Graphical abstract
Keywords: SARS-CoV-2, Molecular dynamics, Umbrella sampling, Amprenavir, Enalaprilat, Plerixafor
Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that leads to coronavirus disease (COVID-19) has put public health at risk in 2020. The spike protein (SP) in SARS-CoV-2 is primarily responsible for the attachment and entry of the virus into the cell, which binds to the angiotensin-converting enzyme 2 (ACE2). Owing to the lack of an effective therapy, drug repositioning is an opportunity to search for molecules with pharmacological potential for the treatment of COVID-19. In this study, three candidates with the potential to destabilize the SP-ACE2 complex are reported. Through molecular docking, 147 drugs were evaluated and their possible binding sites in the interface region of the SP-ACE2 complex and the SP of SARS-CoV-2 were identified. The five best candidate molecules were selected for molecular dynamics studies to observe changes in interactions between SP-ACE2 and ligands with the SP-ACE2 complex. Using umbrella sampling molecular dynamics simulations, the binding energy of SP with ACE2 (−29.58 kcal/mol) without ligands, and in complex with amprenavir (−20.13 kcal/mol), enalaprilat (–23.84 kcal/mol), and plerixafor (−19.72 kcal/mol) were calculated. These drugs are potential candidates for the treatment of COVID-19 as they destabilize the SP-ACE2 complex; the binding energy of SP is decreased in the presence of these drugs and may prevent the virus from entering the cell. Plerixafor is the drug with the greatest potential to destabilize the SP-ACE2 complex, followed by amprenavir and enalaprilat; thus, these three drugs are proposed for future in vitro and in vivo evaluations.
1. Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus that leads to the coronavirus disease (COVID-19) has put public health at risk worldwide in 2020. The first reports of patients with COVID-19 were from Wuhan, China [1]; by April 2020, there were more than 137,000 deaths globally [2] and on March 3, in China, 80,270 confirmed cases of SARS-CoV-2 infection were reported [3]. In the Americas region, 24,035,766 cumulative cases and 690,023 cumulative deaths were reported in November 2020 [4]. Currently, the total number of SARS-CoV-2 infections is underestimated, as there are asymptomatic patients or those with mild symptoms (generally children and young adults), who are not considered in the statistical data [2], [5].
The SARS-CoV-2 infection and the associated destruction of lung cells promote a local immune response, recruiting macrophages and monocytes that respond to infection, releasing cytokines, and preparing the T and B lymphocyte response. CD8+ T cells are important in destroying virus-infected cells, whereas CD4+ T cells regulate the activity of CD8+ and B lymphocytes [6]. In patients with COVID-19, the response of B cells generally begins with the production of antibodies against the nucleocapsid (N) protein, and 4 to 8 days after the onset of symptoms, the production of antibodies against spike protein (SP) begins [7].
SARS-CoV-2 virus belongs to the β-coronaviruses (CoVs) [3]. This virus has a 96% sequence identity with that of the coronavirus identified in bats, which are the biggest coronavirus reservoirs in the world [2], [8]. This virus maintains its RNA sequence covered by a phosphorylated nucleocapsid protein inside the lipid membrane. The membrane is covered by two types of spike proteins; the glycoprotein S or SP, which is a trimer that exists in all CoVs, and the hemagglutinin esterase (HE) protein, the latter is only present in some CoVs. Additionally, other proteins such as the membrane (M) protein and the envelope (E) protein that are found between the SP exist in the lipid membrane (Fig. 1 ) [9].
Fig. 1.
Typical structure of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). a: Viral RNA, b: Nucleocapsid (N)-protein, c: Membrane, d: Membrane (M)-Protein, e: Envelope (E)-protein, and f: Spike (S)-protein.
