Abstract
Novel coronavirus (SARS–CoV–2), turned out to be a global pandemic with unstoppable morbidity and mortality rate. However, till date there is no effective treatment found against SARS–CoV–2. We report on the major in–depth molecular and docking analysis by using antiretroviral (Lopinavir and ritonavir), antimalarial (Hydroxychloroquine), antibiotics (Azithromycin), and dietary supplements (Vitamin C and E) to provide new insight into drug repurposing molecular events involved in SARS–CoV–2. We constructed three drug–target–pathways–disease networks to predict the targets and drugs interactions as well as important pathways involved in SARS–CoV–2. The results suggested that by using the combination of Lopinavir, Ritonavir along with Hydroxychloroquine and Vitamin C may turned out to be the effective line of treatment for SARS–CoV–2 as it shows the involvement of PARP–1, MAPK–8, EGFR, PRKCB, PTGS–2, and BCL–2. Gene ontology biological process analysis further confirmed multiple viral infection–related processes (P < 0.001), including viral life cycle, modulation by virus, C–C chemokine receptor activity, and platelet activation. KEGG pathway analysis involves multiple pathways (P < 0.05), including FoxO, GnRH, ErbB, Neurotrophin, Toll–like receptor, IL–17, TNF, Insulin, HIF–1, JAK–STAT, Estrogen, NF–kappa, Chemokine, VEGF, and Thyroid hormone signaling pathway in SARS–CoV–2. Docking study was carried out to predict the molecular mechanism Thus, the potential drug combinations could reduce viral infectivity, viral replication, and abnormal host inflammatory responses and may be useful for multi–target drugs against SARS–CoV–2.
Keywords: Drug repurposing, SARS–CoV–2, Molecular docking, Molecular mechanism
List of abbreviations
- ADR
Adverse drug reaction
- Bcl-2
B-cell leukemia/lymphoma-2
- COVID
coronavirus disease
- CCR-3
C-C Motif Chemokine Receptor 3
- CXCR3
C-X-C chemokine receptor type 3
- EC
endothelial cell
- EGFR
Epidermal growth factor receptor
- EIF4E
Eukaryotic Translation Initiation Factor 4E
- ErbB
Erb-B2 Receptor Tyrosine Kinase-2
- FoXo
Forkhead box O
- GnRH
Gonadotropin-releasing hormone
- GO
Gene ontology
- HIF-1
Hypoxia-inducible factor 1
- HIV
Human immunodeficiency virus
- IL:
Interleukin
- JAK
Janus Kinase
- KEGG
Kyoto Encyclopaedia of Genes and Genomes
- MAPK
Mitogen-activated protein kinase
- PDB
Protein Data bank
- PPI
Protein-protein interaction
- PRKQ
(A, C, E)
- PTGS2
Prostaglandin-endoperoxide synthase 2
- RNA
Ribonucleic Acid
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- STAT
signal transducer and activator of transcription proteins
- TNF
Tumor necrosis factor
- VEGF
Vascular endothelial growth factor
- VEGFR
Vascular endothelial growth factor receptor
1. Introduction
The current pandemic of SARS–CoV–2 (also known as COVID–19), a respiratory disease that has led to continues and over 6140934 confirmed cases and 373548 confirmed death fatalities in over 216 countries, areas or territories since its emergence in late 2019 (https://www.who.int/emerg, 2019). The SARS–CoV–2 belongs to the β genus which contain at least four structural proteins: spike (S) protein, envelope (E) protein, membrane (M) protein, and nucleocapsid (N) protein. Spike promotes host attachment and virus–cell membrane fusion during virus infection. Therefore, Spike protein is considered as the host range (Li, 2016). Potential anti–coronavirus therapies can be divided into two categories depending on the target, one is acting on the human immune system or human cells, and the other is on coronavirus itself (Wu et al., 2020). In terms of the human immune system, the innate immune system response plays an important role in controlling the replication and infection of coronavirus, and interferon is expected to enhance the immune response (Nikolich-Zugich et al., 2020). In order to show anti–retroviral effects, it is important to block the signaling pathways of human cell required for virus replication (Arts and Hazuda, 2012). The main aspect behind designing a drug for coronavirus includes inhibiting the synthesis of viral RNA through acting on the genetic material of the virus ultimately inhibiting virus replication through acting on critical enzymes of virus. This strategy can block the virus binding to human cell receptors thus, inhibiting the virus's self–assembly process through acting on some structural proteins (Prajapat et al., 2020). There is a strong interest in the use of combinations of drugs in order to increase the therapeutic efficacy caused by individual drugs due to high dose (BayatMokhtari et al., 2017). The important aspect behind combining the drugs is that the drug may target nodes of the pathways in order to elucidate polypharmacological behaviour of the drugs (Schenone et al., 2013).
