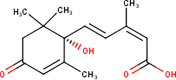
Abstract
Coronavirus disease (COVID-19) is a global pandemic caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). Up to date, there has been no specific cure to treat the disease. Indonesia is one of the countries that is still fighting to control virus transmission. Yet, at the same time, Indonesia has a rich biodiversity of natural medicinal products that potentially become an alternative cure. Thus, this study examined the potency of a natural medicinal product, Sulawesi propolis compounds produced by Tetragonula sapiens, inhibiting angiotensin-converting activity enzyme-2 (ACE-2), a receptor of SARS-CoV-2 in the human body. In this study, molecular docking was done to analyze the docking scores as the representation of binding affinity and the interaction profiles of propolis compounds toward ACE-2. The results illustrated that by considering the docking score and the presence of interaction with targeted sites, five compounds, namely glyasperin A, broussoflavonol F, sulabiroins A, (2S)-5,7-dihydroxy-4′-methoxy-8-prenylflavanone and isorhamnetin are potential to inhibit the binding of ACE-2 and SARS-CoV-2, with the docking score of −10.8, −9.9, −9.5, −9.3 and −9.2 kcal/mol respectively. The docking scores are considered to be more favorable compared to MLN-4760 as a potent inhibitor.
Keywords: COVID-19, Molecular docking, Potent inhibitor, ACE-2, Sulawesi propolis
1. Introduction
As of November 24th, 2020, the world is still struggling to overcome coronavirus disease (COVID-19) global pandemic. The disease is caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), first identified in Wuhan, China, in December 2019. The virus can transmit human-to-human through saliva and respiratory secretion droplets, spreading quickly to many countries (World Health Organization, 2020a). Currently, there have been 1.41 million people died and 59.7 million people infected globally (World Health Organization, 2020b).
SARS-CoV-2 mainly enters the human body through respiratory tract. The infection starts with the binding between the spike protein of SARS-CoV-2 and a protein inside the human body, namely angiotensin-converting enzyme 2 (ACE-2) (Astuti, 2020). The process then continues to the cutting of spike protein initiated by cell surface-associated transmembrane protease serine-2 (TMPRSS-2) and cathepsin, a fusion of virus's membrane and replication sequentially (Alanagreh et al., 2020, Walls et al., 2020). The infection may lead to several symptoms, such as fever, fatigue, sore throat, headache, and, eventually, respiratory failure (Singhal, 2020). Nevertheless, some infected people may also perform no symptoms (Kim et al., 2020).
At present, a specific cure for COVID-19 has not been found (Beigel et al., 2020). Scientists around the world are still looking for drugs that may be effective in treating COVID-19. Some drug candidates are also still on preclinical or clinical trials (Ghasemiyeh and Mohammadi-Samani, 2020). At the same time, in order to inhibit the transmission, regional shutdowns are applied by governments in many countries. This method is proven to be successful in 11 European countries (Flaxman et al., 2020). In China, strict quarantine has led to a significant reduction in the number of cases (Flaxman et al., 2020). Although many countries have been able to control COVID-19 transmission, some countries are still fighting in flattening the curve. Indonesia is one of those countries. Being the fourth most populous country in the world, the number of COVID-19 cases in Indonesia is still rising and has reached more than 506,000 cases up to date (World Health Organization., 2020b, Djalante et al., 2020).
Apart from COVID-19 conditions in Indonesia, the country, in fact, has a rich biodiversity of natural medicinal products that may be potential to become an alternative cure. A resinous bee product, namely propolis, exhibits antiviral activity due to containing caffeic acids, flavonoids, and esters of aromatic acids (Marcucci, 1995). Some research has been conducted to analyze the ability of propolis to become an antiviral agent. Gekker et al. (2005) has proved that propolis may inhibit HIV-1 expression in microglial cell culture (Gekker et al., 2005). Regarding the potency to treat COVID-19, propolis extract and some of its components may reduce the expression of transmembrane serine protease 2 (TMPRSS-2) and inhibit the activity of SARS-CoV-2 main protease (Berretta et al., 2020, Kumar et al., 2020).
Furthermore, recently, Güler et al. (2020) have shown that propolis from the Black Sea region may prevent the binding of SARS-CoV-2 and ACE-2 due to the favorable binding energy and inhibition constant towards ACE-2 (Güler et al., 2020). However, propolis composition from a region is different from another region (Alday et al., 2016). It depends on the plant's type that the bees used as the food source (Miyata et al., 2019, Miyata et al., 2020a, Miyata et al., 2020b). Therefore, a study regarding the potency of Indonesia propolis to become an ACE-2 inhibitor should be conducted.
