Abstract
The COVID-19 pandemic has caused an unprecedented public health, social, and economic crisis. Improving understanding on available tests for detecting COVID-19 is critical for effective management of the pandemic. We proposed that a multidisciplinary expert panel can establish recommendations on ideal use of diagnostic tools, with a focus on RT-PCR and serological high-affinity antibodies (both IgM and IgG) tests for the Latin America region.
Study design
A collaborative multidisciplinary panel of 5 recognized experts in Latin America (an infectious disease specialist, three pathologists, and an immunologist) was convened and supported by Roche Diagnostics to develop standard guidelines and an evidence-based document of best practices on the use of diagnostic tools for COVID-19.
Results
The authors reached consensus on the applicability of diagnostic tools to provide testing algorithms for the use of RT-PCR and serological high-affinity antibodies (both IgM and IgG) tests in three settings: 1) For asymptomatic subjects exposed to a SARS-CoV-2 infected person; 2) For epidemiological purposes and; 3) For symptomatic subjects.
Conclusion
The serological high-affinity SARS-CoV-2 antibodies (both IgM and IgG) tests play a key role in COVID-19 diagnosis. These tests can be applied for suspected false-negative RT-PCR results and for individual determination of response. The use of these tests can also contribute greatly to public health strategies, such as population screening and supporting vaccination planning. Serological status for high-affinity antibodies (both IgM and IgG) should be performed ideally 21 days after potential infectious contact, given that the majority of exposed individuals will have seroconverted.
Keywords: COVID-19, Diagnosis, Serological test, High-affinity antibodies, RT-PCR
Introduction
In December 2019, atypical pneumonia cases caused by a new coronavirus were identified in Wuhan, a city of Hubei Province in China (Zhu et al., 2020). Within days, the virus had spread, resulting in an epidemic throughout China (The Novel Coronavirus Pneumonia Emergency Response Epidemiology Team, 2020). An increasing number of cases were reported in countries around the world in the ensuing weeks (WHO, 2020b). In February 2020, the World Health Organization (WHO) named the disease as COVID-19, which stands for “coronavirus disease 2019” (WHO, 2020c). The virus that causes COVID-19 was then named as Severe Acute Respiratory Syndrome CoronaVirus 2 (SARS-CoV-2) (Coronaviridae Study Group of the International Committee on Taxonomy of Viruses, 2020). COVID-19 has since been declared a global pandemic (Cucinotta and Vanelli, 2020), with 15,301,530 cases worldwide and 625,005 deaths globally. In the Americas, the numbers are also staggering: 11,667,196 confirmed cases with 419,995 deaths (as of August 17th, 2020) (PAHO, 2020).
The initial stage involves an incubation period when SARS-CoV-2 multiplies and establishes itself mainly in the respiratory system. During the second stage, localized inflammation can occur in the lungs. The third (and most severe) stage of the disease can cause the syndrome of extrapulmonary systemic hyperinflammation (Siddiqi and Mehra, 2020).
RT-PCR is a test for diagnosing COVID-19, based on nasopharyngeal swab samples or other upper respiratory tract samples (Wang et al., 2020b). In symptomatic individuals, viral RNA can be detectable early on day one of symptoms and culminate within the first week of symptom onset. By week three, positivity of the test for detecting viral RNA starts to decline (Sethuraman et al., 2020). A downside of this sample collection approach involves false-negative results, largely due to inappropriate timing of sample collection relative to illness onset and poor sampling technique, especially for nasopharyngeal swabs. Given that the design for the RT-PCR test is based on the genome sequence of SARS-CoV-2, its specificity is almost 100%, with few false-positive results (Sethuraman et al., 2020).
Another diagnostic tool for detecting SARS-CoV-2 infection is serological testing which evaluates the host immune response (Loeffelholz and Tang, 2020). It is essential for patients with mild to moderate illness who may present two weeks after illness onset. Serological diagnosis is also becoming an important tool to help understand the extent of COVID-19 in the community (Sethuraman et al., 2020).
