Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a beta coronavirus that uses the human angiotensin-converting enzyme 2 (ACE2) receptor as a point of entry. The present review discusses the origin and structure of the virus and its mechanism of cell entry followed by the therapeutic potentials of strategies directed towards SARS-CoV2-ACE2 binding, the renin-angiotensin system, and the kinin-kallikrein system. SARS-CoV2-ACE2 binding-directed approaches mainly consist of targeting receptor binding domain, ACE2 blockers, soluble ACE2, and host protease inhibitors. In conclusion, blocking or manipulating the SARS-CoV2-ACE2 binding interface perhaps offers the best tactic against the virus that should be treated as a fundamental subject of future research.
Keywords: ACE2, COVID-19, SARS-CoV2, Origin, Therapeutic, Transmission
1. Introduction
In December 2019, Wuhan, a city in China, witnessed the outbreak of a pneumonia disease known as coronavirus disease (COVID-19). The disease spread throughout the world, forcing the World Health Organization (WHO) to declare it a global pandemic on March 11, 2020. During the last seven months, it has infected over 14 million people worldwide, including health care workers, caused nationwide lockdowns, and damaged economies worldwide (Hanaei and Rezaei, 2020; Jabbari et al., 2020; Kafieh et al., 2020).
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is known as the causative agent of COVID-19. It is a beta coronavirus and uses the human angiotensin-converting enzyme 2 (ACE2) receptor as a point of entry (Hoffmann et al., 2020). The distribution of ACE2 over a variety of tissues explains the evidence for the occurrence of infection in multiple organs/systems from the central nervous system to the respiratory, cardiovascular, and digestive systems (Jahanshahlu and Rezaei, 2020a; Lotfi et al., 2020; Saleki et al., 2020). People mostly experience a benign phenotype of the disease; however, one of every five people develops a severe phenotype of the disease. It might be the presentation of individual genetic background (Darbeheshti and Rezaei, 2020; Yousefzadegan and Rezaei, 2020) and immune profile (Fathi and Rezaei, 2020; Nasab et al., 2020; Saghazadeh and Rezaei, 2020a; Sahu et al., 2020; Yazdanpanah et al., 2020). People with severe COVID-19 mostly show evidence of hyper inflammation, mainly managed by cytokines and macrophages (Bahrami et al., 2020), and therefore, a hypothesis emerged that the immune system, either directly or indirectly, manages the lethal pathogenesis of infection (Yazdanpanah et al., 2020). As a result, anti-inflammatory agents are considered along with antivirals and other supportive care (Saghazadeh and Rezaei, 2020b).
To date, there have been records of more than 600,000 deaths related to COVID-19 worldwide, while no specific treatment and prevention are available (Lotfi et al., 2020). Many efforts occurred in solving the issue (Moazzami et al., 2020; Mohamed et al., 2020; Momtazmanesh et al., 2020; Moradian et al., 2020) and adding to the potential of some methods for the treatment of COVID-19 (Basiri et al., 2020; Jahanshahlu and Rezaei, 2020b; Rabiee et al., 2020). The best practical method remained is the understanding of the pathogenesis of the disease. The present review discusses from the origin of the virus and its modes of transmission, how the previously suggested mode of transmission can be reaffirmed using ACE2 expression, to structural analysis, how much the novel coronavirus resembles other beta coronaviruses and how slight structural changes have affected its virulence, and also to possible therapeutic strategies.
2. From SARS-CoV1 to SARS-CoV2: mechanism of pathogenesis and origin
SARS-CoV-2 shares similarities with the other human coronaviruses (hCoVs) at both the structural level and the associated clinical symptoms. In particular, it seems to be most closely connected with SARS-CoV, which itself was the cause of an outbreak in 2002. Studies have revealed that there is about 80% structural identity between SARS-CoV and SARS-CoV-2 (Basiri et al., 2020; Guo et al., 2020b; Zhou et al., 2020a, Zhou et al., 2020b, Zhou et al., 2020c).
Such a high similarity allowed scientists to speculate on the mechanism of cell entry and pathogenesis of SARS-CoV-2. Relevant statements proven by recent studies are as follows. First, it was suggested that like SARS-CoV, SARS-CoV-2 uses the same receptor, i.e., ACE2, to enter cells and cause infection. Zhou et al. (Zhou et al., 2020a, Zhou et al., 2020b, Zhou et al., 2020c) proved that the ACE2 is the cell entry receptor of the SARS-CoV-2 and except mice cells, a variety of animal cells expressing ACE2 receptor and also human cells with ACE2 on their surface can be considered as the target of the virus. Also, they showed that other coronavirus receptors, such as aminopeptidase N (APN) and dipeptidyl peptidase (DPP), do not play a role in the cell entry of the SARS-CoV-2. Further studies also found that SARS-CoV-2 receptor-binding domain (RBD) has more affinity to interact and attach ACE2 receptor on the surface of the target cells, this may be the most likely cause of SARS-CoV-2 being more infectious and pathogenic than SARS-CoV--1 (Procko, 2020; Wan et al., 2020; Wrapp et al., 2020).