Beforehand, some research regarding Indonesia propolis from North Luwu, Sulawesi, have been conducted. Our previous study has shown that Sulawesi propolis may perform an antifungal activity to Candida albicans, C. tropicalis, C. krusei, C. parapsilosis, C. glabrata, and Cryptococcus neoformans (Pratami et al., 2020, Sahlan et al., 2020). It also exhibits antioxidant and anti-inflammatory activity due to the high phenolic content (Christina et al., 2018, Pratami et al., 2018, Sahlan et al., 2019). The pharmacological potency of Sulawesi propolis should be further explored. The use of Sulawesi propolis to treat viral infection has not been reported.
In this research, molecular docking simulation was done to analyze the ability of Sulawesi propolis compounds from North Luwu to bind to ACE-2 (PDB ID: 1R4L). Molecular docking is a computational method that aims to find out the most stable conformation of a small molecule when binds to a macromolecule along with the prediction of the binding affinity. The binding affinity is represented by a docking score (kcal/mol) (Quiroga and Villarreal, 2016). The docking score and the interaction profiles generated by propolis compounds are compared to the data resulting from (S,S)-2-{1-carboxy-2-[3-(3,5-dichlorobenzyl)–3H-imidazol4-yl]-ethylamino}-4-methylpentanoic acid (MLN-4760), a potent inhibitor that may prevent the binding of SARS-CoV-2 and ACE-2 by changing ACE-2 conformation (Towler et al., 2004).
2. Materials and method
2.1. Hardware
A laptop with specifications of processor Intel® Core™ i7-8550U @ 1.80 GHz, operating system 8 GB of RAM, the Windows 10 Home Single Language 64-bit, and graphics processing unit (GPU) an Intel® UHD Graphics 620 was used to perform molecular docking and results in analysis.
2.2. Software
The software used in this research included MarvinSketch (ChemAxon, Budapest, Hungary) (ChemAxon, 2018), Autodock Tools 1.5.6 (The Scripps Research Institute, USA), Autodock Vina (The Scripps Research Institute) (Trott and Olson, 2010), LigPlot+ (EMBL-EBI, UK) (Wallace et al., 1995) and Visual Molecular Dynamics (VMD) (University of Illinois, Urbana-Champaign) (Humphrey et al., 1996).
2.3. Protein preparation
First, the pdb format of crystal structure of ACE-2 in complex with inhibitor MLN-4760 was downloaded from the RCSB Protein Data Bank (http://www.rcsb.org). The complex was available under PDB ID 1R4L. VMD was then used to separate the protein from the inhibitor. At the same time, undesired molecules were also removed. After that, both protein and inhibitor molecules were saved into two different pdb files. The inhibitor molecule file was set aside, while the protein file was loaded into Autodock Tools 1.5.6 for further preparation steps. The steps included the addition of polar hydrogen and Kollman charges, as well as the conversion into pdbqt format (Afriza et al., 2018). The file conversion into pdbqt format was done to allow the file to be loaded in Autodock Vina for molecular docking simulation (Huey et al., 2012).
2.4. Ligand preparation
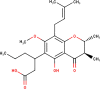
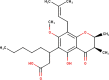
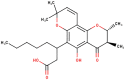
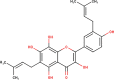
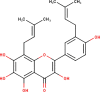
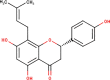
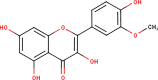
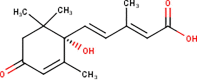
The ligands performed in molecular docking simulation were divided into two types, which are the ligand that previously extracted from the protein structure, in this case, was the inhibitor MLN-4760, and the propolis compounds, which later on will be referred to as the test ligands. The test ligands are Sulawesi propolis compounds from North Luwu that have been identified by Miyata et al. by using the combination of mass spectrometry and nuclear magnetic resonance (NMR) spectroscopy (Miyata et al., 2019, Miyata et al., 2020a, Miyata et al., 2020b). On the paper, it was stated that the propolis was produced by Tetragonula biroi. However, a recent study has found out that it was produced by Tetragonula sapiens (Sayusti et al., 2020). The test ligands are attached in Table 1 . The 2-dimensional structures of the propolis compounds were constructed by using MarvinSketch. The energy was also minimized prior to converting the structures into 3-dimensional forms by using the same program. After the 3-dimensional structures were ready, each ligand was loaded to Autodock Tools 1.5.6. The program was used to optimize the ligands. The steps were adding polar hydrogen, and Gasteiger charges to each ligand, adjusting the number of the rotatable bond, and converting the ligands files into pdbqt format (Huey et al., 2012). The number of rotatable bonds was set as default.
Table 1.
Identified Sulawesi propolis compounds.