Antibodies start to increase from the second week of symptom onset, constituting the earliest and most sensitive serological marker, with IgM and IgG levels peaking in the second and third weeks of illness. Subsequently, IgM decreases by week 5, while IgG remains high beyond 7 weeks (Sethuraman et al., 2020).
These findings together with the plethora of available testing methodologies (CDC, 2020b, Loeffelholz and Tang, 2020), evolving knowledge on the behavior of the virus, and the complexity of the human immune response, have led to the need for guidance on how to use and appropriately interpret results of the available tests.
As the pandemic progresses, it has become clear that the primary transmission pathway is through respiratory aerosols (Bahl et al., 2020) as well as through direct contact of eyes, nose, or mouth with contaminated surfaces (Ong et al., 2020). The virus has also been detected in nonrespiratory samples such as stools, urine, blood, ocular secretions, and semen (Wang et al., 2020b). The risk of transmission of SARS-CoV-2 from an infected person to another appears to vary and depends on the type and duration of exposure, use of preventive measures, and other individual factors (Rosenberg et al., 2020).
Latin America is a large and heterogeneous territory, including well-developed and poor areas with limited resources, in which the pandemic rapidly spreads. The aim of this paper is to provide Latin American clinicians with guidance on the use of RT-PCR and serological high-affinity antibodies (both IgM and IgG) tests.
Methods
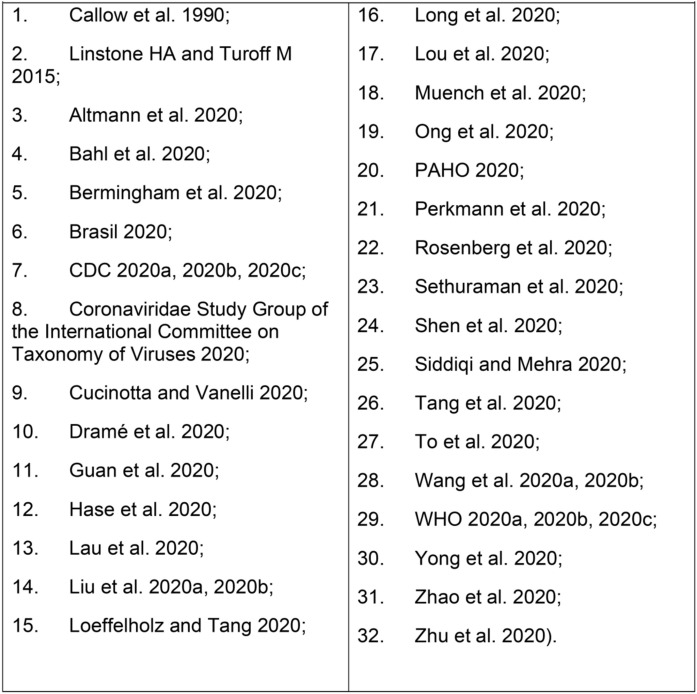
Five recognized experts in Latin America joined an online expert panel and worked collaboratively on an online application (Within3®) from June 12th to 24th, 2020, supported by Roche Diagnostics. Panel members had either clinical or scientific experience in infectious disease or immunology and serological tests. Adopting standard guideline development processes (Linstone and Turoff, 2015), a literature review was performed on serological diagnosis and panelists shared the articles on COVID-19 listed in Chart 1 .
Chart 1.
Articles selected by panelists for discussion during expert pane.
The search was performed on Medline only, from January 2020 until the start date of the expert panel in June 2020.
