission to the hospital and 1 week after admission (95% confidence interval)
| % seropositive | Roche Ig-N | Abbott IgG-N | Euro NCP IgG-N | Mikrogen IgG-N | Maglumi IgG-N/S | Diasorin IgG-S | Euro S1 IgG-S |
|---|---|---|---|---|---|---|---|
| At time of admission (n = 76) | 34.2% (26.0–43.6) | 30.3% (22.4–39.5) | 36.8% (28.3–46.3) | 32.9% (24.7–42.2) | 28.9% (21.2–38.1) | 21.1% (14.4–29.7) | 21.1% (14.4–29.7) |
| Critical (n = 23) | 26.1% (13.9–43.3) | 30.4% (17.3–47.7) | 39.1% (22.1–59.3) | 39.1% (22.1–59.3) | 30.4% (17.3–47.7) | 21.7% (10.7–38.7) | 17.4% (6.4–37.7) |
| Non-critical (n = 53) | 37.7% (25.9–51.2) | 30.2% (19.5–43.6) | 35.8% (24.3–49.3) | 30.2% (19.5–43.6) | 28.3% (17.9–41.7) | 20.8% (11.8–33.6) | 22.6% (13.3–35.7) |
| After 1 week (n = 41)a | 92.7% (79.9–98.2) | 95.1% (83.0–99.5) | 92.7% (79.9–98.2) | 92.7% (79.9–98.2) | 90.2% (76.9–96.7) | 85.4% (71.2–93.5) | 92.7% (79.9–98.2) |
| At admission | 26.8% (15.6–42.0) | 26.8% (15.6–42.0) | 31.7% (19.5–47.1) | 31.7% (19.5–47.1) | 26.8% (15.6–42.0) | 19.5% (10.0–34.3) | 17.1% (8.2–31.6) |
| After 1 week if negative | 27/30 (90.0%) (73.6–97.3) |
28/30 (93.3%) (77.6–0.99) |
25/28 (89.3%) (72.0–97.1) |
25/28 (89.3%) (72.0–97.1) |
25/31 (80.6%) (63.4–91.2) |
27/33 (81.8%) (65.2–91.8) |
31/34 (91.2%) (76.3–97.8) |
Subgroup consisting of those patients for whom a sample was available.
One week after admission, the seropositivity varied between 85.4% (71.2–93.5) and 95.1% (83.0–99.5) for the different assays in patients for whom a sample was available at admission and after 1 week (6–8 days).
Evolution of antibody levels
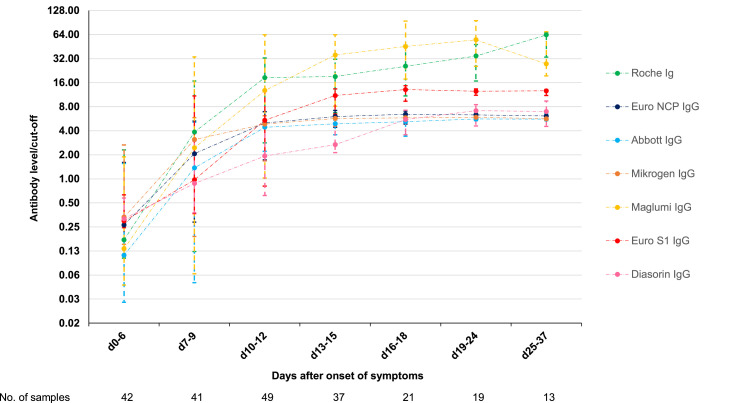
Only one of the seven assays (Diasorin) is intended for the quantitative detection of SARS-CoV-2 antibodies. All assays do, however, provide a signal over cut-off value, which is expected to correlate with the antibody level. The evolution of antibody levels is shown in Fig. 3 . The antibody levels of Abbott, Euro NCP, and Mikrogen reach a plateau 2 weeks after onset of symptoms around six to ten times the cut-off. The antibody levels with Diasorin appear to rise slower, but also reach a plateau during the third week around six to ten times the cut-off. The plateau for these four assays could be the upper limit of quantitation for the assays. None of the manufacturers did, however, provide an upper limit of quantitation. The median antibody levels with Euro S1 reach a plateau around 15 times the cut-off, while the median levels with Maglumi and Roche rise above 50 times the cut-off.
Fig. 3.
Evolution of the antibody levels (P25, median, P75) with the different assays. Results of the individual assays were normalized by dividing the result by the cut-off proposed by the manufacturer (lowest cut-off in case the manufacturer defines an equivocal zone).
Discussion
We evaluated the antibody response and time to seroconversion using four chemiluminescent assays (CLIAs) and three ELISAs for the detection of IgG and total Ig antibodies against SARS-CoV-2 N and/or S protein. During the first 3 weeks, the total Ig anti-N assay of Roche had the best diagnostic performance taking into account both sensitivity and specificity followed by the Abbott IgG anti-N assay. We found that seroconversion for IgG occurred on average 2 days faster for N than for S protein.