2.5. Molecular docking of propolis compounds and ACE-2
Molecular docking simulation performed in this research is categorized as semiflexible docking. The conformation of the protein is remained rigid, while the ligand's conformation may change to find the minimum energy needed to bind to the protein (Pagadala et al., 2017). This method is known as the most common method. It is also considered the most efficient since it only requires simple computation compared to the other method (Fine et al., 2020).
Furthermore, in terms of docking area, the method implemented in this research is categorized as oriented docking. Oriented docking is a method when the ligand is docked into the area, whereas another molecule was previously bound, in this case, was inhibitor MLN-4760. This method is more accurate than blind docking, which uses the whole protein as the docking area. The smaller area provides ease for the program to find the most stable binding pose. Therefore, the error can be minimized (Syahputra, 2014).
In order to find out the specific location where inhibitor MLN-4760 was bound, redocking simulation was conducted. Redocking is a method that aims to dock inhibitor MLN-4760 back into its original site in ACE-2. The center coordinate of the docking area was set to center on ligand, so it automatically adjusted to the previously bound molecule. The location was known to be at the coordinate of x = 40.199, y = −6.024, and z = 29.006. The spacing was set to 1.0 Å, and the optimum search area was 25 × 25 × 25 Å. Afterward, the docking score and root mean square deviation (RMSD) were checked. The area is considered valid if the docking score is negative, and the RMSD is lower than 2,0 Å (Ramírez and Caballero, 2018). The docking score's negative value indicates that the molecule has successfully bound into the protein (Du et al., 2016). In the meantime, the value of RMSD that lower than 2,0 Å expresses that the molecule does not need to change its conformation to fit the cavity significantly. That shows that the molecule binds in the same area where it formerly was. After these parameters were confirmed, the docking area parameters may be used to dock the test ligands into the protein.
2.6. Molecular interaction analysis
Molecular interaction analysis was done to determine whether the test ligands have the same interaction profiles as inhibitor MLN-4760 when binding to ACE-2. The interaction profile determination is defined by the list of amino acids involved in the binding between the ligand and the protein and the types of interaction generated between them. The interaction profile analysis was done to the test ligands with the same or lower docking score than inhibitor MLN-4760. In this step, the structure of ACE-2 that was attached with each selected ligand was loaded to LigPlot+. The program then generated the 2-dimensional figure that illustrates the types of interaction formed along with the amino acid involved. After that, the interactions were recorded, and the binding similarity was calculated. Binding similarity describes how similar the interaction generated between the test ligand and the protein compared to the inhibitor and the protein in the form of a percentage. The percentage calculation was determined by the number of amino acids involved both in the binding and in the interaction between the inhibitor and the protein. This method is based on Flamandita et al. (2020) (Flamandita et al., 2020). The greater the value of binding similarity, the bigger the test ligand's possibility to have the same ability to inhibit the protein as the inhibitor.
3. Results and discussion
3.1. Docking score analysis
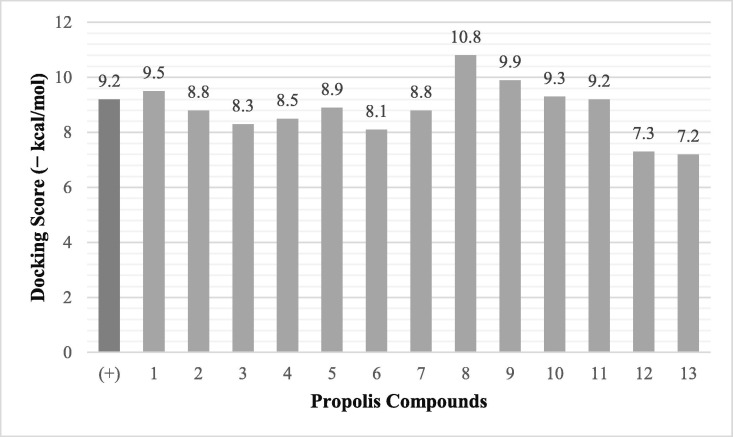
The docking scores of the inhibitor MLN-4760 and the test ligands are available in Fig. 1 . The docking score of the inhibitor MLN-4760 is − 9.2 kcal/mol. The value is different from the result of Güler et al. (2020). It may be due to the program and the docking parameters being used are different (Güler et al., 2020). However, to confirm the docking parameters' accuracy, the interaction profile generated between the inhibitor and the protein in this research was analyzed. The similarity of interaction formed in this research and in the crystallographer's journal indicates that the docking parameters were valid. It is discussed further in the Interaction Profile Analysis section.
Fig. 1.