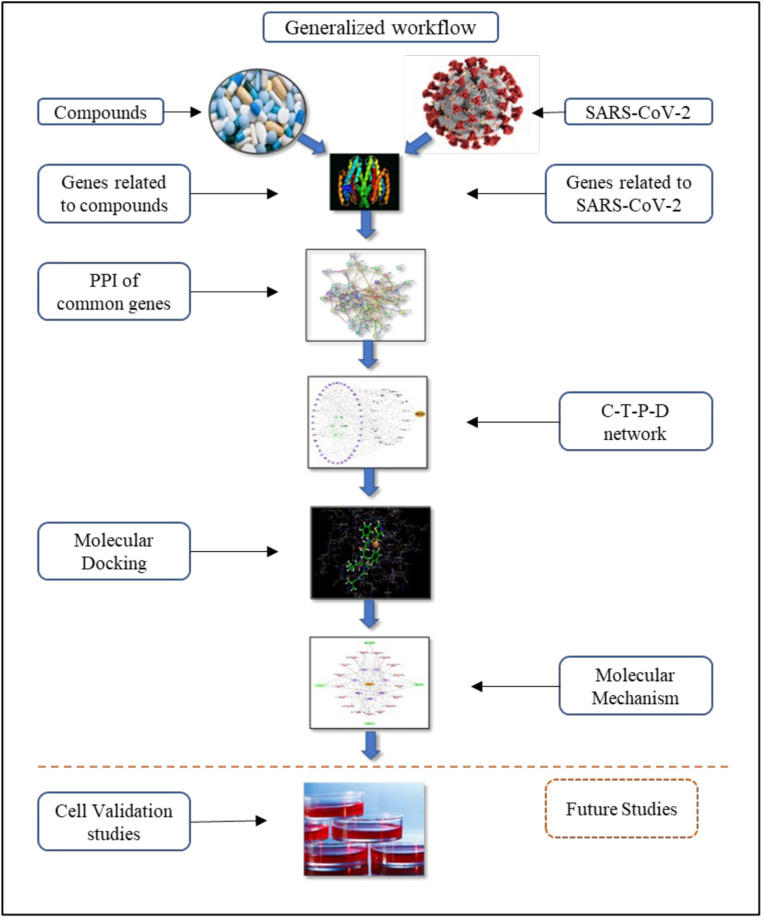
In the fight against coronavirus, we have come up with three strategies for repurposing of the clinically available new drugs. The first strategy is to evaluate existing HIV protease inhibitors i.e., Lopinavir and Ritonavir. The advantages of these therapies are that their metabolic characteristics, dosages used, potential efficacy and side effects which are approved for treating HIV/AIDS infections. Retrospective literature was carried with the patients who has undergone the two treatment as Lopinavir and Ritonavir and Hydroxychloroquine in combination with azithromycin. The study include a total of 199 patients with laboratory confirmed SARS–CoV–2 infection underwent randomization; 99 were assigned to the Lopinavir–ritonavir group, and 100 to the standard–care group. In a modified intention to treat analysis, Lopinavir–ritonavir led to a median time to clinical improvement that was shorter by 1 day than that observed with standard care. Gastrointestinal adverse events were more common in the Lopinavir–ritonavir group, but serious adverse events were more common in the standard care group. Lopinavir–ritonavir treatment was stopped early in 13 patients because of adverse events (ClinicalReferenceGroupSBAR). Second study showed significantly more efficient for virus elimination in case of Azithromycin with Hydroxychloroquine. As the treatment found significantly associated with viral load reduction/disappearance in COVID–19 patients and its effect was secure by azithromycin (Gautret et al., 2020). It is generally recommended to use with other antimalarial drugs and essential dietary supplements. Thus, Hydroxychloroquine was added with Vitamin C and Vitamin E. The second strategy is to use existing molecular databases to screen for molecules that may have therapeutic effect on coronavirus. Computational screening makes this strategy possible, and new functions of many drug molecules can be found through this strategy, for example, the discovery of macrolide anti–biotic drug Azithromycin with the first strategy (Hydroxychloroquine, Vitamin C, and Vitamin E). The third strategy is directly based on combining both the strategies. Theoretically, the drugs found through these therapies would exhibit better anti–coronavirus effects, but the research procedure of new drug might cost several years, or even more. We follow this analysis with bioinformatics validation of drug–induced gene signatures and by constructing the compound– target– pathway– disease network to inspect the postulated mechanism–of–action in a specific SARS–CoV–2 for which we propose repurposing (Fig. 1 ).
Fig. 1.
The integrated strategy and work flow of drug repurposing for SARS-CoV-2.
2. Materials and methods
2.1. Data preparation
SARS–CoV–2–human genes were obtained from Gene card, human gene database (https://www.genecards.org/) which provides information related to all annotated and predicted human genes (https://www.genecards.org). Among the GeneCards categories only protein–coding class was selected for the interactions. Information about the drug i.e., Lopinavir, ritonavir, Hydroxychloroquine, Azithromycin, Vitamin C, and Vitamin E were obtained through PubChem (https://pubchem.ncbi.nlm.nih.gov) (https://pubchem.ncbi.nlm.) and Swiss target prediction (http://new.swisstargetpre). Protein–protein interaction (PPI) is an important aspect to study the involvement of proteins in various biochemical processes as well as to understand the cellular organization, bioprocess, and functions.
2.2. Construction of PPI of selected genes
The genes of the selected components were uploaded to STRING (https://string-db.org/cgi/network.pl) to get the information about PPIs (https://stringdb.org/cgi/). The setting for generating the PPI network was in accordance with ‘Homo Sapiens’ and the confidence in the interaction between the target protein was set to the highest confidence data >0.9. The network nodes represent proteins whereas the edge represents associated protein–protein interaction (Brinda and Vishveshwara, 2005).
2.3. Construction of compound–target–pathway–disease network and enrichment analysis of SARS–CoV–2 targets and gene ontology
The compound–target network helps to understand and analyse the mechanism of the components with target as well as the pathways involved. Basically, three networks i.e., class I (Lopinavir, Ritonavir, Hydroxychloroquine, Vitamin C, and Vitamin E), class II (Azithromycin, Hydroxychloroquine, Vitamin E, and Vitamin C) and class III (Lopinavir, Ritonavir, Hydroxychloroquine, Azithromycin Vitamin C, and Vitamin E) were constructed to understand the in–depth of the genes involved in treatment regime which helps us to find the correlation of the drug and disease.