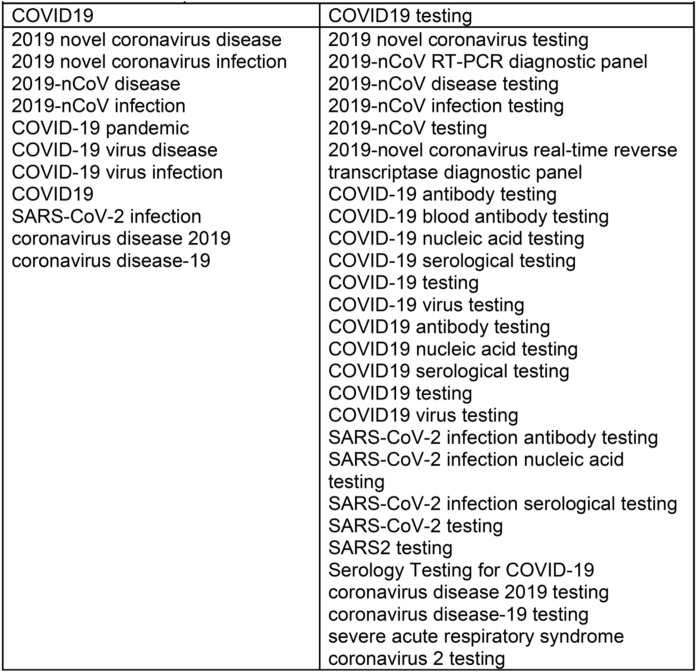
All MESH Terms related to “COVID19” and “COVID19 testing” were used as main descriptor terms by using the Boolean connector “OR” to include all their supplementary concept terms; plus “AND” to connect both. These are shown in Chart 2 .
Chart 2.
All MESH Terms related to “COVID19” and “COVID19 testing” used as main descriptor.
The purpose of the expert panel was to develop a test algorithm for the three situations proposed in the paper, not including antigen or antibody rapid testing, and no search for rapid testing was conducted.
On the basis of the papers retrieved, an infectious disease specialist prepared nine questions (Chart 3 ) and drafts of algorithms testing for SARS-CoV-2, focusing on the use of RT-PCR and serology testing in different settings. Panelists had the opportunity to suggest modifications to these algorithms and were required to propose evidence-based best practices for the RT-PCR and serological diagnosis of COVID-19.
Chart 3.
Narrative summaries of clinical questions addressed by panel to formulate recommendations for algorithms .
These preliminary efforts served as the basis for discussion and to establish the guidelines. The panelists had the opportunity to make further reviews and remarks using the online platform in reaching the consensus for the guidelines presented.
Results
Algorithms emerged in a consensus of the panelists on serologic testing for COVID-19 in Latin America in three different settings:
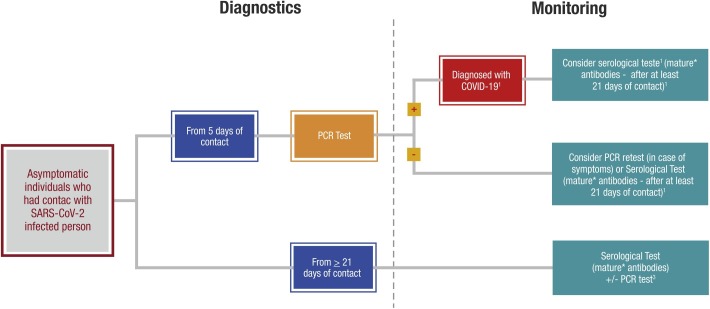
1. Asymptomatic individual exposed to Sars-CoV-2 infected patients – Algorithm 1.
For asymptomatic individuals who had contact with a confirmed case of COVID-19 (Algorithm 1), RT-PCR should be performed preferably after 5 days of contact, while a serology test can be performed to detect mature antibodies against SARS-CoV-2, high-affinity antibodies (both IgM and IgG), ideally 21 days after contact.
Accordingly, because of the lower sensitivity of antibody tests in detecting infection during earlier phases (To et al., 2020), the panelists proposed adoption of a minimum cut-off period after potential infectious contact for performing serological evaluation of asymptomatic individuals (Figure 1 ) (Dramé et al., 2020, Zhao et al., 2020).
Figure 1.