Second, the SARS-CoV-2 may also be transmitted between humans and animals (Wan et al., 2020). Bats and civet cats are recognized as the origin and the intermediate hosts of SARS-CoV (Kakodkar et al., 2020; Wang and Eaton, 2007). Based on the homology between SARS-CoV-2 and SARS-CoV, bats might also play a role as the origin of SARS-CoV-2. Bat-CoV RaTG13 separated from Rhinolophus affinis bat, is the most closely related bat coronavirus to the SARS-CoV-2 with about 96% whole-genome sequencing identity. It reinforces the possibility that bats are the probable reservoir host of the new coronavirus. Despite this similarity, studies have shown that in the Pangolin-CoV, all five key amino acids that belong to RBD part of the S1 subunit of the spike protein which has a role in the RBD/ACE2 interactions are the same as SARS-CoV-2, but in the RaTG13 four of five major residues are different. Pangolin-CoV whole genome is 91.02% similar to SARS-CoV-2 and 90.55% similar to RaTG13. Further research is required to determine the origin and intermediate animals, which would allow us to eliminate virus transmission and prevent further mutations (Andersen et al., 2020; Zhang et al., 2020; Zhou et al., 2020a, Zhou et al., 2020b, Zhou et al., 2020c).
3. SARS-CoV-2 and ACE2 interaction
SARS-CoV-2, like its other cousins SARS-CoV and Middle East Respiratory Syndrome (MERS)-CoV, bind to the ACE2 for entering the cells (Fig. 1 ). In this line, Zhou et al. performed virus infectivity studies. They used two groups of ACE2 expressing and non-expressing HeLa cells from humans, Chinese horseshoe bats, civet, pig, and mouse. As they reported, SARS-CoV-2 used all, but mouse ACE2, as an entry receptor in the ACE2-expressing cells; however, it was unable to enter into the ACE2 non-expressing cells. Interestingly, SARS-CoV-2 did not use aminopeptidase N (APN) and dipeptidyl peptidase 4 (DPPIV), the other coronavirus receptor (Zhou et al., 2020a). Although SARS-CoV-2, SARS-CoV-1, and MERS-CoV have genetic sequence homology, they have some distant sequencing. SARS-CoV-2 S-protein is suggested to have a strong binding affinity to human ACE2. SARS-CoV-2 and SARS-COV-1 share 73.5% identity in the alignment of RBD sequences of spike glycoprotein. Xu et al. assessed the binding free energy of SARS-CoV-2 S-protein in comparison with that of SARS-COV-1 S-protein. They estimated the free energy required for binding of SARS-CoV-2 S-protein to the ACE2 to be about −50.6 kcal/mol, which was significantly lower than that between SARS-CoV S-protein and ACE2 (−78.6 kcal/mol). This relatively higher affinity of SARS-CoV-2 S-protein to the ACE2 can be an ideal target for vaccine design and antiviral drug discovery (Xu et al., 2020b).
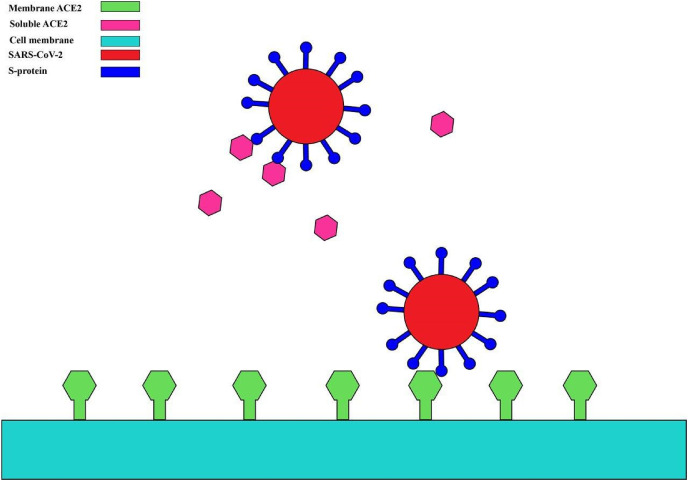
Fig. 1.
The interaction between SARS-CoV-2 S protein and membrane ACE2.
As for other coronaviruses, SARS-CoV-2 possesses a spike (S) glycoprotein, which binds to the cell membrane protein ACE2 to enter human cells. The virus-ACE2 binding results in the release of the viral genome in the host cells. The coronavirus S-protein has two functional units, S1 and S2. During infection, S-protein is a trimeric class I viral fusion protein, which is cleaved into these two subunits (Liu et al., 2020a). SARS-CoV-2 binds to the host receptors by its S1 unit. S1 contains two domains: the N-terminal domain and the C-terminal RBD domain. RBD domain enables coronaviruses to directly bind to the peptidase domain (PD) of the human receptor. S2 subunit is suggested to play a role in membrane fusion (Li, 2012).
Single-cell RNA sequencing (ScRNA) datasets provide evidence that the tissues of the lung, upper respiratory tract, ileum, heart, and kidney express ACE2, and this expression might explain the role of these organs in the pathogenesis of COVID-19 (Zou et al., 2020). Also, the observation of the high expression of ACE2 in the oral cavity, especially on the surface of epithelial cells of the tongue, suggests the oral cavity a favorable site of SARS-CoV-2 transmission (Xu et al., 2020a).