In order to find the ontology terms associated with molecular, cellular, biological, and KEGG pathway enrichment analysis was carried out. The analysis was performed by Cytoscape plug–in called ClueGO and Cluepedia. Pathways and networks were ranked according to the amounts of the molecules participating in pathways and networks, respectively. Pathways and networks shared by targets related to SARS–CoV–2–human and the potential Lopinavir, ritonavir, Hydroxychloroquine, Azithromycin, Vitamin C, and Vitamin E targets were identified.
2.4. Molecular docking of selected genes
Docking was carried out to find the binding affinity as well as orientation of the selected active components by docking the compounds of the selected class. Docking was performed on using Glide v_7.6 program interfaced with Maestro v_11.3 of Schrödinger (2017) (Schrodinger, LLC, New York, NY, USA) of compounds with their respective targets which are screened down relating to SARS–CoV–2–human disease. The structures of compounds were built using Maestro build panel and optimized to lower energy conformers using Ligprep v_3.3. Molecular docking helps us to narrow down the search of target proteins to which our compound has more binding affinity which can prove the potential targets for the disease. The screened data can be used for molecular mechanism deduction.
2.5. Deduction of molecular mechanism
After finalising the important compounds relating to vital genes that may be crucial in curing the SARS–CoV–2 in order to deduce the molecular mechanism of compounds and disease correlating via genes and pathways. Molecular mechanism can show us the action of a compound on a protein relating to disease through the pathway. This can be beneficial in finding the vital pathways to determine the up/down regulation in the diseased state.
3. Results
3.1. Data mining
A total of 332 human genes associated with SARS–CoV–2 were identified in the Gene Bank database. Genes obtained from Swiss target prediction was found to be 100 for Azithromycin, Lopinavir, Ritonavir, Hydroxychloroquine, and Vitamin E; whereas for Vitamin C was 47. After filtering out the common genes between the compounds and disease, 47 genes were used for constructing the network of Azithromycin, Hydroxychloroquine, Vitamin C, and Vitamin E with respect to SARS–CoV–2 and for second network 35 genes of Lopinavir, Ritonavir, Hydroxychloroquine, Vitamin C, and Vitamin E whereas for the third network, the combination of both the treatment regime about 43 genes were filtered out against SARS–CoV–2 (Table 1 ).
Table 1.
Combined common gene list of compounds and disease.
| Sr. No. | Genes | Gene description | UniProt ID |
|---|---|---|---|
| 1 | ACE | Angiotensin I Converting Enzyme | P12821 |
| 2 | ANPEP | Alanyl Aminopeptidase, Membrane | P15144 |
| 3 | ATP1A1 | ATPase Na+/K + Transporting Subunit Alpha 1 | P05023 |
| 4 | BAD | BCL2 Associated Agonist Of Cell Death | Q92934 |
| 5 | BCL2 | BCL2 Apoptosis Regulator | P10415 |
| 6 | BCL2L1 | BCL2 Like 1 | Q07817 |
| 7 | CASP3 | Caspase 3 | P42574 |
| 8 | CASP6 | Caspase 6 | P55212 |
| 9 | CASP8 | Caspase 8 | Q14790 |
| 10 | CCR1 | C-C Motif Chemokine Receptor 1 | P32246 |
| 11 | CCR3 | C-C Motif Chemokine Receptor 3 | P51677 |
| 12 | CDK4 | Cyclin Dependent Kinase 4 | P11802 |
| 13 | CSNK2A2 | Casein Kinase 2 Alpha 2 | P19784 |
| 14 | CTSB | Cathepsin B | P07858 |
| 15 | CTSL | Cathepsin L | P07711 |
| 16 | CXCR3 | C-X-C Motif Chemokine Receptor 3 | P49682 |
| 17 | DPP4 | Dipeptidyl Peptidase 4 | P27487 |
| 18 | EGFR | Epidermal Growth Factor Receptor | P00533 |
| 19 | EIF4E | Eukaryotic Translation Initiation Factor 4E | P06730 |
| 20 | F10 | Coagulation Factor X | P00742 |
| 21 | GAPDH | Glyceraldehyde-3-Phosphate Dehydrogenase | P04406 |
| 22 | HPGDS | Hematopoietic Prostaglandin D Synthase | O60760 |
| 23 | HSP90B1 | Heat Shock Protein 90 Beta Family Member 1 | P14625 |
| 24 | ITGA5 | Integrin Subunit Alpha 5 | P08648 |
| 25 | ITGB1 | Integrin Subunit Beta 1 | P05556 |
| 26 | JAK1 | Janus Kinase 1 | P23458 |
| 27 | LCK | LCK Proto-Oncogene, Src Family Tyrosine Kinase | P06239 |
| 28 | MAPK14 | Mitogen-Activated Protein Kinase 14 | Q16539 |
| 29 | MAPK8 | Mitogen-Activated Protein Kinase 8 | P45983 |
| 30 | NOS2 | Nitric Oxide Synthase 2 | P35228 |
| 31 | NOS3 | Nitric Oxide Synthase 3 | P29474 |
| 32 | PARP1 | Poly(ADP-Ribose) Polymerase 1 | P09874 |
| 33 | PIK3CA | Phosphatidylinositol-4,5-Bisphosphate 3-Kinase Catalytic Subunit Alpha | P42336 |
| 34 | PIK3CB | Phosphatidylinositol-4,5-Bisphosphate 3-Kinase Catalytic Subunit Beta | P42338 |
| 35 | PIK3CD | Phosphatidylinositol-4,5-Bisphosphate 3-Kinase Catalytic Subunit Delta | O00329 |
| 36 | PIK3CG | Phosphatidylinositol-4,5-Bisphosphate 3-Kinase Catalytic Subunit Gamma | P48736 |
| 37 | PPARG | Peroxisome Proliferator Activated Receptor Gamma | P37231 |
| 38 | PRKCA | Protein Kinase C Alpha | P17252 |
| 39 | PRKCB | Protein Kinase C Beta | P05771 |
| 40 | PRKCE | Protein Kinase C Epsilon | Q02156 |
| 41 | PRKCQ | Protein Kinase C Theta | Q04759 |
| 42 | PTGS1 | Prostaglandin-Endoperoxide Synthase 1 | P23219 |
| 43 | PTGS2 | Prostaglandin-Endoperoxide Synthase 2 | P35354 |
3.2. Protein–protein interaction
PPI network was conducted in order to analyse and understand the mechanisms of SARS–CoV–2 based on the study of protein–protein interactions by using STRING software. A total of 43 interrelations, as well as 31 related targets, are obtained in PPI network after setting the confidence level greater than 0.9 and rejecting the target protein independent of the network. The importance prioritization of key proteins is analysed according to the degree of the node exported from STRING database. Among them, the EGFR value (degree = 26) and MAPK–8 value (degree = 24) is much higher than that of other protein nodes, which indicates that this protein might play a role of bridge to connect other nodes in PPI network (Supplementary Figure S1). The PPI network combined scores were showed in supplementary information file (Supplementary Table S1). So, PPI network analysis and pathway analysis of novel genes were carried out for the recognition of critical genes related to the SARS–CoV–2.