Asymptomatic individual exposed to Sars-Cov-2 infected patients — Algorithm 1.
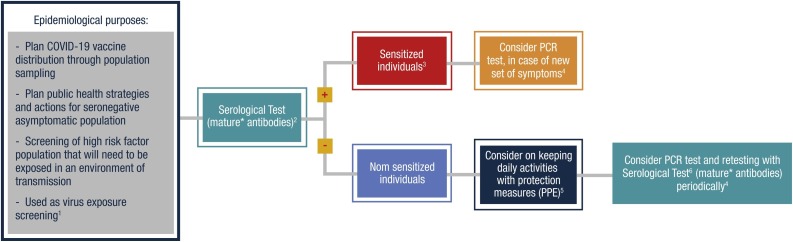
2. Epidemiological purposes – Algorithm 2.
Serological testing also has utility for epidemiological purposes such as virus exposure screening, especially of high-risk populations (police and military personnel, food market suppliers, traffic agents, medical personnel), planning public health strategies and actions for seronegative asymptomatic populations, and planning vaccine distribution through population sampling. In these situations, the use of mature antibodies serological tests can help understand those who are sensitized to SARS-CoV-2. For those, PCR testing should be considered in the case of a new set of symptoms. In such cases, social distancing and other precautionary measures according to local health authority decisions should be taken. For nonsensitized individuals, performing daily activities using personal protection equipment (PPE) should be considered as well as regular retesting with PCR and serological tests (Figure 2 ).
Figure 2.
Epidemiological purposes — Algorithm 2.
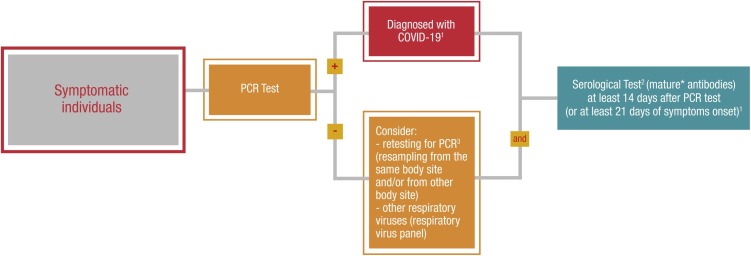
3. Symptomatic individuals – Algorithm 3.
For symptomatic individuals, the gold standard is the RT-PCR test performed predominantly on nasopharynx and/or oropharyngeal swab samples. If patients present with a negative PCR test, retesting for PCR (same and/or other body site) should be considered, as well as performing a respiratory virus panel. In all symptomatic people, a serology test to measure high-affinity antibodies (both IgM and IgG) could be performed at least 14 days after PCR test or 21 days after symptom onset (Figure 3 ).
Figure 3.
Symptomatic individuals — Algorithm 3.
The rationale for this timeframe relies on a study that reported the proportion of patients with positive virus-specific IgG reached 100% approximately 17 to 19 days after the onset of symptoms (Long et al., 2020).
4. Special situations – no algorithm proposed.
Some clinical conditions were not discussed in this expert panel, such as patients with or without symptoms that have long-term SARS-CoV-2 RNA detection in nasopharyngeal swabs, developing weak or no clinical signs, and undetectable antibodies in serum 15–20 days after manifesting clinical symptoms, and the rare clinical condition of patients suspected to have SARS-CoV-2 reinfection.
A decrease in IgG antibodies in patients with SARS-CoV-2 has been documented. A recent study showed a drop of 26.5% in detectable IgG antibodies over 3 months by using lateral flow assay. In the population aged 75+ years, the drop was 39%. These data suggest that the possibility of decreasing population immunity over time could increase the risk of reinfection (Ward et al., 2020). Currently the importance of antibody decline for reinfection by SARS-CoV-2 is not yet answered and further longitudinal studies are needed. SARS-CoV-2 vaccine studies will also support the clarification of this issue.
Lastly, the participants explained the rationale for their recommendations until a final consensus was reached (Figure 4 ).
