Abstract
Objectives
Aside from the outbreak of the coronavirus disease 2019 (COVID-19), serological tests are not well known for their diagnostic value. We assessed the performance of serological tests using stored sera from patients with a variety of pathologic conditions, collected before the 2020 pandemic in Italy.
Methods
Rapid lateral flow tests and Enzyme-Linked Immunosorbent Assays (ELISA) that detect Immunoglobulin M (IgM) and Immunoglobulin G (IgG) antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) were carried out using 1150 stored human serum samples that had been collected in 2018 and 2019. The tests were also run using samples from 15 control patients who had positive or negative oral swab test results, as assessed using real-time reverse transcription-polymerase chain reaction (rRT-PCR). The urea dissociation test was employed to rule out false-positive reactivity in the two antibody detection methods.
Results
The lateral flow tests revealed 21 positive samples from the stored sera: 12 for IgM, four for IgG, and five for IgM/IgG. Among the nine rRT-PCR- positive controls, six individuals presented IgG and three IgM/IgG positivity. Using the urea (6 mol/L) dissociation test, two of the twelve stored samples that had shown IgM positivity were confirmed to be positive. The ELISA test detected four IgM-positive and three IgG-positive specimens. After treatment with 4 mol/L urea, the IgM-positive samples became negative, whereas the IgG positivity persisted. All of the rRT-PCR-positive controls were found to retain IgM or IgG positivity following the urea treatment.
Conclusions
Our findings highlight the limited utility of serological testing for the SARS-CoV-2 virus based on the results of specimens collected before the outbreak of the infection.
Keywords: SARS-CoV-2, COVID-19, Virus, Serological tests, IgM/IgG antibodies
Introduction
Tests for the coronavirus disease 2019 (COVID-19) are instrumental in the management of the pandemic caused by the novel betacoronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Laboratory diagnostic tests currently fall into two major categories: molecular assays that detect SARS-CoV-2 viral RNA, and serological assays that detect antibodies in individuals previously exposed to the virus. The dominant technique remains the real-time reverse transcription-polymerase chain reaction (rRT-PCR) performed on respiratory samples. However, there are major challenges that relate to this complex technique. These include the frequency of false-negative results that lead to early infections being missed, the set-up required to ensure the accurate collection and handling of samples, the long turnaround times for testing, and the need for skilled personnel. Serological tests may avoid some of these difficulties, and the results can complement the information provided by nucleic acid tests that diagnose the COVID-19 infection.
Serological tests for COVID-19 detect antibodies against SARS-CoV-2 antigens. Several assays have been developed that detect Immunoglobulin G (IgG) and Immunoglobulin M (IgM) based on Enzyme-Linked Immunosorbent Assays (ELISA) and lateral flow immunoassays. The assays appear to differ in terms of their overall performance, as well as in their sensitivity, specificity, and ability to measure IgM, IgG, or both simultaneously. However, an investigation that pooled data from 38 studies with a total of 7848 individuals reported a high specificity for the different assays, with some of them reaching a value of 99% (Kontou et al., 2020). Serological tests can assist in determining the immune status of individuals, irrespective of whether or not they have a current infection, and they can be used to estimate herd immunity. They can also be used to indicate when an infection occurred, as IgM antibodies can be a sign of recent infection, while IgG antibodies indicate a later time point (Tang et al., 2020, Zhang et al., 2020, Padoan et al., 2020).
However, there are several important issues that relate to the use of serological tests. For instance, it is currently unclear whether a positive serological test indicates a previous encounter with the virus (Valenti et al., 2020), expresses only a false-positive laboratory result, or indicates a cross reaction with other endemic coronaviruses (Meyer et al., 2014, Okba et al., 2020, Patrick et al., 2006, Lv et al., 2020). Given this uncertainty, caution should be advised when screening programs are carried out based on the seroprevalence of IgM and IgG antibodies against SARS-CoV-2. These programs track the viral outbreak in different countries, support epidemiological investigations, and inform disease prevention policies, and so it is important that the data collected is accurate. Aside from pandemics, the significance of positive serological test results within communities with a low or null viral infection rate has been less well evaluated. It is currently unknown whether serological testing still retains the potential to identify infected individuals among persons with unknown exposure history.
This study aims to assess the performance of serological tests, used for diagnosing a COVID-19 infection, in a large number of serum samples that were stored in 2018 and 2019, well before the 2020 pandemic in Italy.