Docking score between the test ligands and ACE-2. (+) MLN-4760, (1) sulabiroins A, (2) sulabiroins B, (3) 2′,3′-dihydro-3′-hydroxypapuanic acid, (4) (−)-papuanic acid, (5) (−)-isocalolongic acid, (6) isopapuanic acid, (7) isocalopolyanic acid, (8) glyasperin A, (9) broussoflavonol F, (10) (2S)-5,7-dihydroxy-4′-methoxy-8-prenylflavanone, (11) isorhamnetin, (12) (1′S)-2-trans, 4-trans-abscisic acid, (13) (1′S)-2-cis, 4-trans-abscisic acid.
In the meantime, the docking scores of the test ligands are all negative. It indicates that all identified propolis compounds are able to bind to ACE-2. The test ligands with docking scores similar or lower than the inhibitor MLN-4760 are isorhamnetin, (2S)-5,7-dihydroxy-4′-methoxy-8-prenylflavanone, sulabiroins A, broussoflavonol F, and glyasperin A, with the value of −9.2, −9.3, −9.5, −9.9 and −10.8 kcal/mol respectively. The more negative the docking score's value indicates that the ligand may bind stronger and considered as more stable (Du et al., 2016). The interaction profiles between these compounds and ACE-2 were then analyzed.
3.2. Interaction profile analysis
Interaction profile analysis mainly focused on hydrogen bonds and hydrophobic interaction. Hydrogen bonds greatly influence the value of binding affinity (Arora and Tchertanov, 2012). In the docking score computation using Autodock Vina, hydrogen bonds become the primary factor in the docking score (Trott and Olson, 2010). The strength of hydrogen bonds can be divided based on the distance of the interaction. Hydrogen bonds with a distance of 2.2–2.5 Å are categorized as strong, 2.5–3.2 Å are intermediate, and 3.2–4.0 Å are weak (Jeffrey and Jeffrey, 1997).
Meanwhile, hydrophobic interactions also play an important role in many chemical and biological mechanisms. In terms of the interaction between the protein and its ligand, hydrophobic interactions are the most dominant (Bronowska, 2011). Although hydrophobic interactions are not as strong as hydrogen bonds, their existence within the catalytic sites may indicate that the ligand can proceed with inhibitory activity. Thus, since hydrogen bonds are stronger and contribute significantly to the docking score's value, their existence becomes the main concern in this study.
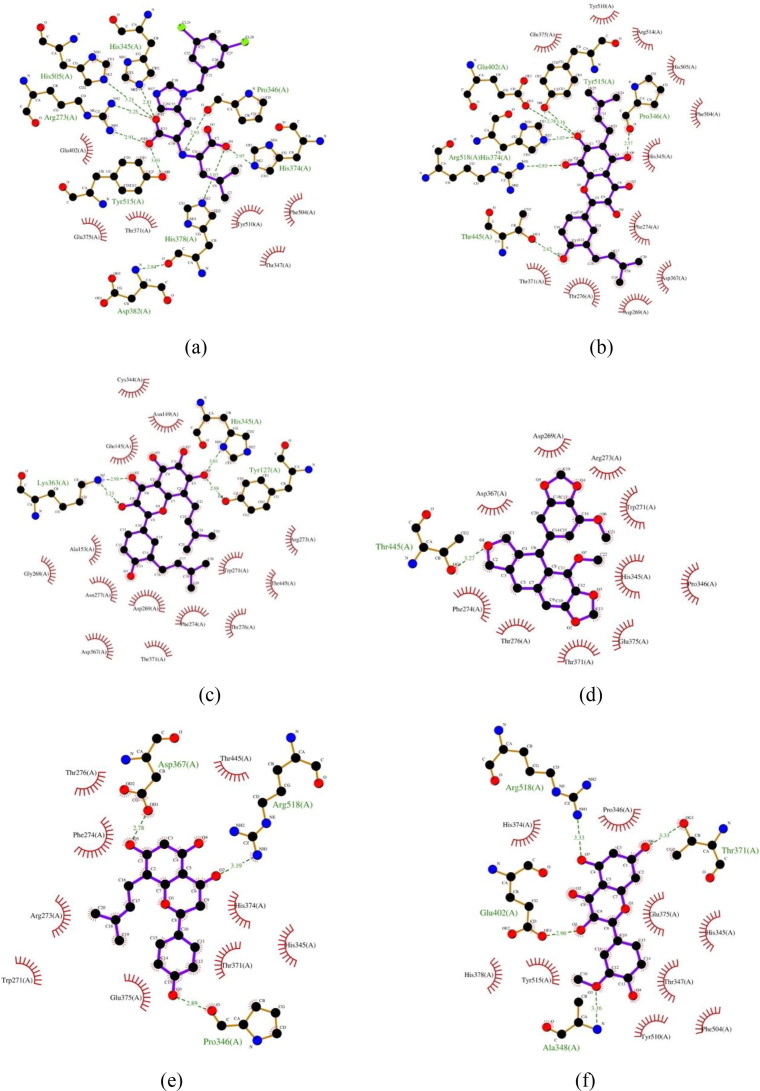
The 2-dimensional illustrations of the interaction between inhibitor MLN-4760 and selected propolis compounds with ACE-2 severally are attached in Fig. 2 . According to the figure, MLN-4760 formed hydrogen bonds with Arg273, His345, Pro346, His378, His505, and Tyr515. In the meantime, compared to the data provided by the crystallographer, MLN-4760 was able to form hydrogen bonds with Arg273, His345, Pro346, Thr371, and His505 (Towler et al., 2004). It can be seen that this study was unable to capture hydrogen bonding with Thr371. Nonetheless, the interaction with Thr371 was detected as hydrophobic interaction. The absence of zinc, chloride, and N-acetylglucosamine might give rise to a slight difference. The similarity of amino acids interact with the inhibitor MLN-4760 might also validate the docking parameters used in the simulation.