4. Therapeutic potentials
4.1. SARS-CoV2-ACE2 binding-directed approaches
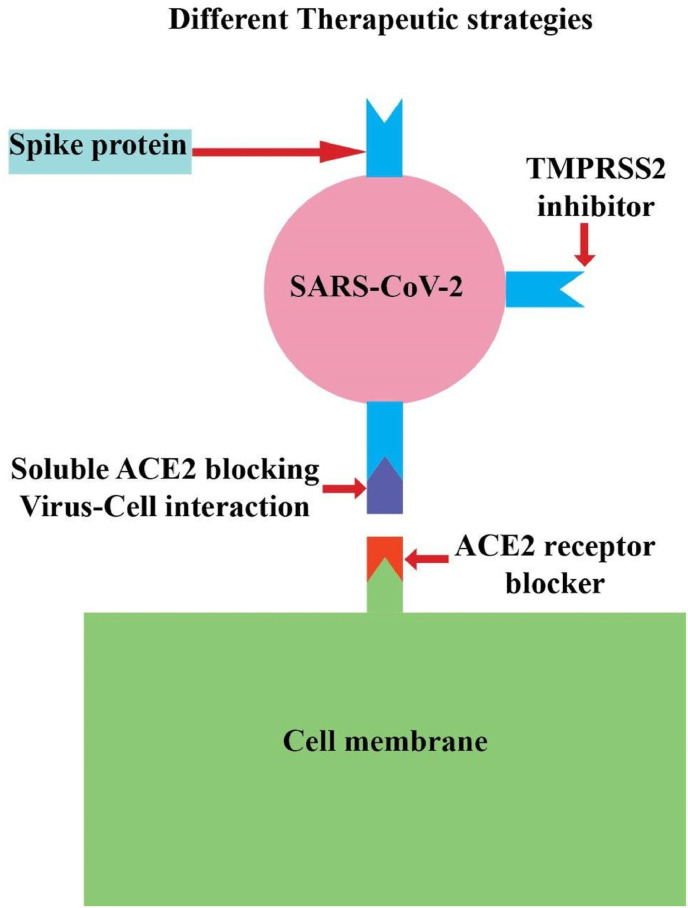
Fig. 2 presents a schematic illustration of different therapeutic strategies directed towards SARS-CoV2-ACE2 binding.
Fig. 2.
Different therapeutic strategies directed towards SARS-CoV-2 binding to membrane ACE2.
4.1.1. Receptor binding domain
The S protein of SARS-CoV-2 serves as an essential component of the virus for cellular attachment, fusion, and viral entry. The RBD fragment of SARS-CoV-2 is located in the middle of the S1 domain. The RBD domain attaches to ACE2 with a high affinity. The spike glycoprotein consists of two S1 and S2 domains. S1 domain contributes to the virus binding to the receptor in target cells (He et al., 2004), and the S2 domain mediates fusion between viral and target cell membranes (Babcock et al., 2004; Wong et al., 2004; Xiao et al., 2003). Before binding of the virus to ACE2, blocking the RBD can prevent virus infection. A possible way to stop the virus infection is the use of antibodies, or molecular inhibitors were tested for SARS-CoV with N-(2-aminoethyl)-1 aziridine-ethanamine as a novel ACE2 inhibitor. Novel ACE inhibitors (ACEI) like captopril, perindopril, ramipril, lisinopril, benazepril, and moexipril are used to treat hypertension and target ACE, a homolog of ACE2 with 42% sequence identity and 61% sequence similarity in the catalytic domain. It will be worthy of testing over ACE inhibitors their ability to block the RBD/ACE2 interaction (Morse et al., 2020). Also, the CR3022 is a SARS-CoV-specific human monoclonal antibody. The epitope of CR3022 does not overlap with the ACE2 binding site within SARS-CoV-2 RBD. However, the CR3022 might have the possibility to be advanced as candidate treatment, alone or in association with other counteracting antibodies, for the prevention and treatment of SARS-CoV-2 infection.
4.1.2. ACE2 blockers (anti-ACE2 monoclonal antibodies)
SARS-CoV-2 attaches to the same host cell receptor using their Spike (S) protein. Hence, there is a structural similarity between the SARS-CoV-2 and SARS-CoV-1, and MERS-CoV antibodies targeting S protein in these two viruses or antibodies blocking ACE2 could be potentially useful on blocking virus entry to cells (Shanmugaraj et al., 2020). In a study by Han et al. (Han and Král, 2020), they have designed a peptide inhibiting SARS-CoV-2 binding to ACE2, which could theoretically block SARS-CoV-2, and this agent can easily be used by inhalation. Currently, few studies are addressing this therapeutic method, so further studies, including clinical trials, are necessary.
4.1.3. Soluble ACE2
It offers another ACE2-directed approach in the context of COVID-19. The SARS-CoV-2 attaches to the extracellular part of ACE2 in the cell membrane. So, it can be hypothesized that using a soluble form of ACE2, which lacks membrane part, can inhibit viral attachment, entry, and replication in the cells by competing with full-length ACE2 (Batlle et al., 2020). Using human recombinant soluble ACE2 showed benefits in inhibiting SARS-CoV-2 infection in vitro (Monteil et al., 2020). This therapeutic method can be improved by attaching the Fc region of the human antibody attaching to the ACE2 extracellular domain (Lei et al., 2020). Another proposed method for improving soluble ACE2 is using cyclodextrin. As Sun et al. (Sun et al.) hypothesized, using cyclodextrin compounded with soluble ACE2 can increase its water solubility, and designing an inhalable drug might block SARS-CoV-2 infectivity. However, these methods need to be studied in further studies, including clinical trials.