3.3. Compound–target network and network based drug re–purposing for HCoVs and gene ontology (GO) enrichment and network analysis
3.3.1. Construction of compound–target network
The compound and disease interaction network was constructed which elucidate the mechanisms of action of drugs in the treatment of SARS–CoV–2. The connectivity of a drug in the networks represents the drug's effects, either therapeutic, in the context of the pathway networks, or toxic, in the case of the ADR network. As far as drug–target network is considered the important genes which correlates with SARS–CoV–2 are CXCR3, CCR–3, NOS, MAPK–8, EGFR, PRKQ (A, C, E), BCL–2, EIF4E, and PTGS2. Inhibition of these targets may turn out to be an important aspect of network regulation of SAR–CoV-2 as it involves regulation, inhibition and modulation of the proteins which are important to overcome SARS–CoV. The rationale behind using the class III network is that it contains the genes which plays an important role in SARS–CoV–2. Also, from the introduction we can say that the treatment needs to have one of all categories, like anti–retroviral, anti–malarial and dietary supplements.
3.3.2. Gene ontology and KEGG pathway enrichment analysis
KEGG pathway enrichment analysis revealed multiple significant biological pathways (adjusted P value < 0.05), including measles, RNA transport, HIF signaling pathway, NF–kappa B signaling, Epstein–Barr virus infection, and influenza. Gene ontology (GO) biological process enrichment analysis further confirmed multiple viral infection–related processes (adjusted P value < 0.001), including viral life cycle, modulation by virus of host morphology or physiology, viral process, positive regulation of viral life cycle, transport of virus, and virion attachment to host cell.
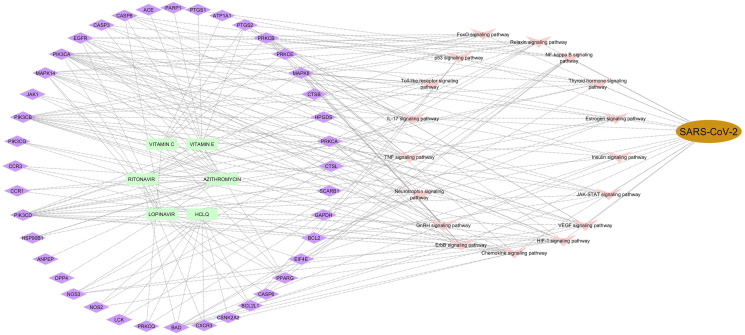
3.3.3. Construction of compound–Target–Pathway–Disease network
Among 43 genes only 19 genes were pooled out for construction of the Compound–Target–Pathway–Disease network (Fig. 2 ). Pathways were selected from the results of gene enrichment analysis and KEGG analysis. The pathways mainly responsible for SARS–CoV–2 are NF–kappa B, FoxO signaling pathway, p5, Toll–like receptor, IL–17, TNF, Neurotrophin, GnRH, ErbB, Chemokine, HIF–1, VEGF, JAK–STAT, Insulin, Estrogen, Thyroid hormone, and Relaxin signaling pathways (Table 2 ).
Fig. 2.
Compound-target-pathway-disease network.
Table 2.
Enriched KEGG Pathways related to genes.