Fig. 2.
Visualization of the molecular interactions of ACE-2 with various ligands. (a) MLN-4760, (b) glyasperin A, (c) broussoflavonol F, (d) sulabiroins A, (e) (2S)-5,7-dihydroxy-4′-methoxy-8-prenylflavanone, (f) isorhamnetin. The purple lines denote the ligand structure, whereas the brown lines denote the structure of amino acid residues. The molecular interactions are reflected as dashed lines and arcs. The green dashed lines between atoms represent hydrogen bonds, and the numbers above these lines indicate the length of the bond. In the meantime, the arcs with spokes radiating toward the ligand atoms represent hydrophobic interactions. The atoms involve in hydrophobic interactions are indicated by the presence of spokes radiating back (Wallace et al., 1995).
In the interaction profile analysis between the selected propolis compounds and ACE-2, the information regarding the catalytic sites of ACE-2 is relatively unknown. Therefore, the amino acids involved in forming hydrogen bonds with the inhibitor MLN-4760 based on the crystallographer's data were considered as the target sites. Glyasperin A was able to form hydrogen bonds with Pro346 and His374 and interacted hydrophobically with Thr371 and His505. The interatomic distance between glyasperin A and Pro346 is 2.57 Å. The interaction is stronger than that with inhibitor MLN-4760.
Broussoflavonol F formed a hydrogen bond with His345 and generated hydrophobic interactions with Arg273 and Thr371. The hydrogen bonds formed are not as strong as the hydrogen bonds formed with inhibitor MLN-4760. Meanwhile, sulabiroins A did not form hydrogen bonds with any targeted sites. However, it interacted hydrophobically with Arg273, His345, Pro346, and Thr371. For (2S)-5,7-dihydroxy-4′-methoxy-8-prenylflavanone, similar to glyasperin A, it generated a stronger hydrogen bond with Pro346 distance of 2.89 Å. It also formed hydrophobic interactions with Arg273, His345, and Thr371. Lastly, isorhamnetin generated hydrogen bond with Thr371 and interacted hydrophobically with His345 and Pro346. The summary of interatomic distances formed between each propolis compounds and ACE-2 is available in Table 3 .
Table 3.
Hydrogen bonds between ACE-2 and test ligands.
| No. | Compounds | Hydrogen bonds distance (Å) | Interacting amino acid | Binding ligand group | Binding amino acid group |
|---|---|---|---|---|---|
| 1 | *MLN-4760 | 2.81 | His345 | –OH | –NH |
| 2.91 | Arg273 | –O | –NH | ||
| 2.93 | Pro346 | –NH | –O | ||
| 2.97 | His374 | –O | –NH | ||
| 3.03 | His378 | –O | –NH | ||
| 3.03 | Tyr515 | –O | –OH | ||
| 3.21 | His505 | –OH | –NH | ||
| 3.25 | Arg273 | –OH | –NH2 | ||
| 2 | Glyasperin A | 2.57 | Pro346 | –OH | –O |
| 2.62 | Thr445 | –OH | –OH | ||
| 2.78 | Glu402 | –OH | –OH | ||
| 2.82 | Arg518 | –OH | –NH | ||
| 3.07 | His374 | –OH | –NH | ||
| 3.16 | Tyr515 | –OH | –OH | ||
| 3 | Broussoflavonol F | 2,98 | Tyr127 | –OH | –OH |
| 2,98 | Lys363 | –O | –NH2 | ||
| 3,03 | His345 | –OH | –N | ||
| 3,23 | Lys363 | –OH | –NH2 | ||
| 4 | Sulabiroins A | 3.27 | Thr445 | –O | –OH |
| 5 | (2S)-5,7-dihydroxy-4′-methoxy-8-prenylflavanone | 2.78 | Asp367 | –OH | –O |
| 2.89 | Pro346 | –OH | –OH | ||
| 3.19 | Arg518 | –O | –NH | ||
| 6 | Isorhamnetin | 2.90 | Glu402 | –OH | –O |
| 3.16 | Ala348 | –O | –NH2 | ||
| 3.33 | Thr371 | –OH | –OH | ||
| 3.33 | Arg518 | –OH | –NH |
Native ligand as a control.