Material and methods
Specimens
Between January 2018 and December 2019, human serum samples had been obtained from subjects with a variety of pathologic conditions and stored in the Research Laboratory of the Division of Gastroenterology, IRCCS ‘Casa Sollievo della Sofferenza’, in San Giovanni Rotondo, Italy. The blood had been centrifuged at 3000 rpm for 10 min at room temperature, and the serum aliquots had been stored at −80 °C in NUNC cryovial tubes. The samples used in this study were rendered anonymous. Patients’ identities were only held by certain laboratory staff members, and the results of the analyses were not made known to the patients. Two sets of control sera were also analyzed, serving as internal procedural controls. The samples in the first control set were collected from confirmed COVID-19-infected patients who had positive oral swab test results (rRT-PCR positive). These samples were obtained during the April 2020 outbreak in the Apulia region of southern Italy. The samples in the second control set were obtained from blood donated by individuals who had negative oral swab test results (rRT-PCR negative). Rheumatoid factor (RF) values were obtained for all of the individuals who were found to have positive serology for SARS-CoV-2. All of the patients had given their written informed consent for several research projects, which were all approved by the Ethics Committee at our institution.
SARS-CoV-2 IgM and IgG serological tests
The primary aim of this investigation was to assess the diagnostic accuracy of serological tests (qualitative and semi-quantitative) in a population which had not been exposed to the virus.
Qualitative test
The serum samples were first tested using the POCT Diagnostic Kit for SARS-CoV-2 IgM/IgG antibodies (Colloidal Gold), developed by KHB (Shanghai Kehua Bio-engineering Co., Shanghai, China; Cod. REF-423-25-C-CE). This COVID-19 rapid test is a qualitative lateral flow assay for the detection of antibodies against the nucleocapsid (N) protein of SARS-CoV-2. The tests were carried out according to the manufacturer’s instructions. The kit showed no significant cross-reaction between the solid phase antigen and antibodies against other pathogens (hepatitis B, hepatitis C, HIV, and syphilis); in addition, the presence of potentially-interfering endogenous substances, including an RF up to a concentration of 4470 IU/mL, did not affect the performance of the kit. Moreover, as acclaimed by the Italian producer, the test results have a clinical sensitivity of 95.1%, and a clinical specificity of 99.3%, based on an Italian population of 407 subjects, which included 135 samples that were rRT-PCR positive and 272 samples that were rRT-PCR negative (Technogenetics KHB Group POCT Validation Report VR11- POCT clinical study for SARS-CoV-2 IgM/IgG antibodies).
Semi-quantitative test
The semi-quantitative detection of antibodies was performed by means of the ELISA technique, using a commercially available kit to detect IgM and IgG antibodies against the nucleocapsid (N) protein of SARS-CoV-2 (SARS-CoV-2 ELISA kit for IgM and IgG: Cat. No. 30177448, and Cat. No. 30177447, respectively; Tecan, IBL International GmbH, Hamburg, Germany). The tests were carried out according to the manufacturer’s instructions. The results were positive, negative, or borderline. The latter were interpreted as positive for the purposes of this study.
Urea dissociation test of qualitative and semi-quantitative assays
In order to assess the reliability of the positive results from the qualitative and semi-quantitative assays and to reduce the risk of false positives, we tested the positive samples following urea treatment. The urea dissociation test, recently proposed by Wang et al., is based on the principle that urea dissociates the antigen-antibody reactions. The test has been used to evaluate the affinity of IgM and IgG for different pathogens (Wang et al., 2019, Wang et al., 2020). The urea treatment was performed using sera from the rRT-PCR-positive controls, as well as the samples that had tested positive for IgM and/or IgG in either test. For the rapid test, the urea dissociation concentration (6 mol/L) and duration (25 min) were in line with previous studies (Wang et al., 2019, Wang et al., 2020); for ELISA, two different concentrations were tested in different wells: 4 mol/L, previously reported to be the best concentration (Wang et al., 2020), and 6 mol/L. Results were expressed as the Affinity Index (AI) and interpreted according to the threshold values calculation method described by Wang et al. (2020).