| Sr. No. | KEGG Pathway ID | KEGG Pathway Description | Associated genes |
|---|---|---|---|
| 1 | hsa04064 | NF-kappa B signaling pathway | BCL2, BCL2L1, CSNK2A2, LCK, PARP1, PRKCB, PRKCQ, PTGS2 |
| 2 | hsa04068 | FoxO signaling pathway | EGFR, MAPK14, MAPK8, PIK3CA, PIK3CB, PIK3CD |
| 3 | hsa04115 | p53 signaling pathway | BCL2, BCL2L1, CASP3, CASP8 |
| 4 | hsa04620 | Toll-like receptor signaling pathway | CASP8, MAPK14, MAPK8, PIK3CA, PIK3CB, PIK3CD |
| 5 | hsa04657 | IL-17 signaling pathway | CASP3, CASP8, HSP90B1, MAPK14, MAPK8, PTGS2 |
| 6 | hsa04668 | TNF signaling pathway | CASP3, CASP8, MAPK14, MAPK8, PIK3CA, PIK3CB, PIK3CD, PTGS2 |
| 7 | hsa04722 | Neurotrophin signaling pathway | BAD, BCL2, MAPK14, MAPK8, PIK3CA, PIK3CB, PIK3CD |
| 8 | hsa04912 | GnRH signaling pathway | EGFR, MAPK14, MAPK8, PRKCA, PRKCB |
| 9 | hsa04012 | ErbB signaling pathway | BAD, EGFR, MAPK8, PIK3CA, PIK3CB, PIK3CD, PRKCA, PRKCB |
| 10 | hsa04062 | Chemokine signaling pathway | BAD, CCR1, CCR3, CXCR3, PIK3CA, PIK3CB, PIK3CD, PIK3CG, PRKCB |
| 11 | hsa04066 | HIF-1 signaling pathway | BCL2, EGFR, EIF4E, GAPDH, NOS2, NOS3, PIK3CA, PIK3CB, PIK3CD, PRKCA, PRKCB |
| 12 | hsa04370 | VEGF signaling pathway | BAD, MAPK14, NOS3, PIK3CA, PIK3CB, PIK3CD, PRKCA, PRKCB, PTGS2 |
| 13 | hsa04630 | JAK-STAT signaling pathway | BCL2, BCL2L1, EGFR, JAK1, PIK3CA, PIK3CB, PIK3CD |
| 14 | hsa04910 | Insulin signaling pathway | BAD, EIF4E, MAPK8, PIK3CA, PIK3CB, PIK3CD |
| 15 | hsa04915 | Estrogen signaling pathway | BCL2, EGFR, HSP90B1, NOS3, PIK3CA, PIK3CB, PIK3CD |
| 16 | hsa04919 | Thyroid hormone signaling pathway | ATP1A1, BAD, PIK3CA, PIK3CB, PIK3CD, PRKCA, PRKCB |
| 17 | hsa04926 | Relaxin signaling pathway | EGFR, MAPK14, MAPK8, NOS2, NOS3, PIK3CA, PIK3CB, PIK3CD, PRKCA |
3.4. Molecular docking analysis
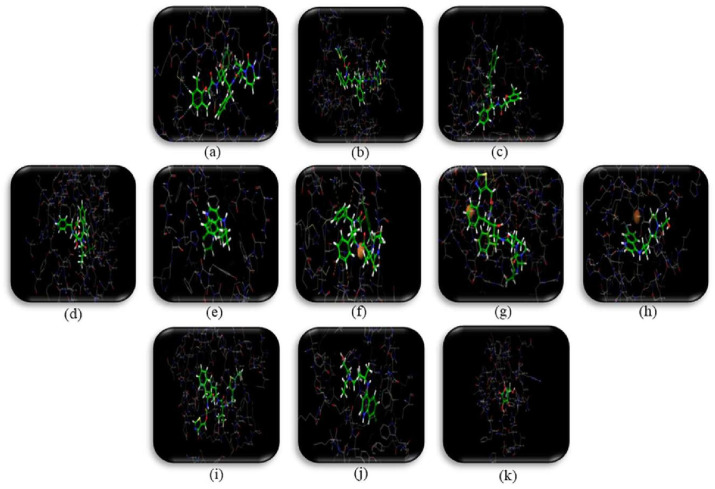
The mechanism of the selected drugs was reflected by interaction of compound and target through molecular docking. The crystal structures for CASP-8 (PDB ID:4PRZ), BCL2L1 (PDB ID:6RNU), BCL2 (PDB ID:5M54), PARP1 (PDB ID:1UK1), EGFR(PDB ID:5FEQ), MAPK14 (PDB ID:4TYH), PRKCA (PDB ID:4RA4), MAPK8 (PDB ID:2XS0), LCK (PDB ID:4REW), EIF4E (PDB ID: 5EKV), PRKCB (PDB ID:2IOE), PTGS2 (PDB ID:5KIR), NOS2 (PDB ID:4CX7), GAPDH (PDB ID:6ADE), and NOS3 (PDB ID:1M9R) were taken from RCSB Protein Data Bank and prepared for docking using ‘protein preparation wizard’ (Table 3 ). Water molecules were removed then the receptor was prepared for docking. In order to enhance the docking score, the protein constraints were defined using protein preparation wizard. We found hydrogen bonding and π–π stacking the main forms of interaction. Molecular docking scores were obtained and used for further screening of compounds for determining the molecular mechanism. A cut–off value less than –7.49 was used to screen down the compound–target interactions. (Table 4 and Fig. 3 ).
Table 3.
Molecular Docking Score of compounds against screened targets.