In addition, for further consideration, binding similarities were calculated and recorded in Table 2. Based on the results, glyasperin A, broussoflavonol F, sulabiroins A, (2S)-5,7-dihydroxy-4′-methoxy-8-prenylflavanone, and isorhamnetin bound to ACE-2 with the similarity of 77%, 23%, 38%, 46%, and 85%, respectively. As can be seen, only 2 out of 6 compounds that generate binding similarity with the value of greater than 50%. The binding similarity may illustrate the potency of a compound to have the same inhibitory pathway as the potent inhibitor.
Table 2.
Interaction profiles between ACE-2 and test ligands.
| No. | Compounds | Hydrogen bonds | Hydrophobic interactions | Number of interactions | Binding similarity |
|---|---|---|---|---|---|
| 1 | *MLN-4760 | Arg273, His345, Pro346, His374, His378, His505 | Thr347, Thr371, Glu375, Glu402, Phe504, Tyr510, Tyr515 | 13 | 100% |
| 2 | Glyasperin A | Pro346, His374, Glu402, Thr445, Tyr515, Arg518 | Asp269, Thr276, Phe274, His345, Asp367, Thr371, Glu375, Phe504, His505, Tyr510, Arg514 | 17 | 77% |
| 3 | Broussoflavonol F | Tyr127, His345, Lys363 | Glu145, Asn149, Ala153, Gly268, Asp269, Trp271, Arg273, Phe274, Thr276, Asn277, Cys344, Asp367, Thr371, Thr445 | 17 | 23% |
| 4 | Sulabiroins A | Thr445 | Asp269, Trp271, Arg273, Phe274, Thr276, His345, Pro346, Asp367, Thr371, Glu375 | 11 | 38% |
| 5 | (2S)-5,7-dihydroxy-4′-methoxy-8-prenylflavanone | Pro346, Asp367, Arg518 | Trp271, Arg273, Phe274, Thr276, His345, Thr371, His374, Glu375, Thr445 | 12 | 46% |
| 6 | Isorhamnetin | Ala348, Thr371, Glu402, Arg518 | His345, Pro346, Thr347, His374, Glu375, His378, Phe504, Tyr510, Tyr515 | 13 | 85% |
Native ligand as a control.
4. Conclusion
In conclusion, considering the docking scores and the existence of interactions with five targeted sites, five propolis compounds, namely glyasperin A, broussoflavonol F, sulabiroins A, (2S)-5,7-dihydroxy-4′-methoxy-8-prenylflavanone, and isorhamnetin are potential to inhibit the binding of ACE-2 and SARS-CoV-2, with the docking score of −10.8, −9.9, −9.5, −9.3 and −9.2 kcal/mol respectively. However, by taking into account the value of binding similarity, two compounds are considered to be the most potential. These compounds are isorhamnetin and glyasperin A, with the binding similarity values of 85% and 77% accordingly. Molecular dynamics simulation involving both ACE-2 and SARS-CoV-2 with each selected propolis compounds should be conducted to confirm whether the alter of ACE-2 conformation caused by each selected propolis compounds may inhibit SARS-CoV-2 to bind to the protein.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
The authors would like to thank the DRPM Universitas Indonesia for financial support through Grant Publikasi Terindeks Internasional (PUTI) Q2 2020 No: NKB-1713/UN2.RST/HKP.05.00/2020 and the authors extend their appreciation to the Researchers Supporting Project number (RSP-2020/283), King Saud University, Riyadh, Saudi Arabia.
Footnotes
Peer review under responsibility of King Saud University.
References
- Afriza D., Suriyah W.H., Ichwan S.J.A. In silico analysis of molecular interactions between the anti-apoptotic protein survivin and dentatin, nordentatin, and quercetin. J. Phys. Conf. Ser. 2018;1073:032001. doi: 10.1088/1742-6596/1073/3/032001. [DOI] [Google Scholar]
- Alanagreh Lo’ai, Alzoughool F., Atoum M. The human coronavirus disease COVID-19: its origin, characteristics, and insights into potential drugs and its mechanisms. Pathogens. 2020;9(5):331. doi: 10.3390/pathogens9050331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alday, E., Navarro-Navarro, M., Garibay-Escobar, A., Robles-Zepeda, R., Hernandez, J., Velazquez, C., 2016. Advances in pharmacological activities and chemical composition of propolis produced in Americas. Beekeeping and Bee Conservation—Advances in Research.