4.1.4. Host proteases
Viral entry requires S protein priming by cellular proteases. Transmembrane protease serine 2 (TMPRSS2) mediates S1/S2 cleavage (Hoffmann et al., 2020). Not only SARS-CoV-2, but also other coronaviruses and influenza viruses are dependent on TMPRSS2 for entry. TMPRSS2 is widely expressed in the epithelial respiratory tract, especially type 2 pneumocytes and bronchial epithelial cells (Stopsack et al., 2020). SARS-CoV employs endosomal cysteine proteases cathepsin B and L (CatB/L) in addition to TMRSS2. A recent in vitro study has revealed that SARS-CoV-2 possibly uses both CatB/L as well as TMPRSS2. Thus TMPRSS2 inhibitors could have promising effects in inhibiting viral entry (Hoffmann et al., 2020). Blocking TMPRSS2 and CatB/L inhibited SARS-CoV entry completely, however, could not totally prohibit SARS-CoV-2 entry in an in vitro study (Hoffmann et al., 2020; Kawase et al., 2012). It suggests another cellular protease for SARS-CoV-2 priming. The proprotease convertase furin has been announced to be a candidate to cleavage S glycoprotein mediating viral entry. Furin also serves as a cellular protease for MERS-CoV but not SARS-CoV (Lukassen et al., 2020). Bestle et al. reported that replication of SARS-CoV-2 was prohibited to a great extent with two peptide mimetic inhibitors of TMPRRS2, MI-432, and MI-1900, and aprotinin (a broad range serine protease inhibitor) (Bestle et al., 2020). Moreover, MI-1851, a synthetic furin inhibitor, reduced viral replication significantly. A combination of MI-1851 with other TMPRSS2 protease inhibitors could show much more effective results. However, applying E64d, an inhibitor of endosomal cathepsin, did not affect viral replication (Bestle et al., 2020). Impairing protease activity of TMPRSS2 with drugs including camostat, nafamostat, and aerosolized aprotinin has been shown to attenuate TMPRSS2 protease activity (Stopsack et al., 2020).
TMPRSS2 was first discovered in prostate cancer (Lin et al., 1999). This protease is expressed in the luminal side of epithelial cells. TMPRSS2 expression is increased in prostate cancer tissue when compared with non-cancerous tissue (Lucas et al., 2008), and the levels of TMPRSS2 have been observed to increase in response to androgens notably. A great response in TMPRSS2 expression under androgen signaling could suggest one of the reasons why men are more susceptible to COVID-19. Androgen administration to adenocarcinoma cell line could significantly upregulate TMPRSS2 transcription whereas, androgen antagonists downregulated its expression (Mikkonen et al., 2010; Stopsack et al., 2020). Thus, the theory of targeting TMPRSS2 expression with anti-androgens such as enzalutamide, apalutamide, and darolutamide (used to treat prostate cancer) could have promising effects to reduce COVID-19 severity (Stopsack et al., 2020).
4.2. The renin-angiotensin system
The renin-angiotensin system (RAS) has an essential role in developing hypertension. Angiotensin II (Ang II) AT1 receptor blockers (ARBs) and ACEIs are the most commonly used drugs in this regard. ACE2 hydrolyzes Ang I and Ang II to Ang1-9 and Ang1-7, respectively. Administration of ARBs has been observed to significantly increase the levels of ACE2 substrates, Ang1-7, and Ang1-9. Ang1-9 can be transformed to Ang1-7 by ACE. Ang1-7 plays different protective roles, including anti-inflammatory, anti-hypertrophic, anti-cell proliferative, endothelial protective, and anti-fibrosis effects. Experimental investigations provide evidence that ACEI, as well as ARBs, induce the expression of ACE2 in a variety of tissues, such as the heart, kidney, aorta, and the small intestine (Chappel and Ferrario, 2006; Ferrario and Varagic, 2010; Igase et al., 2005; Vuille-dit-Bille et al., 2015).
Many studies have proclaimed that taking RAS inhibitors upregulate ACE2 expression, thus, is beneficial in controlling blood pressure. It has been evidenced that the downregulation of ACE2 causes an increase in the activation of the RAS. More clearly, following the downregulation of ACE2, the conversion of Ang I and Ang II to Ang 1–7 decreases correspondingly, and therefore, the plasma concentrations of Ang I and Ang II would increase. Increasing concentrations of Ang I and Ang II, which are the effector molecules of the RAS, result in the activation of RAS (Silhol et al., 2020). The increased activation of RAS positively correlates with lung injury in SARS-CoV infection (Li et al., 2020; Mortensen et al., 2008; Rossi et al., 2020). Moreover, when the ACE2 expression is upregulated, the risk for COVID-19 seems to be increased (Fang et al., 2020; Yang et al., 2020). The levels of Ang II increase in patients with COVID-19. Meanwhile, a positive correlation has been found between Ang II levels and lung injury (Liu et al., 2020b). Besides, RAS activation results in multi-organ damage and endothelial dysfunction. RAS inhibition by ACEI and ARBs increases ACE2 expression in heart, kidney, and plasma, whereas its effect on ACE2 expression in airway epithelial cells has yet to be known. It has been announced in a study that using ARBs or ACEI could downregulate ACE2 expression but did not influence its activity. However, whether RAS inhibitors make patients more susceptible to SARS-CoV-2 infection has not been fully understood, and it could lead to a different area for cardiologists regarding treatment whether to keep patients on ACEIs or ARBs (Guo et al., 2020a).