|
Active constituents |
Docking Score with PDB ID |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAPK-8 (PDB ID: 2XS0) |
CASP-8 (PDB ID: 4PRZ) |
EGFR (PDB ID: 5FEQ) |
PARP1 (PDB ID: 1UK1) |
MAPK-14 (PDB ID: 4TYH) |
BCL-2 (PDB ID: 5M54) | BCL2L1 (PDB< ID: 6RNU) |
PRKCA (PDB ID: 4RA4) | LCK (PDB ID: 4REW) | EIF4E (PDB ID: 5EKV) | PRKCB (PDB ID: 2I0E) | PTGS2 (PDB ID: 5KIR) | NOS2 (PDB ID: 4CX7) | GAPDH (PDB ID: 6ADE) |
NOS3 (PDB ID: 1M9R) | |
| Lopinavir (PubChem CID: 92727) | -7.692 | -4.557 | -7.126 | -9.455 | -6.750 | -4.727 | -5.664 | -6.404 | -6.701 | -4.776 | -6.780 | 4.867 | -4.425 | -6.431 | -5.805 |
| Ritonavir (PubChem CID: 392622) | -8.512 | -6.402 | -8.109 | -8.298 | -6.616 | -5.879 | -6.298 | -6.748 | -5.106 | -4.712 | -7.153 | -4.853 | -5.925 | -6.254 | -6.803 |
| HCQL (PubChem CID: 3652) | -6.887 | -5.103 | -7.035 | -7.243 | -6.370 | – | -5.044 | -5.112 | -3.664 | -3.848 | -5.803 | -7.557 | -5.569 | -6.388 | -6.927 |
| Vitamin E (PubChem CID: 14985) | -3.617 | -4.097 | -5.571 | -6.491 | -5.887 | -3.635 | -2.563 | -4.111 | -3.463 | -2.881 | -4.518 | – | -3.713 | -5.622 | -4.033 |
| Vitamin C (PubChem CID:54670067) | -4.286 | -4.249 | -4.659 | 3.843 | -5.736 | -8.157 | -4.389 | -4.213 | -5.097 | -4.055 | -5.270 | -3.243 | -3.467 | -4.887 | -4.583 |
| Azithromycin (PubChem CID: 447043) | – | – | – | – | – | -2.655 | – | – | -2.27 | – | – | – | – | -4.115 | – |
Table 4.
Molecular Docking Score with interacting amino acid residues.
| Sr.No | Tyrosine Kinase | Active constituent | Docking Score | Amino acid residues involved in H-bond formation |
|---|---|---|---|---|
| 1 | MAPK-8 | Lopinavir | -7.692 | ASN114, SER155, LYS55, LYS153 |
| Ritonavir | -8.512 | ASN114, SER34, LYS55, SER155, LYS153 | ||
| 2 | PARP-1 | Lopinavir | -8.298 | TYR907, ARG878, HIE909 |
| Ritonavir | -9.455 | GLU763, MET890, TYR907 | ||
| Hydroxychloroquine | -7.243 | TYR907, GLY863, ASP770 | ||
| 3 | EGFR | Lopinavir | -7.126 | ARG841, CYS797, LYS745 |
| Ritonavir | -8.109 | ASN842, ASP837, ASP855 | ||
| Hydroxychloroquine | -7.035 | GLN 791, ASP855, PHE723 | ||
| 4 | PRKCB | Ritonavir | -7.153 | ASP470, ASP427, LYS486, ASN471 |
| 5 | PTGS-2 | Hydroxychloroquine | -7.557 | TRP387, ARG120 |
| 6 | BCL-2 | Vitamin C | -8.157 | ASP98, GLU71, GLU11 |
Fig. 3.
Molecular docking visualization of (a) MAPK8-Lopinavir (b) MAPK8-Ritonavir (c) PARP1-Lopinavir (d) PARP1-Ritonavir (e) PARP1-Hydroxychloroquine (f)EGFR-Lopinavir (g) EGFR-Ritonavir (h) EGFR-Hydroxychloroquine (i) PKRKCB-Ritonavir (j) PTGS2 Hydroxychloroquine (k) BCL2-Vitamin C.
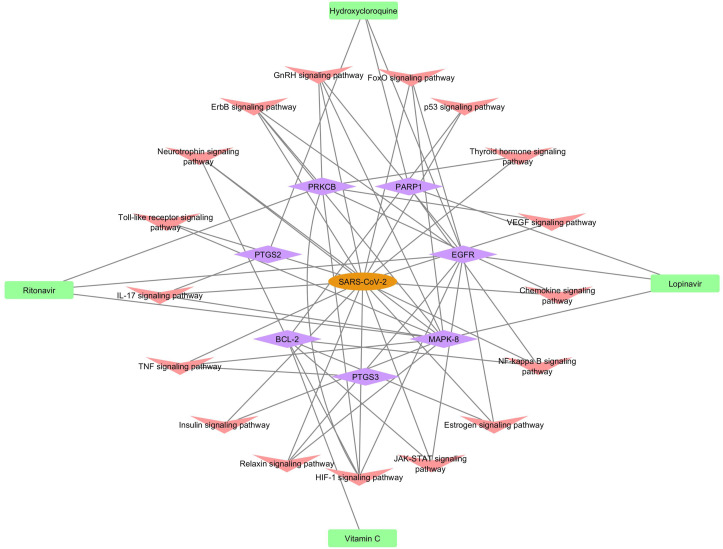
3.5. Molecular mechanism
We received the final screened compounds and targets after molecular docking. These compounds act on the specified targets which can play a major role in regulation of coronavirus disease through the specified pathway. This molecular mechanism can help us to further investigate the pathways which are involved in the up/down regulation of SARS–CoV–2 (Fig. 4 ).
Fig. 4.
Molecular mechanism.
4. Discussion
The origin for the proposed network–based drug repurposing methodologies is based on the concepts that the proteins associated functionally govern the viral infection which are localized in the within the comprehensive protein–protein interaction network in human cells (Zhou et al., 2020). For a drug with multiple targets to be effective against SARS–CoV–2, its target proteins should be within or in the corresponding sub–network in the human protein–protein interactome, as we demonstrated using multiple drugs through network–based strategy.