Results
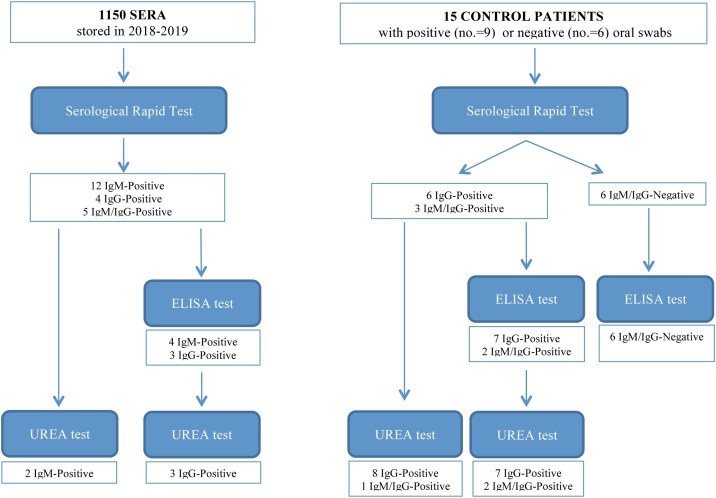
In total, 1150 serum samples were analyzed that had been stored in 2018 and 2019 (531 males and 619 females, with a median age of 58 years [range = 18–96 years]). Control serum samples were collected from nine symptomatic individuals with a SARS-CoV-2 infection (rRT-PCR positive) who were admitted to the IRCCS ‘Casa Sollievo della Sofferenza’ Hospital in March 2020 and April 2020 during the COVID-19 pandemic. Control samples were also collected from six individuals who were rRT-PCR negative. The study procedures and results are depicted in Figure 1 .
Figure 1.
Study procedures and results.
Serological tests
Positive results were obtained for 21 of the 1150 samples (1.83%). Twelve samples (1.04%) tested positive for IgM, four samples (0.35%) for IgG, and five samples (0.43%) for IgM/IgG. For two patients, serum samples had been collected twice during the years 2018–2019, both 1 year apart. Both of the first serum samples tested positive for IgM, and this result was repeated for the second serum samples. Among the rRT-PCR-positive controls, six individuals were IgG positive, and the other three patients were IgM/IgG positive. All of the rRT-PCR-negative controls had no detectable IgG or IgM antibodies (Table 1; Figure 1).
Table 1.
SARS-CoV-2 IgM/IgG antibodies detected using rapid serological tests and ELISA.
| Sample ID | Rapid test |
ELISA |
RF (IU/mL) | ||
|---|---|---|---|---|---|
| IgM | IgG | IgM | IgG | ||
| Sample 1 | + | + | – | – | <10 |
| Sample 2 | + | + | – | – | 294 |
| Sample 3 | + | – | – | – | <10 |
| Sample 4 | + | – | + | – | <10 |
| Sample 5 | + | – | + | – | <10 |
| Sample 6 | + | – | – | – | 18.5 |
| Sample 7 | + | – | – | – | <10 |
| Sample 8 | + | – | – | – | <10 |
| Sample 9 | + | – | – | – | <10 |
| Sample 10 | + | – | – | + | 29.8 |
| Sample 11 | + | + | + | – | >600 |
| Sample 12 | + | – | – | – | <10 |
| Sample 13 | + | – | + | – | <10 |
| Sample 14 | + | – | – | – | <10 |
| Sample 15 | – | + | – | + | <10 |
| Sample 16 | – | + | – | – | <10 |
| Sample 17 | + | + | – | + | 15.8 |
| Sample 18 | – | + | – | – | <10 |
| Sample 19 | + | – | – | – | <10 |
| Sample 20 | + | + | – | – | <10 |
| Sample 21 | – | + | – | – | <10 |
| Positive control 1 | – | + | – | + | <10 |
| Positive control 2 | – | + | – | + | <10 |
| Positive control 3 | – | + | – | + | <10 |
| Positive control 4 | – | + | – | + | <10 |
| Positive control 5 | + | + | + | + | <10 |
| Positive control 6 | + | + | – | + | <10 |
| Positive control 7 | + | + | + | + | <10 |
| Positive control 8 | – | + | – | + | <10 |
| Positive control 9 | – | + | – | + | <10 |
| Negative control 1 | – | – | – | – | <10 |
| Negative control 2 | – | – | – | – | <10 |
| Negative control 3 | – | – | – | – | <10 |
| Negative control 4 | – | – | – | – | <10 |
| Negative control 5 | – | – | – | – | <10 |
| Negative control 6 | – | – | – | – | <10 |
Abbreviations: RF: rheumatoid factor; IgM: Immunoglobulin M; IgG: Immunoglobulin G.