4.3. The kinin–kallikrein system
The role of the kinin–kallikrein system (KKS) in decreasing blood pressure in dogs has been observed (Abelous, 1909). This system consists of high molecular mass kininogen (HMMK). Kallikrein proteolyzes HMMK to bradykinin (BK), an essential pro-inflammatory peptide, and [des-Arg973]-BK (the DABK-the active metabolite of bradykinin). The active bradykinin metabolite DABK binds to the bradykinin B1 receptor (BKB1R). While BK binds to BKB2R (Tolouian et al., 2020), BKB1R is barely expressed on baselines and can be induced by inflammation. It has been shown that BKB1R plays an important role in the pathogenesis of inflammatory diseases (Qadri and Bader, 2018). One of the roles of ACE2 is to hydrolyze and deactivate the bradykinin metabolite DABK (Kaushik et al 2002). BK system activation mediates high levels of inflammatory mediators leading to acute respiratory distress syndrome, increased capillary permeability, and multiple organ failure (Murugesan et al., 2016). A downregulation and reduced activity of ACE2, which may probably be induced by the SARS-CoV-2 results in unopposed activation of DABK. It promotes DABK signaling through the BKB1R resulting in leukocyte infiltration and fluid extravasation to the lungs. Thus, blocking either bradykinin or its receptors may show promising therapeutic effects in the treatment of COVID-19 induced acute respiratory distress syndrome (ARDS) (Tolouian et al., 2020).
5. Conclusion
Studies centered around the ACE2 receptor have provided us with valuable information in the context of understanding the origin and modes of transmission of COVID-19. The novel coronavirus, SARS-CoV-2, is highly homologous with its cousin SARS-CoV-1. They both could be traced to bats. Also, there are studies pointing out that the virus might be originated from pangolins. Because the virus has a high affinity to connect with ACE2 receptors found in other species, animal to human transmission is possible and can be another point of interest for future studies. SARS-CoV-2 is a beta coronavirus that has many of the structural features of its family members, as a spike glycoprotein for interaction with its host cells. However, subtle differences were observed in this structure between SARS-CoV-2 and other coronaviruses. The most important one is that the SARS-CoV-2 S glycoprotein has a higher binding affinity with ACE2 compared to SARS-CoV, and this can explain its higher pathogenicity.
The suggested mode of transmission of SARS-CoV-2 is through the respiratory tract and the fact that ACE2 is expressed in that area only solidifies the latter claim, but as ACE2 is found in other organs like the oral cavity or the gastrointestinal tract, this fact allows us to assume that the virus can use these systems for transmission as well and further studies can help in proving or disproving them.
RAS inhibitors are considered for therapeutic utilization against SARS-CoV-2. Here, we looked upon their mechanism and how they can increase ACE2 expression and therefore exacerbate the severity of the disease. ACEIs and ARBs, which are commonly used drugs in patients suffering from cardiovascular diseases. However, further studies and examination are needed to fully assess the contribution that these drugs can have in treating COVID-19. Other than ACE2, the virus needs proteases to infect host cells, and TMPRSS2 is one discovered protease serving SARS-CoV-2. Drugs exploiting this protease have shown great prospects in dealing with COVID-19 and should be a high priority subject in future studies and trials.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and materials
Not applicable.
Funding
There is no funding for the present study.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Not applicable.