To improve the quality and completeness of the human protein interactome network, we integrated PPIs and further constructed out the drug– target– pathway– disease network with three types of experimental data:
-
(1)
Lopinavir, ritonavir, Hydroxychloroquine, Vitamin C, and Vitamin E (Class I);
-
(2)
Azithromycin, Hydroxycloroquine, Vitamin C, and Vitamin E, (Class II);
-
(3)
Lopinavir, ritonavir, Azithromycin, Hydroxycloroquine, Vitamin C, and Vitamin E; (Class III).
In this study, we presented a network–based methodology for systematic identification of repurposable drugs and drug combinations in order to predict the molecular mechanism for potential treatment of SARS–CoV–2. Integration of compound–target–pathway–disease networks, and human protein–protein network are essential for such identification. Based on comprehensive evaluation, we prioritized 6 candidate repurposable drugs and 4 potential drug combinations for targeting SARS–CoV–2. According to the stated output, PARP-1, MAPK-8, EGFR, PRKCB, PTGS-2 and BCL-2 are found to be essentially related and involvement in SARS–CoV–2. PARPs have both antiviral and immunomodulatory roles. PARPs have evolved rapidly, which may reflect their involvement in virus infections (Grunewald et al., 2019). Consistent with this, several PARPs are known ISGs (interferon stimulated genes), and many PARPs have been shown to be antiviral. PARP1 inhibitors restrict the replication of several viruses such as herpes viruses, adenoviruses, and HIV (Fehr et al., 2020). Mitogen–activated protein kinases (MAPKs) are signal transducers that respond to extracellular stimulation by cytokines, growth factors, viral infection, and stress, and in turn regulate cell differentiation, proliferation, survival, and apoptosis (Hommes et al., 2003). In particular, p38 MAPK is strongly activated by stress and inflammatory cytokines (Cargnello and Roux, 2011). EGFR signaling is a key regulator of SARS-CoV-induced lung damage and that targeting this response could protect against the development of pulmonary fibrosis caused by respiratory viruses such as SARS-CoV (Bachstetter and Van Eldik, 2010). The transcription factor nuclear factor kappa–light–chain–enhancer of activated B cells (NF–κB) is a master regulator of pro–inflammatory response and innate immunity (Venkataraman et al., 2017; Liu et al., 2017). NF–κB is required for the induction of pro–inflammatory cytokines during RNA virus infection. Interestingly, induction of TNF–α, IL–6 and IL–8 has been detected in cells over–expressing the spike protein of SARS–CoV via the NF–κB pathway (Wang et al., 2007). Thus, it is interesting to consider the involvement of NF–κB pathway during coronavirus infection. BCL–2 protein inhibits the caspase–dependent apoptosis induced by SARS–CoV infection without affecting viral replication kinetics BCL–2 protein inhibits the caspase–dependent apoptosis induced by SARS–CoV infection without affecting viral replication kinetics (Bordi et al., 2006). As the selected drugs regime is mainly followed in India but the results are not been in favour of the line of treatment thus the present study highlights the use of drug combinations which includes major genes for targeting SARS–CoV–2. Our methodology remain to be a useful network–based tool for prediction of combining multiple drugs toward exploring the pathways as well as the molecular mechanism of multiple drugs and targets with the SARS-CoV-2. Thus, combination of the existing anti–retroviral drugs with the network–predicted repurposable drugs or drug combinations may improve coverage of current network–based methodologies by utilizing multi–layer network framework (Sanders et al., 2020; Singh et al., 2020; Lima et al., 2020).
5. Limitations and future work
In this study, we presented a computational approach for identification of the potential compounds for treatment against SARS–CoV–2, along with predictive mechanism. We have acknowledged several limitations in the current study. Although sizeable efforts were made for assembling large scale, experimentally reported drug–target networks from publicly available databases, the network data may be incomplete and some drug–target interactions may be biologically associated. Comprehensive identification of the virus–host interactome for SARS–CoV–2, with specific biological effects using functional genomics assays will significantly improve the accuracy of the proposed network–based methodologies further. Preclinical studies are warranted to evaluate in vivo efficiency and side effects by cell validation studies, before clinical trials. Therefore, further experimental validation on the prediction of network pharmacology is needed to support the presented hypothesis in the future work.
6. Conclusion
In conclusion, this study offers a powerful, integrative network–based systems pharmacology methodology for rapid identification of repurposable drugs and drug combinations for the potential treatment of SARS–CoV–2. Our approach can minimize the translational gap between preclinical testing results and clinical outcomes, which is a significant problem in the rapid development of efficient treatment strategies for the emerging SARS–CoV–2 outbreak. From the current outlook, if broadly applied, the network tools developed here could help develop effective treatment strategies for other emerging viral infections and other human complex diseases and disorders as well.
Declarations
Ethics approval and consent to participate
This research did not require an ethical approval as it does not involve any human or animal experiment.
Human and animal rights
No Animals/Humans were used for studies that are base of this research.
Consent for publication
Not applicable.
Availability of data and materials
Not applicable.
Funding
This research received no grant from any funding agency.
Conflict of interest
The authors declare no conflict of interest, financial or otherwise.
CRediT authorship contribution statement
Shweta A. More: Methodology, Software, Resources, Data curation, Writing - review & editing, Writing - original draft. Akshay S. Patil: Software, Data curation, Writing - review & editing. Nikhil S. Sakle: Conceptualization, Data curation, Writing - review & editing. Santosh N. Mokale: Validation, Investigation, Data curation, Writing - review & editing, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors thankful to the Mrs. Fatima Rafiq Zakaria, Chairman Maulana Azad Educational Trust, Dr. Rafiq Zakaria Campus, Aurangabad 431001(MS), India for providing the laboratory facility. This work was supported by the Department of Science and Technology (DST), New Delhi, India [Project File No. EEQ/2016/00055].