The 21 stored samples with positive serological rapid test results underwent further testing using ELISA for IgM and IgG. Of these, seven samples tested positive (33.33%): four for IgM (19%) and three for IgG (14.3%). The sera that had been tested twice from samples taken 1 year apart were no longer found to be IgM positive. For the rRT-PCR-positive control patients, seven samples were IgG positive and two were IgM/IgG positive. All of the rRT-PCR-negative controls had negative results for both IgM and IgG (Table 1; Figure 1). The RF results were also examined in order to identify any potential effects on the results of these assays. As shown in Table 1, only two sera presented a concentration of this factor >70 IU/mL, the threshold considered to potentially confound the serological test results (Wang et al., 2020). Both of these samples had positive results for IgM/IgG using the rapid test, while only one of these was confirmed to be positive for IgM using ELISA (Table 1).
Comparison of results before and after urea dissociation treatment
Serological rapid test
The dissociation test was carried out for the 21 IgM and/or IgG positive serum samples and for the nine rRT-PCR-positive controls. Two of the IgM positive stored samples (9.6%) remained positive (Table 2 ), while the remaining specimens were found to be negative. Among the rRT-PCR-positive controls, all of the IgG positive results were confirmed, while only one of the three samples that had tested IgM/IgG positive remained so; the other two tested IgG positive (Table 2).
Table 2.
SARS-CoV-2 IgM/IgG antibodies detected using rapid serological tests and ELISA after urea dissociation in samples that had previously tested positive.
| Sample ID | Rapid test |
ELISA |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Before dissociation |
Urea 6M |
Before dissociation |
Urea 4M |
Urea 6M |
||||||
| IgM | IgG | IgM | IgG | IgM | IgG | IgM | IgG | IgM | IgG | |
| Sample 4 | + | – | + | – | + | – | – | nd | – | nd |
| Sample 5 | + | – | + | – | + | – | – | nd | – | nd |
| Sample 10 | + | – | – | – | – | + | nd | + | nd | + |
| Sample 11 | + | + | – | – | + | – | – | nd | – | nd |
| Sample 13 | + | – | – | – | + | – | – | nd | – | nd |
| Sample 15 | – | + | – | – | – | + | nd | + | nd | + |
| Sample 17 | + | + | – | – | – | + | nd | + | nd | + |
| Positive control 1 | – | + | – | + | – | + | nd | + | nd | – |
| Positive control 2 | – | + | – | + | – | + | nd | + | nd | + |
| Positive control 3 | – | + | – | + | – | + | nd | + | nd | + |
| Positive control 4 | – | + | – | + | – | + | nd | + | nd | + |
| Positive control 5 | + | + | – | + | + | + | + | + | + | + |
| Positive control 6 | + | + | – | + | – | + | nd | + | nd | – |
| Positive control 7 | + | + | + | + | + | + | + | + | – | + |
| Positive control 8 | – | + | – | + | – | + | nd | + | nd | – |
| Positive control 9 | – | + | – | + | – | + | nd | + | nd | + |
Abbreviations: IgM: Immunoglobulin M; IgG: Immunoglobulin G; nd.
ELISA
To determine the urea concentration needed to dissociate the immune complexes during the ELISA procedure, the dissociation test was carried out with urea at both 4 mol/L and 6 mol/L. Samples were tested that had positive results for IgM (n = 4) and IgG (n = 3) using ELISA before urea treatment, as well as all of the rRT-PCR-positive controls (n = 9). When the dissociation concentration of urea was 6 mol/L, the AI threshold values were 1.389 and 4.230 for IgM and IgG, respectively. Accordingly, the results became negative for one of the two IgM-positive controls (positive control 7; Table 2), and for three of the nine IgG-positive controls (positive control 1, 6, and 8; Table 2). Conversely, all of the controls who had tested positive for IgM or IgG antibodies remained positive after treatment with urea at 4 mol/L, with AI threshold values of 0.666 and 1.980 for IgM and IgG, respectively (Table 2).
Discussion
Serological tests are universally considered to be accurate and convenient for identifying infected asymptomatic and symptomatic individuals, and for monitoring the immune status of convalescent people recovering from an acute COVID-19 infection (Winter and Hegde, 2020). However, this widely accepted notion only refers to people with a clinical presentation compatible with COVID-19, who undergo testing at the time of an outbreak. The utility of these tests is limited when applied to asymptomatic, healthy subjects with no history of exposure to SARS-CoV-2; in this case, positive serology may only indicate a false-positive result, thus highlighting the need for caution when testing is not within the context of the COVID-19 pandemic.