- Arora, R., Tchertanov, L., 2012. The HIV-1 Integrase: Modeling and Beyond. INTECH
- Astuti I. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes Metab. Syndrome: Clinical Res. Rev. 2020;14(4):407–412. doi: 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S. Remdesivir for the treatment of Covid-19—preliminary report. New England J. Med. 2020 doi: 10.1056/NEJMc2022236. [DOI] [PubMed] [Google Scholar]
- Berretta A.A., Silveira M.A.D., Capcha J.M.C., De Jong D. Propolis and its potential against SARS-CoV-2 infection mechanisms and COVID-19 disease. Biomed. Pharmacother. 2020;110622 doi: 10.1016/j.biopha.2020.110622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronowska, A.K., 2011. Thermodynamics of ligand-protein interactions: implications for molecular design. In Thermodynamics-Interaction Studies-Solids, Liquids and Gases. IntechOpen
- ChemAxon, 2018. Marvin was used for drawing, displaying and characterizing chemical structures, substructures and reactions, Marvin 18.28, 2018, ChemAxon (http://www.chemaxon.com). In. (Version 18.28).
- Christina D., Hermansyah H., Wijanarko A., Rohmatin E., Sahlan M., Pratami D.K., Mun'im A. AIP Conference Proceedings. 2018. Selection of propolis Tetragonula sp. extract solvent with flavonoids and polyphenols concentration and antioxidant activity parameters. [Google Scholar]
- Djalante, R., Lassa, J., Setiamarga, D., Mahfud, C., Sudjatma, A., Indrawan, M., Haryanto, B., Sinapoy, M.S., Rafliana, I., Djalante, S., 2020. Review and analysis of current responses to COVID-19 in Indonesia: Period of January to March 2020. Progress in Disaster Science, 100091. [DOI] [PMC free article] [PubMed]
- Du X., Li Y., Xia Y.-L., Ai S.-M., Liang J., Sang P., Ji X.-L., Liu S.-Q. Insights into protein–ligand interactions: mechanisms, models, and methods. Int. J. Mol. Sci. 2016;17(2):144. doi: 10.3390/ijms17020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine J., Konc J., Samudrala R., Chopra G. CANDOCK: Chemical atomic network-based hierarchical flexible docking algorithm using generalized statistical potentials. J. Chem. Inf. Model. 2020;60(3):1509–1527. doi: 10.1021/acs.jcim.9b00686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flamandita D., Lischer K., Pratami D.K., Aditama R., Sahlan M. AIP Conference Proceedings. 2020. Molecular docking analysis of podophyllotoxin derivatives in Sulawesi propolis as potent inhibitors of protein kinases. [Google Scholar]
- Flaxman S., Mishra S., Gandy A., Unwin H.J.T., Mellan T.A., Coupland H., Whittaker C., Zhu H., Berah T., Eaton J.W. Estimating the effects of non-pharmaceutical interventions on COVID-19 in Europe. Nature. 2020;584(7820):257–261. doi: 10.1038/s41586-020-2405-7. [DOI] [PubMed] [Google Scholar]
- Gekker G., Hu S., Spivak M., Lokensgard J.R., Peterson P.K. Anti-HIV-1 activity of propolis in CD4+ lymphocyte and microglial cell cultures. J. Ethnopharmacol. 2005;102(2):158–163. doi: 10.1016/j.jep.2005.05.045. [DOI] [PubMed] [Google Scholar]
- Ghasemiyeh P., Mohammadi-Samani S. COVID-19 outbreak: challenges in pharmacotherapy based on pharmacokinetic and pharmacodynamic aspects of drug therapy in patients with moderate to severe infection. Heart Lung. 2020;49(6):763–773. doi: 10.1016/j.hrtlng.2020.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güler H.I., Tatar G., Yildiz O., Belduz A.O., Kolayli S. An investigation of ethanolic propolis extracts: their potential inhibitor properties against ACE-II receptors for COVID-19 treatment by Molecular Docking Study. ScienceOpen Preprints. 2020 doi: 10.1007/s00203-021-02351-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huey R., Morris G.M., Forli S. The Scripps Research Institute Molecular Graphics Laboratory; 2012. Using AutoDock 4 and AutoDock Vina with AutoDockTools: A Tutorial. [Google Scholar]
- Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Jeffrey G.A., Jeffrey G.A. vol. 12. Oxford University Press; New York: 1997. (An introduction to hydrogen bonding). [Google Scholar]