The sensitivity of IgG antibodies in COVID-19 patients found in this study is in line with other recent publications. Long et al. showed a 100% seroconversion of IgG 19 days after onset of symptoms [7]. To et al. already saw a 100% IgG seropositivity 14 days after onset of symptoms [6] and Padoan et al. 12 days after onset of fever [4]. It is important to note that some studies suggest the sensitivity never reaches 100% in asymptomatic individuals who are positive for SARS-CoV-2 with PCR [[21], [22], [23]]. We did not observe a significant difference in seroconversion time of IgG between critical and non-critical (moderate and severe) patients, confirming the results of previous studies [3,6,16].
We found that seroconversion for anti-N occurs significantly faster than for anti-S in COVID-19 patients. This is similar to the response after SARS-CoV-1 and other HCoV infections where it has been described that anti-N antibodies appear before anti-S antibodies [12]. Several authors have suggested that assays using full-length N protein might be more prone to false positives since it has several conserved regions with high sequence homology to other HCoVs such as common cold viruses HCoV-229E, -NL63, -OC43 and -HKU1 [24]. For example Okba et al. have described cross-reactivity of a home-made anti-SARS-CoV-2 antibody ELISA with MERS and SARS-CoV-1 antibodies [18]. In our study, however, two of the three assays with the highest specificity (Roche, Abbott, and Diasorin) were assays detecting anti-N antibodies and none of the seven assays had a false-positive result for any of the 24 samples of patients with a non-SARS-CoV-2 HCoV infection.
Note that the ELISA and CLIA assays described in this article do not directly measure neutralizing anti-SARS-CoV-2 antibodies. However, studies have shown that both IgG anti-N and IgG anti-S antibody titres correlate with microneutralization and plaque reduction neutralization tests in vitro [6,18].
While the diagnostic performance of a number of rapid tests for detection of IgG anti-SARS-CoV-2 antibodies is good [19,25], the availability of automated assays for the detection of anti-SARS-CoV-2 antibodies opens the possibility for largescale testing. There are, however, still a number of important questions. First, it is uncertain how long antibodies persist after infection. A recent study reported that 12.9% of symptomatic and 40% of asymptomatic individuals became seronegative two to three months after infection [23]. Second, there are currently no studies which demonstrated that antibodies are protective against reinfection in humans. For these reasons, the WHO does not recommend the use of immunity passports at this moment [26]. We therefore recommend to use serological assays for SARS-CoV-2 as a complementary diagnostic tool and for epidemiologic purposes, rather than as a means to determine immunity.
The use of assays from different manufacturers for anti-N and anti-S strengthens our conclusions as this reduces the risk that our observations could be influenced by the quality of one of the assays used. This risk is particularly present when home-made assays are used as was the case in all previously published peer-reviewed studies comparing the antibody response to different SARS-CoV-2 antigens [6,13,15,17,18].
There are a number of limitations to our study. First, we only included a limited number of samples from patients with frequent respiratory infections such as influenza, Mycoplasma pneumoniae, and Chlamydophila pneumoniae and no samples from patients with a SARS-CoV-1 or MERS-CoV infection. Second, the samples selected for specificity were challenging and most likely underestimate the specificity in a routine laboratory setting. Finally, we only evaluated the diagnostic performance in patients with moderate to critical COVID-19 and did not study the antibody response in asymptomatic persons and patients with mild COVID-19.
In conclusion, the specificity of the assays varied between 94.7% (88.7–97.8) and 100% (96.1–100) in a challenging set of pre-COVID control samples. Seroconversion occurred within 3 weeks after onset of symptoms with all assays and on average 2 days earlier for assays detecting IgG or total Ig anti-N antibodies than for assays detecting IgG anti-S. The assay detecting both anti-N and anti-S showed an intermediate time to seropositivity. Our results demonstrate that commercial automated assays and ELISAs are suitable for the detection of IgG and total Ig antibodies against SARS-CoV-2.
Transparency declaration
Pieter Vermeersch reports personal fees from Roche, outside the submitted work. Katrien Lagrou reports personal fees and non-financial support from Pfizer, personal fees and non-financial support from MSD, personal fees from SMB Laboratoires, personal fees from Gilead, and personal fees from FUJIFILM Wako, outside the submitted work. The other authors state no conflicts of interests. The research did not receive any specific grant from funding agencies in the public, commercial or not-for-profit sectors.
Author contributions
P.V. devised the study, collected data and drafted the manuscript. J.V.E. collected data and drafted the manuscript. All other authors aided in collecting data and critically reviewed the manuscript.
Acknowledgements
P.V. is a senior clinical investigator of the FWO-Vlaanderen. We thank Ine Empsen, Jeroen Vandersmissen, Mirte Tonsenst, Marie-Christine Clukkers, Katrijn Overloop, Kat Delaat and Hannes Briels for their expert technical assistance.