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virol.2020.12.006.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Arts E.J., Hazuda D.J. HIV-1 antiretroviral drug therapy. Cold Spring Harb. Perspect. Med. 2012;2(4) doi: 10.1101/cshperspect.a007161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachstetter A.D., Van Eldik L.J. The p38 MAP kinase family as regulators of proinflammatory cytokine production in degenerative diseases of the CNS. Aging Dis. 2010;1(3):199–211. [PMC free article] [PubMed] [Google Scholar]
- Bayat Mokhtari R., Homayouni T.S., Baluch N., et al. Combination therapy in combating cancer. Oncotarget. 2017;8(23) doi: 10.18632/oncotarget.16723. 38022‐38043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordi L., Castilletti C., Falasca L., et al. Bcl-2 inhibits the caspase-dependent apoptosis induced by SARS-CoV without affecting virus replication kinetics. Arch. Virol. 2006;151(2):369–377. doi: 10.1007/s00705-005-0632-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinda K.V., Vishveshwara S. A network representation of protein structures: implications for protein stability. Biophys. J. 2005;89(6):4159–4170. doi: 10.1529/biophysj.105.064485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargnello M., Roux P.P. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases [published correction appears in Microbiol Mol Biol Rev. Microbiol. Mol. Biol. Rev. 2011;75(1):50–83. doi: 10.1128/MMBR.00031-10. 2012 Jun;76(2):496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ClinicalReferenceGroupSBAR TherapiesforCOVID19. http://www.bccdc.ca/HealthProfessionalsSite/Documents/Guidelines_Unproven_Therapies_COVID-19.pdf
- Fehr A.R., Singh S.A., Kerr C.M., Mukai S., Higashi H., Aikawa M. The impact of PARPs and ADP-ribosylation on inflammation and host-pathogen interactions. Genes Dev. 2020;34(5–6):341–359. doi: 10.1101/gad.334425.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautret P., Lagier J.C., Parola P., et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial [published online ahead of print, 2020 Mar 20] Int. J. Antimicrob. Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald M.E., Chen Y., Kuny C., et al. The coronavirus macrodomain is required to prevent PARP-mediated inhibition of virus replication and enhancement of IFN expression. PLoS Pathog. 2019;15(5) doi: 10.1371/journal.ppat.1007756. Published 2019 May 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hommes D.W., Peppelenbosch M.P., van Deventer S.J. Mitogen activated protein (MAP) kinase signal transduction pathways and novel anti-inflammatory targets. Gut. 2003;52(1):144–151. doi: 10.1136/gut.52.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- http://new.swisstargetprediction.ch/
- https://pubchem.ncbi.nlm.nih.gov/
- https://stringdb.org/cgi/input.pl?sessionId=7dz41kE6UWdR&input_page_active_form=multiple_identifiers
- https://www.genecards.org/
- https://www.who.int/emergencies/diseases/novel-coronavirus-2019 visited on 02-06-2020; 15:08.
- Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3(1):237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima W.G., Brito J.C.M., Overhage J., Nizer W.S.D.C. The potential of drug repositioning as a short-term strategy for the control and treatment of COVID-19 (SARS-CoV-2): a systematic review. Arch. Virol. 2020;165(8):1729–1737. doi: 10.1007/s00705-020-04693-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Zhang L., Joo D., Sun S.C. NF-κB signaling in inflammation. Signal Transduct. Target Ther. 2017;2 doi: 10.1038/sigtrans.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolich-Zugich J., Knox K.S., Rios C.T., Natt B., Bhattacharya D., Fain M.J. SARS-CoV-2 and COVID-19 in older adults: what we may expect regarding pathogenesis, immune responses, and outcomes. GeroScience. 2020:1–10. doi: 10.1007/s11357-020-00186-0. [published online ahead of print, 2020 Apr 10] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajapat M., Sarma P., Shekhar N., et al. Drug targets for corona virus: a systematic review. Indian J. Pharmacol. 2020;52(1):56–65. doi: 10.4103/ijp.IJP_115_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders J.M., Monogue M.L., Jodlowski T.Z., Cutrell J.B. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a review. J. Am. Med. Assoc. 2020;323(18):1824–1836. doi: 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- Schenone M., Dančík V., Wagner B.K., Clemons P.A. Target identification and mechanism of action in chemical biology and drug discovery. Nat. Chem. Biol. 2013;9(4):232–240. doi: 10.1038/nchembio.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh T.U., Parida S., Lingaraju M.C., Kesavan M., Kumar D., Singh R.K. Drug repurposing approach to fight COVID-19 [published online ahead of print, 2020 Sep 5] Pharmacol. Rep. 2020:1–30. doi: 10.1007/s43440-020-00155-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman T., Coleman C.M., Frieman M.B. Overactive epidermal growth factor receptor signaling leads to increased fibrosis after severe acute respiratory syndrome coronavirus infection. J. Virol. 2017;91(12) doi: 10.1128/JVI.00182-17. 17. Published 2017 May 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Ye L., Ye L., et al. Up-regulation of IL-6 and TNF-alpha induced by SARS-coronavirus spike protein in murine macrophages via NF-kappaB pathway. Virus Res. 2007;128(1–2):1–8. doi: 10.1016/j.virusres.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Liu Y., Yang Y., et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods [published online ahead of print, 2020 Feb 27] Acta Pharm. Sin. B. 2020 doi: 10.1016/j.apsb.2020.02.008. 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14. doi: 10.1038/s41421-020-0153-3. Published 2020 Mar 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.