Figure 4.
Important primary considerations when defining any testing approach.
Discussion
RT-PCR for SARS-CoV-2, based on samples obtained preferably from the upper nasopharynx (Loeffelholz and Tang, 2020) remains the gold standard for the diagnosis of the acute phase of COVID-19 (Loeffelholz and Tang, 2020, Wang et al., 2020b). However, there are some drawbacks of this technique, namely its variability in accuracy depending on the specimen (Wang et al., 2020a), hazards in collecting samples (Shen et al., 2020), and sensitivity concerns (Hase et al., 2020, Shen et al., 2020). For instance, negative tests in patients with SARS-CoV-2 lower respiratory tract infection and minimum upper respiratory symptoms are not uncommon (Hase et al., 2020, Liu et al., 2020a). Given the limitations of the RT-PCR, in the context of urgent need for accurate detection of infected subjects and their subsequent isolation as a pivotal step for effective prevention of the spread of the SARS-CoV-2 virus (Guan et al., 2020), serological testing plays an essential role in differential diagnostic and epidemiological settings (CDC, 2020b, Lou et al., 2020, Yong et al., 2020).
Nevertheless, relying solely on IgM serological detection for the diagnosis of acute disease is not a suitable strategy, particularly for the early acute phase. In one study (Loeffelholz and Tang, 2020), all 39 patients had both IgM and IgG after 5-7 days of symptom onset. In another Chinese study of 285 patients, three seroconversion patterns were observed: synchronous seroconversion of both IgM and IgG, IgM seroconversion earlier than IgG (expected pattern), and IgM seroconversion after IgG. The proportion of patients with virus-specific IgM peaked at 94.1% in approximately 20–22 days, whereas for IgG, 100% reached a peak 17–19 days after symptom onset (Long et al., 2020). However, antibody responses against SARS-CoV-2 are not fully understood (Callow et al., 1990), and the neutralizing activities of detected IgG antibodies have yet to be determined (Lou et al., 2020).
According to the North American Centers for Disease Control (CDC-USA), the results of serological tests should not be used as a single diagnostic test for an acute infection, excluding, or diagnosing SARS-CoV-2 infections (CDC, 2020b). Moreover, the US Food and Drug Administration (FDA) recommends use of serological tests to detect SARS-CoV-2 antibodies by health professionals, as this may help identify individuals exposed to or who have recovered from COVID-19 infection (CDC, 2020a). In Latin America, the Brazilian Ministry of Health recommends the use of laboratory tests, RT-PCR until the eighth day of symptom onset and immunological, which detects the presence of antibodies in samples collected from the eighth day of symptom onset in patients presenting with a flu-like syndrome or Severe Acute Respiratory Syndrome (SARS) (Brasil, 2020). Despite the increased knowledge on the utility of serological tests, a recent survey by the Royal College of Physicians highlighted misinterpretation issues, where 40% of respondents considered patients to have “cleared COVID-19” in cases with active symptoms and IgM–IgG + serologies (Bermingham et al., 2020).
Serological testing for SARS-CoV-2 high-specificity antibodies can also be used as an additional diagnostic tool for suspected false-negative RT-PCR results (Perkmann et al., 2020) or for individual determination of antibody levels to trace who has been infected in the past (Perkmann et al., 2020, Yong et al., 2020). In some situations, the use of serological testing may also be applied to determine the immunity status of asymptomatic subjects with an epidemiological history of a high risk of exposure to people with COVID-19 (Lou et al., 2020). In such settings, serologic testing at appropriate intervals following contact with infected subjects might result in relatively fewer false-positive results (Liu et al., 2020b).
Serological testing also plays a pivotal role in population-based seroepidemiological studies. It provides essential data about SARS-CoV-2 transmission dynamics and allows interventions to reduce transmission of the disease. Moreover, this testing can be used to assess seroprevalence overall or in specific groups, thereby helping to estimate core characteristics of the pandemic and to plan intervention measures such as vaccination of populations (Altmann et al., 2020, Tang et al., 2020, Yong et al., 2020).