References
- Abelous J. Les substances hypotensives de l'urine humaine normale. CR Soc Biol. 1909;66:511–512. [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock G.J., Esshaki D.J., Thomas W.D., Ambrosino D.M. Amino acids 270 to 510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor. J. Virol. 2004;78:4552–4560. doi: 10.1128/JVI.78.9.4552-4560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrami A., Vafapour M., Moazzami B., Rezaei N. Hyperinflammatory shock related to COVID-19 in a patient presenting with multisystem inflammatory syndrome in children: first case from Iran. J. Paediatr. Child Health. 2020 doi: 10.1111/jpc.15048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basiri A., Pazhouhnia Z., Beheshtizadeh N., Hoseinpour M., Saghazadeh A., Rezaei N. Regenerative medicine in COVID-19 treatment: real opportunities and range of promises. Stem cell reviews and reports. 2020:1–13. doi: 10.1007/s12015-020-09994-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batlle D., Wysocki J., Satchell K. Soluble angiotensin-converting enzyme 2: a potential approach for coronavirus infection therapy? Clin. Sci. (Lond.) 2020;134:543–545. doi: 10.1042/CS20200163. [DOI] [PubMed] [Google Scholar]
- Bestle D., Heindl M.R., Limburg H., Pilgram O., Moulton H., Stein D.A., Hardes K., Eickmann M., Dolnik O., Rohde C. bioRxiv; 2020. TMPRSS2 and Furin Are Both Essential for Proteolytic Activation and Spread of SARS-CoV-2 in Human Airway Epithelial Cells and Provide Promising Drug Targets. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappel M.C., Ferrario C.M. ACE and ACE2: their role to balance the expression of angiotensin II and angiotensin-(1–7) Kidney Int. 2006;70:8–10. doi: 10.1038/sj.ki.5000321. [DOI] [PubMed] [Google Scholar]
- Darbeheshti F., Rezaei N. Genetic predisposition models to COVID-19 infection. Med. Hypotheses. 2020;142:109818. doi: 10.1016/j.mehy.2020.109818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L., Karakiulakis G., Roth M. Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? The Lancet. Respir. Med. 2020 doi: 10.1016/S2213-2600(20)30116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fathi N., Rezaei N. Lymphopenia in COVID-19: therapeutic opportunities. Cell Biol. Int. 2020 doi: 10.1002/cbin.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrario C.M., Varagic J. The ANG-(1–7)/ACE2/mas axis in the regulation of nephron function. Am. J. Physiol. Ren. Physiol. 2010;298:F1297–F1305. doi: 10.1152/ajprenal.00110.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Huang Z., Lin L., Lv J. Coronavirus disease 2019 (COVID‐19) and cardiovascular disease: a viewpoint on the potential influence of angiotensin‐converting enzyme inhibitors/angiotensin receptor blockers on onset and severity of severe acute respiratory syndrome coronavirus 2 infection. J.Am. Heart Assoc. 2020;9 doi: 10.1161/JAHA.120.016219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.-R., Cao Q.-D., Hong Z.-S., Tan Y.-Y., Chen S.-D., Jin H.-J., Tan K.-S., Wang D.-Y., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak–an update on the status. Military Medical Research. 2020;7:1–10. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Král P. Computational design of ACE2-based peptide inhibitors of SARS-CoV-2. ACS Nano. 2020 doi: 10.1021/acsnano.0c02857. acsnano. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaei S., Rezaei N. COVID-19: developing from an outbreak to a pandemic. Arch. Med. Res. 2020 doi: 10.1016/j.arcmed.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Zhou Y., Liu S., Kou Z., Li W., Farzan M., Jiang S. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine. Biochem. Biophys. Res. Commun. 2004;324:773–781. doi: 10.1016/j.bbrc.2004.09.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igase M., Strawn W.B., Gallagher P.E., Geary R.L., Ferrario C.M. Angiotensin II AT1 receptors regulate ACE2 and angiotensin-(1–7) expression in the aorta of spontaneously hypertensive rats. Am. J. Physiol. Heart Circ. Physiol. 2005;289:H1013–H1019. doi: 10.1152/ajpheart.00068.2005. [DOI] [PubMed] [Google Scholar]
- Jabbari P., Jabbari F., Ebrahimi S., Rezaei N. 2020. COVID-19: A Chimera of Two Pandemics. Disaster Medicine and Public Health Preparedness; pp. 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahanshahlu L., Rezaei N. Archives of medical research; 2020. Central Nervous System Involvement in COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahanshahlu L., Rezaei N. Monoclonal antibody as a potential anti-COVID-19. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2020;129:110337. doi: 10.1016/j.biopha.2020.110337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafieh R., Arian R., Saeedizadeh N., Minaee S., Yadav S.K., Vaezi A., Rezaei N., Javanmard S.H. medRxiv; 2020. COVID-19 in Iran: A Deeper Look into the Future. [Google Scholar]
- Kakodkar P., Kaka N., Baig M. A comprehensive literature review on the clinical presentation, and management of the pandemic coronavirus disease 2019 (COVID-19) Cureus. 2020;12 doi: 10.7759/cureus.7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik V.C.H.P., Tang V.D.L.G.J., Godbout K., Parsons T., Baronas E., Hsieh F. Hydrolysis of biological peptides by human angiotensin-converting enzyme-related carboxypeptidase. J. Biol. Chem. 2002;277:14838–14843. doi: 10.1074/jbc.M200581200. J. [DOI] [PubMed] [Google Scholar]