The present investigation found that 21 out of a total of 1150 serum samples tested positive using the COVID-19 IgM/IgG rapid test kit. These samples had been stored in 2018 and 2019, well before the 2020 pandemic in Italy. As shown in Figure 1, twelve of these samples were IgM positive, four were IgG positive, and five were IgM/IgG positive. Although the total proportion of positive results in our stored sera was rather low (1.8%), and the rate is in accordance with the specificity reported for this kit, the results should not be taken as indicative of an ‘occult’ infection in an otherwise asymptomatic population. Several findings from this study support our claim that the positive results found using the stored sera are false positives. There was concern about the finding that two individuals had IgM-positive results for each of two samples taken 1 year apart. This contrasts sharply with evidence that IgM is only detected in the serum for a couple of weeks in response to an infection, followed by a switch to IgG (Zhang et al., 2020, Xiang et al., 2020). Secondly, it was of some surprise that the majority of positive samples drawn from patients 1 or 2 years before the 2020 pandemic tested positive for IgM and negative for IgG, despite the absence of present or past symptoms reminiscent of a COVID-19 infection. Thirdly, it was found that of the 21 positive rapid test results, only seven produced a positive result using ELISA. This latter observation is indicative of a potential approach to curtail the false positivity of the lateral flow immunochromatographic test. However, although false positivity was reduced with the ELISA assay, it was not completely eliminated, so that a certain amount of uncertainty still remains.
To account for the positive serological results, we should consider that both assays detect antibodies against the nucleocapsid (N) protein of the SARS-CoV-2 virus (personal communication from the Italian seller); consequently, the inconsistent results between the two assays are likely to result from an interference with certain unexplored factors in the assay procedures. Several interfering substances have been reported to potentially affect the results of the POCT Diagnostic Kit for SARS-CoV-2 IgM/IgG antibodies (Colloidal Gold); of these, we explored the relevance of RF. From the available literature, it is known that high (>70 IU/mL) serum concentrations of this factor may affect the detection of SARS-CoV-2 IgM (Wang et al., 2020). As shown in Table 1, only two of the 21 positive samples were found to have concentrations of RF above the relevant threshold, thus suggesting that interference from this factor is minimal, at least in the case of the present study.
In order to avoid unnecessary concern in the case of asymptomatic individuals who test positive for SARS-Co-V-2 antibodies, the specificity of the assays needs to be improved. In the meantime, we evaluated whether the urea dissociation test could help to distinguish true-positive from false-positive results. When the dissociation concentration was 6 mol/L and the duration was 25 min, the urea dissociation test decreased the rate of false-positive results. This test could therefore be adopted by laboratories that run serological tests, and they have the advantage of being low cost as well as rapid. As shown in Table 2, the four stored samples with IgM positive results, detected using ELISA, all tested negative after treatment with 4 mol/L of urea; in contrast, the two IgM positive results from the rRT-PCR-positive patients remained positive even after the urea treatment. As expected, the urea treatment did not affect IgG detection, as this class of antibody has a stronger affinity for the antigen than the IgM class (Mäkelä et al., 1970). In some of our subjects, positive serology may reflect a prior or current infection with other non-SARS-CoV-2 coronavirus strains (Okba et al., 2020) due to some sequence similarities, as has previously been seen in individuals with negative SARS-CoV-2 rRT-PCR results and no symptoms of COVID-19 (Lassaunière et al., 2020).
In conclusion, serological test results should not be interpreted in isolation, but rather in combination with the results from other laboratory tests (e.g., PCR-based tests), the clinical history, and the clinical presentation. Our data show the limited utility of serological tests for the SARS- CoV-2 infection, based on results obtained using sera collected before the COVID-19 outbreak. In order to avoid misdiagnoses due to interfering factors and cross-reactivity with other coronaviruses, it is highly advisable to confirm preliminary positive results by using urea dissociation of the sera followed by an ELISA test. This approach could avoid unnecessary concern over positive serological test results, as well as the need for further invasive testing.
Contributions
AL, FT, AP, and OP designed the study, performed the data analyses, and drafted the article; GAN, RF, and MG recruited patients for the study; TL, GC, DG, and AG performed the experiments; NA, GB, FB, MC, GM, and LdM revised the article for intellectual content. All the authors approved the final version of the manuscript.