- Kim G.-u., Kim M.-J., Ra S.H., Lee J., Bae S., Jung J., Kim S.-H. Clinical characteristics of asymptomatic and symptomatic patients with mild COVID-19. Clin. Microbiol. Infect. 2020;26(7):948.e1–948.e3. doi: 10.1016/j.cmi.2020.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V., Dhanjal J.K., Kaul S.C., Wadhwa R., Sundar D. Withanone and caffeic acid phenethyl ester are predicted to interact with main protease (Mpro) of SARS-CoV-2 and inhibit its activity. J. Biomol. Struct. Dyn. (Just-accepted) 2020;1–17 doi: 10.1080/07391102.2020.1772108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci M.C. Propolis: chemical composition, biological properties and therapeutic activity. Apidologie. 1995;26(2):83–99. doi: 10.1051/apido:19950202. [DOI] [Google Scholar]
- Miyata R., Sahlan M., Ishikawa Y., Hashimoto H., Honda S., Kumazawa S. Propolis components from stingless bees collected on South Sulawesi, Indonesia, and their xanthine oxidase inhibitory activity. J. Nat. Prod. 2019;82(2):205–210. doi: 10.1021/acs.jnatprod.8b00541. [DOI] [PubMed] [Google Scholar]
- Miyata R., Sahlan M., Ishikawa Y., Hashimoto H., Honda S., Kumazawa S. Correction to propolis components from stingless bees collected on south sulawesi, indonesia, and their xanthine oxidase inhibitory activity. J. Nat. Prod. 2020;83(4):1356. doi: 10.1021/acs.jnatprod.0c00282. [DOI] [PubMed] [Google Scholar]
- Miyata R., Sahlan M., Ishikawa Y., Hashimoto H., Honda S., Kumazawa S. Propolis components and biological activities from stingless bees collected on South Sulawesi, Indonesia. HAYATI J. Biosci. 2020;27(1):82. [Google Scholar]
- Pagadala N.S., Syed K., Tuszynski J. Software for molecular docking: a review. Biophys. Rev. 2017;9(2):91–102. doi: 10.1007/s12551-016-0247-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratami D.K., Indrawati T., Istikomah I., Farida S., Pujianto P., Sahlan M. Antifungal activity of microcapsule propolis from Tetragonula spp. to Candida albicans. Commun. Sci. Technol. 2020;5(1):16–21. [Google Scholar]
- Pratami D.K., Mun’im A., Sundowo A., Sahlan M. Phytochemical profile and antioxidant activity of propolis ethanolic extract from Tetragonula Bee. PJ. 2018;10(1):128–135. doi: 10.5530/pj.2018.1.23. [DOI] [Google Scholar]
- Quiroga R., Villarreal M.A. Vinardo: a scoring function based on autodock vina improves scoring, docking, and virtual screening. PLoS ONE. 2016;11(5) doi: 10.1371/journal.pone.0155183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez D., Caballero J. Is it reliable to take the molecular docking top scoring position as the best solution without considering available structural data? Molecules. 2018;23(5):1038. doi: 10.3390/molecules23051038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahlan M., Devina A., Pratami D.K., Situmorang H., Farida S., Munim A., Kusumoputro B., Yohda M., Faried A., Gozan M. Anti-inflammatory activity of Tetragronula species from Indonesia. Saudi J. Biol. Sci. 2019;26(7):1531–1538. doi: 10.1016/j.sjbs.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahlan M., Mandala D.K., Pratami D.K., Adawiyah R., Wijarnako A., Lischer K., Fauzi A. Exploration of the antifungal potential of Indonesian propolis from Tetragonula biroi bee on Candida sp. and Cryptococcus neoformans. Evergr. J. 2020;7:118–125. [Google Scholar]
- Sayusti T., Raffiudin R., Kahono S., Nagir T. Stingless bees (Hymenoptera: Apidae) in South and West Sulawesi, Indonesia: morphology, nest structure, and molecular characteristics. J. Apic. Res. 2020:1–14. [Google Scholar]
- Singhal T. A review of coronavirus disease-2019 (COVID-19) Indian J. Pediatrics. 2020:1–6. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syahputra G. Simulasi docking kurkumin enol, bisdemetoksikurkumin dan analognya sebagai inhibitor enzim12-lipoksigenase. Jurnal Biofisika. 2014;10(1) [Google Scholar]
- Towler P., Staker B., Prasad S.G., Menon S., Tang J., Parsons T., Ryan D., Fisher M., Williams D., Dales N.A. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J. Biol. Chem. 2004;279(17):17996–18007. doi: 10.1074/jbc.M311191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace A.C., Laskowski R.A., Thornton J.M. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. Des. Sel. 1995;8(2):127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020 doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. 2020a. Q&A: How is COVID-19 transmitted? Retrieved October 1st from http://www.who.int
- World Health Organization. 2020b. Retrieved November 25th from https://covid19.who.int/.