- Kawase M., Shirato K., van der Hoek L., Taguchi F., Matsuyama S. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J. Virol. 2012;86:6537–6545. doi: 10.1128/JVI.00094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei C., Fu W., Qian K., Li T., Zhang S., Ding M., Hu S. bioRxiv; 2020. Potent Neutralization of 2019 Novel Coronavirus by Recombinant ACE2-Ig. 2020.2002.2001.929976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Evidence for a common evolutionary origin of coronavirus spike protein receptor-binding subunits. J. Virol. 2012;86:2856–2858. doi: 10.1128/JVI.06882-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Hu R., Zhang X. Antihypertensive treatment with ACEI/ARB of patients with COVID-19 complicated by hypertension. Hypertens. Res. 2020:1–3. doi: 10.1038/s41440-020-0433-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B., Ferguson C., White J.T., Wang S., Vessella R., True L.D., Hood L., Nelson P.S. Prostate-localized and androgen-regulated expression of the membrane-bound serine protease TMPRSS2. Canc. Res. 1999;59:4180–4184. [PubMed] [Google Scholar]
- Liu W., Morse J.S., Lalonde T., Xu S. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019‐nCoV. Chembiochem. 2020 doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Yang Y., Zhang C., Huang F., Wang F., Yuan J., Wang Z., Li J., Li J., Feng C., Zhang Z., Wang L., Peng L., Chen L., Qin Y., Zhao D., Tan S., Yin L., Xu J., Zhou C., Jiang C., Liu L. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci. China Life Sci. 2020;63:364–374. doi: 10.1007/s11427-020-1643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotfi M., Hamblin M.R., Rezaei N. COVID-19: transmission, prevention, and potential therapeutic opportunities. Clin. Chim. Acta. Int. J. Clin. Chem. 2020;508:254–266. doi: 10.1016/j.cca.2020.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas J.M., True L., Hawley S., Matsumura M., Morrissey C., Vessella R., Nelson P.S. The androgen‐regulated type II serine protease TMPRSS2 is differentially expressed and mislocalized in prostate adenocarcinoma. J. Pathol.: J. Pathol.Soc. G. B. Ireland. 2008;215:118–125. doi: 10.1002/path.2330. [DOI] [PubMed] [Google Scholar]
- Lukassen S., Chua R.L., Trefzer T., Kahn N.C., Schneider M.A., Muley T., Winter H., Meister M., Veith C., Boots A.W. SARS‐CoV‐2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020 doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkonen L., Pihlajamaa P., Sahu B., Zhang F.-P., Jnne O.A. Androgen receptor and androgen-dependent gene expression in lung. Mol. Cell. Endocrinol. 2010;317:14–24. doi: 10.1016/j.mce.2009.12.022. [DOI] [PubMed] [Google Scholar]
- Moazzami B., Razavi-Khorasani N., Dooghaie Moghadam A., Farokhi E., Rezaei N. COVID-19 and telemedicine: immediate action required for maintaining healthcare providers well-being. J. Clin. Virol.: Off. Publ.Pan.Am. Soc.Clin. Virol. 2020;126:104345. doi: 10.1016/j.jcv.2020.104345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed K., Rodríguez-Román E., Rahmani F., Zhang H., Ivanovska M., Makka S.A., Joya M., Makuku R., Islam M.S., Radwan N., Rahmah L., Goda R., Abarikwu S.O., Shaw M., Zoghi S., Irtsyan S., Ling I., Cseprekal O., Faten A.B., Hazar Sayar E., Soloukey C., Grancini G., Rezaei N. Borderless collaboration is needed for COVID-19-A disease that knows no borders. Infect. Control Hosp. Epidemiol. 2020:1–2. doi: 10.1017/ice.2020.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momtazmanesh S., Ochs H.D., Uddin L.Q., Perc M., Routes J.M., Vieira D.N., Al-Herz W., Baris S., Prando C., Rosivall L., Abdul Latiff A.H., Ulrichs T., Roudenok V., Aldave Becerra J.C., Salunke D.B., Goudouris E., Condino-Neto A., Stashchak A., Kryvenko O., Stashchak M., Bondarenko A., Rezaei N. All together to fight COVID-19. Am. J. Trop. Med. Hyg. 2020;102:1181–1183. doi: 10.4269/ajtmh.20-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteil V., Kwon H., Prado P., Hagelkrüys A., Wimmer R.A., Stahl M., Leopoldi A., Garreta E., del Pozo C.H., Prosper F. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020 doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradian N., Ochs H.D., Sedikies C., Hamblin M.R., Camargo C.A., Jr., Martinez J.A., Biamonte J.D., Abdollahi M., Torres P.J., Nieto J.J., Ogino S., Seymour J.F., Abraham A., Cauda V., Gupta S., Ramakrishna S., Sellke F.W., Sorooshian A., Wallace Hayes A., Martinez-Urbistondo M., Gupta M., Azadbakht L., Esmaillzadeh A., Kelishadi R., Esteghamati A., Emam-Djomeh Z., Majdzadeh R., Palit P., Badali H., Rao I., Saboury A.A., Jagan Mohan Rao L., Ahmadieh H., Montazeri A., Fadini G.P., Pauly D., Thomas S., Moosavi-Movahed A.A., Aghamohammadi A., Behmanesh M., Rahimi-Movaghar V., Ghavami S., Mehran R., Uddin L.Q., Von Herrath M., Mobasher B., Rezaei N. The urgent need for integrated science to fight COVID-19 pandemic and beyond. J. Transl. Med. 2020;18:205. doi: 10.1186/s12967-020-02364-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse J.S., Lalonde T., Xu S., Liu W.R. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. Chembiochem. 2020;21:730–738. doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen E.M., Restrepo M.I., Copeland L.A., Pugh J.A., Anzueto A. Association of hydrophilic versus lipophilic angiotensin-converting enzyme inhibitor use on pneumonia-related mortality. Am. J. Med. Sci. 2008;336:462–466. doi: 10.1097/MAJ.0b013e31817149ed. [DOI] [PubMed] [Google Scholar]
- Murugesan P., Jung B., Lee D., Khang G., Doods H., Wu D. Kinin B1 receptor inhibition with BI113823 reduces inflammatory response, mitigates organ injury, and improves survival among rats with severe sepsis. J. Infect. Dis. 2016;213:532–540. doi: 10.1093/infdis/jiv426. [DOI] [PubMed] [Google Scholar]