Ethical approval
The study was conducted with the approval of the Local Ethics Committee.
Conflict of interest
The authors declares that they have no conflict of interest.
Acknowledgments
We would like to express our very great appreciation to Dr. Angelo Andriulli for his valuable suggestions, and for his effort to critically revise the final manuscript. The study was supported by the “5 × 1000” voluntary contribution to the Fondazione IRCCS ‘Casa Sollievo della Sofferenza’ Hospital, San Giovanni Rotondo (FG), Italy.
References
- Kontou P.I., Braliou G.G., Dimou N.L., Nikolopoulos G., Bagos P.G. Antibody tests in detecting SARS-CoV-2 infection: a meta-analysis. Diagnostics (Basel) 2020;10(5):319. doi: 10.3390/diagnostics10050319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassaunière R., Frische A., Zitta B., Harboe Z.B., Alex C.Y., Nielsen A.C.Y., et al. Evaluation of nine commercial SARS-CoV -2 immunoassays. medRxiv. 2020 doi: 10.1101/2020.04.09.20056325. [DOI] [Google Scholar]
- Lv H., Wu N.C., Tsang O.T., Yuan M., Perera R.A.P.M., Leung W.S., et al. Cross-reactive antibody response between SARS-CoV-2 and SARS-CoV infections. Cell Rep. 2020;31(9):107725. doi: 10.1016/j.celrep.2020.107725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkelä O., Ruoslahti E., Seppälä I.J. Affinity of IgM and IgG antibodies. Immunochemistry. 1970;7(11):917–932. doi: 10.1016/0019-2791(70)90053-4. [DOI] [PubMed] [Google Scholar]
- Meyer B., Drosten C., Müller M.A. Serological assays for emerging coronaviruses: challenges and pitfalls. Virus Res. 2014;194:175–183. doi: 10.1016/j.virusres.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okba N.M.A., Müller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M., et al. Severe acute respiratory syndrome coronavirus 2üspecific antibody responses in coronavirus disease patients. Emerg Infect Dis. 2020;26(7):1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padoan A., Cosma C., Sciacovelli L., Faggian D., Mario Plebani M. Analytical performances of a chemiluminescence immunoassay for SARS-CoV-2 IgM/IgG and antibody kinetics. Clin Chem Lab Med. 2020;58(7):1081–1088. doi: 10.1515/cclm-2020-0443. [DOI] [PubMed] [Google Scholar]
- Patrick D.M., Petric M., Skowronski D.M., Guasparini Booth R.T.F., Krajden M., et al. An outbreak of human coronavirus OC43 infection and serological cross-reactivity with SARS coronavirus. Can J Infect Dis Med Microbiol. 2006;17(6):330–336. doi: 10.1155/2006/152612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y.W., Schmitz J.E., Persing D.H., Charles W., Stratton C.W. Laboratory diagnosis of COVID-19: current issues and challenges. J Clin Microbiol. 2020;58(6):e00512–e00520. doi: 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenti L., Bergna A., Pelusi S., Facciotti F., Lai A., Tarkowski M., et al. SARS-CoV-2 seroprevalence in healthy blood donors during the COVID -19 Milan outbreak. medRxiv. 2020 doi: 10.1101/2020.05.11.20098442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Lei Y., Lu X., Wang G., Du Q., Guo X., et al. Urea-mediated dissociation alleviate the false-positive Treponema pallidum-specific antibodies detected by ELISA. PLoS One. 2019;14:e0212893. doi: 10.1371/journal.pone.0212893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Du Q., Guo B., Mu D., Lu X., Ma Q., et al. A method to prevent SARS-CoV-2 IgM false positives in gold immunochromatography and enzyme-linked immunosorbent assays. J Clin Microbiol. 2020;58(6) doi: 10.1128/JCM.00375-20. e00375–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter A.K., Hegde S.T. The important role of serology for COVID-19 control. Lancet Infect Dis. 2020;20(7):758–759. doi: 10.1016/S1473-3099(20)30322-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang J., Yan M., Li H., Liu T., Lin C., Huang S., et al. Evaluation of enzyme-linked immunoassay and colloidal gold-immunochromatographic assay kit for detection of novel coronavirus (SARS Cov-2) causing an outbreak of pneumonia (COVID -19) medRxiv. 2020 2020.02.27.20028787. [Google Scholar]
- Zhang W., Du R.H., Li B., Zheng X.S., Yang X.L., Hu B., et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 2020;9(1):386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]