The World Health Organization (WHO) states that seroepidemiological investigation can help understand and provide robust estimates of clinical, epidemiological, and virological characteristics of COVID- 19 (WHO, 2020a).
In Latin America, given the high prevalence of SARS-CoV-2 infection, a serological test can be used to determine the level of exposure and identify people who may be sensitized. The latter tests should ideally provide high specificity, with a small confidence interval, detection of high-affinity antibodies and no cross-reactivity with other coronaviruses (Lau et al., 2020, Muench et al., 2020).
Finally, the CDC Interim Guidelines for COVID-19 Antibody Testing (CDC, 2020b) recommend the use of serological assays in some other scenarios: (a) as a method to support the diagnosis of acute COVID-19 illness for persons who present late onset, for whom serologic testing is offered in addition to RT-PCR; (b) as a method to support establishing a diagnosis when patients present with late complications of COVID-19 illness; and (c) as a method to reduce false-positive results in high prevalence settings.
Limitations
Although based on well-established consensus formation techniques and drawing on panelist’s expertise, these recommendations do not constitute a statement from the institutions or associations to which these professionals are affiliated. The main limitations of this expert panel consensus are selection bias, observer bias, confirmation bias, publication bias, and cohort effects (different features and pace of the COVID-19 pandemic in each country of Latin America).
Implications
This expert panel consensus can help clinicians to apply testing for SARS-CoV-2 on an individual level. Moreover, the guidance can also support decision-making stakeholders when acting on public health measures such as seroprevalence studies and business reopening. Lastly, the consensus can support payers from both private and public settings with a more straightforward tool for evaluating the use of a specific test (or sequence of tests).
Conclusion
In conclusion, serological testing and studies are of great importance for public health strategies, such as population screening, and will prove pivotal to support planning of vaccination strategies. Serological status for high-affinity antibodies (both IgM and IgG) should be determined 21 days after potential infectious contact to allow appropriate time for sensitization to SARS-CoV-2 following exposure.
Conflict of interests
Antonio Condino-Neto: no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. Pablo E. Bonvehi: My only conflict of interest statement on this issue is that I was convened to participate in this white paper by Roche. Juan Carlos Gómez de la Torre: Speaker for Sanofi - Vaccines, Speaker for MSD - Vaccines, Speaker for Cepheid - Molecular diagnostics in infectious diseases. Klever Vinicio Sáenz-Flor: I do not have any financial interests or personal relationships which may be considered as potential competing interests. Carlos Eduardo Ferreira: I participate as a speaker of company events: Ortho Clinical Diagnostics, Roche Diagnostics, Siemens Healthineers, Snibe and Abbott Diagnostics. I participated as a member of the Advisory Board: Roche Diagnostics. Associative Activity: President of the Brazilian Society of Clinical Pathology / Laboratory Medicine (SBPC / ML) - Biennium 2020-2021
Funding source
This paper was supported by Roche Diagnostics.
Ethical approval
Not applicable.
Acknowledgment
This paper was supported by Roche Diagnostics. The data were collected by the sponsor and analyzed in conjunction with the authors. All of them contributed to writing the article and approved to submit for publication. Medical writing assistance was provided by CoreBox Medical Communications.