Editor: F. Allerberger
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2020.07.038.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Huang A.T., Garcia-Carreras B., Hitchings M.D.T., Yang B., Katzelnick L., Rattigan S.M., et al. A systematic review of antibody mediated immunity to coronaviruses: antibody kinetics, correlates of protection, and association of antibody responses with severity of disease. MedRxiv. 2020 doi: 10.1101/2020.04.14.20065771. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xie J., Ding C., Li J., Wang Y., Guo H., Lu Z., et al. Characteristics of patients with coronavirus disease (COVID-19) confirmed using an IgM-IgG antibody test. J Med Virol. 2020 doi: 10.1002/jmv.25930. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qu J., Wu C., Li X., Zhang G., Jiang Z., Li X., et al. Profile of IgG and IgM antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa489. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Padoan A., Cosma C., Sciacovelli L., Faggian D., Plebani M. Analytical performances of a chemiluminescence immunoassay for SARS-CoV-2 IgM/IgG and antibody kinetics. Clin Chem Lab Med. 2020;58:1081–1088. doi: 10.1515/cclm-2020-0443. [DOI] [PubMed] [Google Scholar]
- 5.Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol. 2020;92:1518–1524. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.To K.K.-W., Tsang O.T.-Y., Leung W.-S., Tam A.R., Wu T.-C., Lung D.C., et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Long Q.-X., Liu B.-Z., Deng H.-J., Wu G.-C., Deng K., Chen Y.-K., et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 8.Zhang W., Du R.H., Li B., Zheng X.S., Lou Yang X., Hu B., et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Padoan A., Sciacovelli L., Basso D., Negrini D., Zuin S., Cosma C., et al. IgA-Ab response to spike glycoprotein of SARS-CoV-2 in patients with COVID-19: a longitudinal study. Clin Chim Acta. 2020;507:164–166. doi: 10.1016/j.cca.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perera R.A., Mok C.K., Tsang O.T., Lv H., Ko R.L., Wu N.C., et al. Serological assays for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), March 2020. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.es.2020.25.16.2000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer B., Drosten C., Müller M.A. Serological assays for emerging coronaviruses: challenges and pitfalls. Virus Res. 2014;194:175–183. doi: 10.1016/j.virusres.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grzelak L., Temmam S., Planchais C., Demeret C., Huon C., Guivel F., et al. SARS-CoV-2 serological analysis of COVID-19 hospitalized patients, pauci-symptomatic individuals and blood donors. MedRxiv. 2020 doi: 10.1101/2020.04.21.20068858. (Epub ahead of print) [DOI] [Google Scholar]
- 14.Wu F., Wang A., Liu M., Wang Q., Chen J., Xia S., et al. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. MedRxiv. 2020 doi: 10.1101/2020.03.30.20047365. (Epub ahead of print) [DOI] [Google Scholar]
- 15.Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W., et al. Evaluation of nucleocapsid and spike protein-based ELISAs for detecting antibodies against SARS-CoV-2. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00461-20. e00461–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa344. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ni L., Ye F., Chen M.-L., Feng Y., Deng Y.-Q., Zhao H., et al. Characterization of anti-viral immunity in recovered individuals infected by SARS-CoV-2. MedRxiv. 2020 doi: 10.1101/2020.03.17.20036640. (Epub ahead of print) [DOI] [Google Scholar]
- 18.Okba N.M.A., Muller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M., et al. SARS-CoV-2 specific antibody responses in COVID-19 patients. Emerg Infect Dis. 2020;26:1478–1488. doi: 10.1101/2020.03.18.20038059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Elslande J., Houben E., Depypere M., Brackenier A., Desmet S., André E., et al. Diagnostic performance of 7 rapid IgG/IgM antibody tests and the Euroimmun IgA/IgG ELISA in COVID-19 patients. Clin Microbiol Infect. 2020;26:1082–1087. doi: 10.1016/J.CMI.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yongchen Z., Shen H., Wang X., Shi X., Li Y., Yan J., et al. Different longitudinal patterns of nucleic acid and serology testing results based on disease severity of COVID-19 patients. Emerg Microbes Infect. 2020;9:833–836. doi: 10.1080/22221751.2020.1756699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee Y.L., Liao C.H., Liu P.Y., Cheng C.Y., Chung M.Y., Liu C.E., et al. Dynamics of anti-SARS-Cov-2 IgM and IgG antibodies among COVID-19 patients. J Infect. 2020;81:55–58. doi: 10.1016/j.jinf.2020.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long Q.-X., Tang X., Shi Q., Li Q., Deng H.-J., Yuan D., et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 24.Yamaoka Y., Jeremiah S.S., Miyakawa K., Saji R., Nishii M., Takeuchi I., et al. Whole nucleocapsid protein of SARS-CoV-2 may cause false positive results in serological assays. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa637/5843448. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Decru B., Van Elslande J., Weemaes M., Houben E., Empsen I., André E., et al. Comparison of the diagnostic performance with whole blood and plasma of four rapid antibody tests for SARS-CoV-2. Clin Chem Lab Med. 2020 doi: 10.1515/cclm-2020-0817. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 26.World Health Organization “Immunity passports” in the context of COVID-19. https://www.who.int/news-room/commentaries/detail/immunity-passports-in-the-context-of-covid-19 Available from:
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.