- Nasab M.G., Saghazadeh A., Rezaei N. Archives of medical research; 2020. SARS-CoV-2-A Tough Opponent for the Immune System. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procko E. bioRxiv; 2020. The Sequence of Human ACE2 Is Suboptimal for Binding the S Spike Protein of SARS Coronavirus 2. [Google Scholar]
- Qadri F., Bader M. Kinin B1 receptors as a therapeutic target for inflammation. Expert Opin. Ther. Targets. 2018;22:31–44. doi: 10.1080/14728222.2018.1409724. [DOI] [PubMed] [Google Scholar]
- Rabiee N., Rabiee M., Bagherzadeh M., Rezaei N. COVID-19 and picotechnology: potential opportunities. Med. Hypotheses. 2020;144:109917. doi: 10.1016/j.mehy.2020.109917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi G.P., Sanga V., Barton M. Potential harmful effects of discontinuing ACE-inhibitors and ARBs in COVID-19 patients. Elife. 2020;9 doi: 10.7554/eLife.57278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghazadeh A., Rezaei N. Immune-epidemiological parameters of the novel coronavirus - a perspective. Expet Rev. Clin. Immunol. 2020;16:465–470. doi: 10.1080/1744666X.2020.1750954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghazadeh A., Rezaei N. Towards treatment planning of COVID-19: rationale and hypothesis for the use of multiple immunosuppressive agents: anti-antibodies, immunoglobulins, and corticosteroids. Int. Immunopharm. 2020;84:106560. doi: 10.1016/j.intimp.2020.106560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu K.K., Siddiqui A.D., Rezaei N., Cerny J. Challenges for management of immune thrombocytopenia during COVID-19 pandemic. J. Med. Virol. 2020 doi: 10.1002/jmv.26251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleki K., Banazadeh M., Saghazadeh A., Rezaei N. The involvement of the central nervous system in patients with COVID-19. Rev. Neurosci. 2020;1 doi: 10.1515/revneuro-2020-0026. [DOI] [PubMed] [Google Scholar]
- Shanmugaraj B., Siriwattananon K., Wangkanont K., Phoolcharoen W. Perspectives on monoclonal antibody therapy as potential therapeutic intervention for Coronavirus disease-19 (COVID-19) Asian Pac. J. Allergy Immunol. 2020;38:10–18. doi: 10.12932/AP-200220-0773. [DOI] [PubMed] [Google Scholar]
- Silhol F., Sarlon G., Deharo J.-C., Vaïsse B. Downregulation of ACE2 induces overstimulation of the renin–angiotensin system in COVID-19: should we block the renin–angiotensin system? Hypertens. Res. 2020;43:854–856. doi: 10.1038/s41440-020-0476-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopsack K.H., Mucci L.A., Antonarakis E.S., Nelson P.S., Kantoff P.W. 2020. TMPRSS2 and COVID-19: Serendipity or Opportunity for Intervention? Cancer Discovery. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, P., Lu, X., Xu, C., Wang, Y., Sun, W., Xi, J., CD-sACE2 inclusion compounds: an effective treatment for coronavirus disease 2019 (COVID-19). Journal of Medical Virology n/a. [DOI] [PubMed]
- Tolouian R., Vahed S.Z., Ghiyasvand S., Tolouian A., Ardalan M. COVID-19 interactions with angiotensin-converting enzyme 2 (ACE2) and the kinin system; looking at a potential treatment. J. Ren. Inj. Prev. 2020;9 [Google Scholar]
- Vuille-dit-Bille R.N., Camargo S.M., Emmenegger L., Sasse T., Kummer E., Jando J., Hamie Q.M., Meier C.F., Hunziker S., Forras-Kaufmann Z. Human intestine luminal ACE2 and amino acid transporter expression increased by ACE-inhibitors. Amino Acids. 2015;47:693–705. doi: 10.1007/s00726-014-1889-6. [DOI] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94 doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.-F., Eaton B.T. Springer; 2007. Bats, Civets and the Emergence of SARS, Wildlife and Emerging Zoonotic Diseases: the Biology, Circumstances and Consequences of Cross-Species Transmission; pp. 325–344. [Google Scholar]
- Wong S.K., Li W., Moore M.J., Choe H., Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004;279:3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Chakraborti S., Dimitrov A.S., Gramatikoff K., Dimitrov D.S. The SARS-CoV S glycoprotein: expression and functional characterization. Biochem. Biophys. Res. Commun. 2003;312:1159–1164. doi: 10.1016/j.bbrc.2003.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Zhong L., Deng J., Peng J., Dan H., Zeng X., Li T., Chen Q. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral Sci. 2020;12 doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., Zhong W., Hao P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Yu Y., Xu J., Shu H., Liu H., Wu Y., Zhang L., Yu Z., Fang M., Yu T. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. The Lancet Respiratory Medicine. 2020 doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdanpanah F., Hamblin M.R., Rezaei N. Life Sciences; 2020. The Immune System and COVID-19: Friend or Foe? 117900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefzadegan S., Rezaei N. Case report: death due to COVID-19 in three brothers. Am. J. Trop. Med. Hyg. 2020;102:1203–1204. doi: 10.4269/ajtmh.20-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Wu Q., Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr. Biol. 2020 doi: 10.1016/j.cub.2020.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. BioRxiv. 2020 [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020:1–4. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X., Chen K., Zou J., Han P., Hao J., Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020 doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.