References
- Altmann D.M., Douek D.C., Boyton R.J. What policy makers need to know about COVID-19 protective immunity. Lancet. 2020;395(10236):1527–1529. doi: 10.1016/S0140-6736(20)30985-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahl P., Doolan C., de Silva C., Chughtai A.A., Bourouiba L., MacIntyre C.R. Airborne or droplet precautions for health workers treating COVID-19? J Infect Dis. 2020 doi: 10.1093/infdis/jiaa189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham W.H., Wilding T., Beck S., Huissoon A. SARS-CoV-2 serology: test, test, test, but interpret with caution! Clin Med (Lond) 2020;20(4):365–368. doi: 10.7861/clinmed.2020-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasil . BVS; Brasília: 2020. Ministério da Saúde. Guia de Vigilância Epidemiológica: emergência de saúde pública de importância nacional pela doença pelo Coronavírus 2019.https://portalarquivos.saude.gov.br/images/pdf/2020/April/07/GuiaDeVigiEpidemC19-v2.pdf [Google Scholar]
- Callow K.A., Parry H.F., Sergeant M., Tyrrell D.A. The time course of the immune response to experimental coronavirus infection of man. Epidemiol Infect. 1990;105(2):435–446. doi: 10.1017/s0950268800048019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC . 2020. Frequently asked questions about coronavirus (COVID-19) for laboratories.https://www.cdc.gov/coronavirus/2019-ncov/lab/faqs.html [Google Scholar]
- CDC . 2020. Interim guidelines for COVID-19 antibody testing.https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antibody-tests-guidelines.html [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5(4):536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucinotta D., Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020;91(1):157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dramé M., Tabue Teguo M., Proye E., Hequet F., Hentzien M., Kanagaratnam L., et al. Should RT-PCR be considered a gold standard in the diagnosis of COVID-19? J Med Virol. 2020;92:2312–2313. doi: 10.1002/jmv.25996. 10.1002/jmv.25996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.-J., Chen R.-C., Zhong N.-S. Strategies for the prevention and management of coronavirus disease 2019. Eur Respir J. 2020;55(4) doi: 10.1183/13993003.00597-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase R., Kurita T., Muranaka E., Sasazawa H., Mito H., Yano Y. A case of imported COVID-19 diagnosed by PCR-positive lower respiratory specimen but with PCR-negative throat swabs. Infect Dis (Lond) 2020;52(6):423–426. doi: 10.1080/23744235.2020.1744711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau C.S., Hoo S.P., Yew S.F., Ong S.K., Lum L.T., Heng P.Y., et al. Evaluation of the roche elecsys anti-SARS-CoV-2 assay. medRxiv. 2020 doi: 10.1101/2020.06.28.20142232. posted 2020 June 29. [DOI] [Google Scholar]
- Linstone H.A., Turoff M. New Jersey Institute of Technology; Addison Wesley Newark, NJ: 2015. The Delphi method: techniques and applications. [Google Scholar]
- Liu R., Han H., Liu F., Lv Z., Wu K., Liu Y., et al. Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin Chim Acta. 2020;505:172–175. doi: 10.1016/j.cca.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Wu S., Tao H., Zeng G., Zhou F., Guo F., et al. Prevalence of IgG antibodies to SARS-CoV-2 in Wuhan — implications for the ability to produce long-lasting protective antibodies against SARS-CoV-2. medRxiv. 2020 doi: 10.1101/2020.06.13.20130252. 2020.06.13.20130252. [DOI] [Google Scholar]
- Loeffelholz M.J., Tang Y.-W. Laboratory diagnosis of emerging human coronavirus infections — the state of the art. Emerg Microbes Infect. 2020;9(1):747–756. doi: 10.1080/22221751.2020.1745095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.-X., Liu B.-Z., Deng H.-J., Wu G.-C., Deng K., Chen Y.-K., et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26(6):845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- Lou B., Li T.-D., Zheng S.-F., Su Y.-Y., Li Z.-Y., Liu W., et al. Serology characteristics of SARS-CoV-2 infection since exposure and post symptom onset. Eur Respir J. 2020;56(2) doi: 10.1183/13993003.00763-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muench P., Jochum S., Wenderoth V., Ofenloch-Haehnle B., Hombach M., Strobl M., et al. Development and validation of the Elecsys Anti-SARS-CoV-2 immunoassay as a highly specific tool for determining past exposure to SARS-CoV-2. J Clin Microbiol. 2020;58(10) doi: 10.1128/JCM.01694-20. https://jcm.asm.org/content/early/2020/07/31/JCM.01694-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S.W.X., Tan Y.K., Chia P.Y., Lee T.H., Ng O.T., Wong M.S.Y., et al. Air, surface environmental, and personal protective equipment contamination by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) from a symptomatic patient. JAMA. 2020;323(16):1610–1612. doi: 10.1001/jama.2020.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAHO . PAHO/WHO; Washington: 2020. PAHO/WHO response. 20 July 2020. Report no.: 17. https://iris.paho.org/handle/10665.2/52518. [Accessed 26 June 2020] [Google Scholar]
- Perkmann T., Perkmann-Nagele N., Breyer M.-K., Breyer-Kohansal R., Burghuber O.C., Hartl S., et al. Side by side comparison of three fully automated SARS-CoV-2 antibody assays with a focus on specificity. Clin Chem. 2020;66(11):1405–1413. doi: 10.1093/clinchem/hvaa198/5890466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg E.S., Dufort E.M., Blog D.S., Hall E.W., Hoefer D., Backenson B.P., et al. COVID-19 testing, epidemic features, hospital outcomes, and household prevalence, New York State—March 2020. Clin Infect Dis. 2020;71(8):1953–1959. doi: 10.1093/cid/ciaa549/5831986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethuraman N., Jeremiah S.S., Ryo A. Interpreting diagnostic tests for SARS-CoV-2. JAMA. 2020;323(22):2249. doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- Shen M., Zhou Y., Ye J., Abdullah Al-Maskri A.A., Kang Y., Zeng S., et al. Recent advances and perspectives of nucleic acid detection for coronavirus. J Pharm Anal. 2020;10(2):97–101. doi: 10.1016/j.jpha.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi H.K., Mehra M.R. COVID-19 illness in native and immunosuppressed states: a clinical-therapeutic staging proposal. J Heart Lung Transplant. 2020;39(5):405–407. doi: 10.1016/j.healun.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y.-W., Schmitz J.E., Persing D.H., Stratton C.W. Laboratory diagnosis of COVID-19: current issues and challenges. J Clin Microbiol. 2020;58(6) doi: 10.1128/jcm.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Novel Coronavirus Pneumonia Emergency Response Epidemiology Team The epidemiological characteristics of an outbreak of 2019 Novel Coronavirus Diseases (COVID-19) — China, 2020. China CDC Weekly. 2020;2(8):113–122. doi: 10.46234/ccdcw2020.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K.-W., Tsang O.T.-Y., Leung W.-S., Tam A.R., Wu T.-C., Lung D.C., et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward H., Cooke G., Atchison C., Whitaker M., Elliott J., Moshe M., et al. Declining prevalence of antibody positivity to SARS-CoV-2: a community study of 365,000 adults. medRxiv. 2020 doi: 10.1101/2020.10.26.20219725. 10.26.20219725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . WHO; Washington: 2020. Laboratory testing for coronavirus disease (COVID-19) in suspected human cases: interim guidance.https://apps.who.int/iris/handle/10665/331329 [Google Scholar]
- WHO . WHO; Washington: 2020. Novel coronavirus (2019 n-Cov). Situation report — 10.https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200130-sitrep-10-ncov.pdf?sfvrsn=d0b2e480_2 [Google Scholar]
- WHO . 2020. WHO Director-General’s remarks at the media briefing on 2019-nCoV on 11 February 2020.https://www.who.int/dg/speeches/detail/who-director-general-s-remarks-at-the-media-briefing-on-2019-ncov-on-11-february-2020 [Google Scholar]
- Yong G., Yi Y., Tuantuan L., Xiaowu W., Xiuyong L., Ang L., et al. Evaluation of the auxiliary diagnostic value of antibody assays for the detection of novel coronavirus (SARS-CoV-2) J Med Virol. 2020;92:1975–1979. doi: 10.1002/jmv.25919. 10.1002/jmv.25919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/nejmoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]