SP is primarily responsible for the attachment and entry of the virus into the cell, which binds to its molecular target in the host, the angiotensin-converting enzyme 2 (ACE2) [10]. SP is a homotrimeric glycoprotein of 180–200 kDa that belongs to class I fusion proteins [11]. This protein consists of an extracellular N-terminus, a transmembrane domain attached in viral membrane, and a short intracellular C-terminal segment and exists in a metastable perfusion conformation. Once the virus interacts with the host cell, a rearrangement of SP occurs [12]. During the infection process, SP is cleaved into two subunits (S1 and S2); the S1 subunit is released in the transition to the post-fusion conformation between the virus membrane and the membrane of the infected cell [13].
The S1 subunit contains a receptor-binding domain (RBD), which binds to the peptidase (PD) domain of ACE2. These RBDs are more exposed on the viral surface in the S1 subunit than in the S2 subunit. The structural features of the RBD binding to ACE2 has been determined by X-ray crystallography, showing that the RBD of SARS-CoV-2 contains a twisted five-stranded antiparallel β sheet with connected helices and loops to build the core of the RBD [11].
The S2 subunit is responsible for the fusion of membranes, this subunit contains a fusion peptide, heptapeptide repeat sequence 1 and 2 (HR1, 2), a transmembrane domain, and a cytoplasmic domain. The fusion peptide domain is fundamental to the membrane by disrupting and connecting lipid bilayers of the host cell membrane. HR1 and HR2 are essential to viral fusion and the entry function of the S2 subunit [12]. The SARS-CoV-2 SP binds to ACE2 with a 10 to 20 times greater affinity than that of the SP of the SARS-CoV virus [11], [13].
In contrast, ACE2 is a single-pass transmembrane protein, which is expressed in the pulmonary alveolar epithelial cells, heart, kidney, and enterocytes of the small intestine [13]. ACE2 consists of a PD at the N-terminal, and a collectrin-like domain at the C-terminal [14]. The SARS-CoV-2 recognizes the PD of ACE2 through an interaction between residue K31 and the RBD region in SP [10]. The crystallography of the SP-ACE2 complex has been reported, with the following interactions identified between these proteins: K417-D30, Y453-H34, F486-M82, Q498-Y41, T500-Q42, and N501-K353 and R357, mainly through hydrogen bonds and hydrophobic interactions [15].
Current treatments for COVID-19 focus on symptomatic and respiratory support [13]. Corticosteroids are only used in conjunction with life support to prevent acute respiratory syndrome in patients with severe COVID-19 [3]. Ribavirin, lopinavir/ritonavir, and nelfinavir have been reported to inhibit the replication of SARS-CoV in vitro and they are suggested as a possible treatment for SARS-CoV-2 infection [16], [17], [18]. In addition to the report of in vitro antiviral activity of remdesivir against SARS-COV-2 [19], the use of recombinant ACE2 (rhACE2), mesenchymal stem cells, PD-1 blocking antibodies, bevacizumab injection, and immunoglobulins from cured patients has also been reported for the treatment of COVID-19 [3]. Among others, anti-human immunodeficiency virus drugs, anti-hepatitis C virus drugs, nonsteroidal anti-inflammatory drugs (NSAIDs), antimalarials, and corticosteroids have been evaluated through the repositioning of drugs using computational techniques such as molecular docking and molecular dynamics to find effective therapies against COVID-19 [20], [21], [22], [23], [24], [25], [26].
Owing to the lack of an effective therapy, drug repositioning presents an opportunity to rapidly search for molecules with pharmacological potential for the treatment of COVID-19. Compared to the discovery and development of new molecules, this strategy reduces development time and costs, as the former requires 12 to 16 years and an investment of 1,000 to 2,000 million USD to achieve regulatory approval; in contrast, repositioning a drug takes 6.5 years on average to obtain approval and an investment of $300 million [27]. Because of the urgency in finding therapeutical options to this pandemic with a focus on the inhibition of the first step of SARS-CoV-2 infection to prevent its dissemination, the aim of this study is to report potential candidates for the treatment of COVID-19 that inhibit and destabilize the SP-ACE2 complex analyzed using molecular docking, molecular dynamics simulations, and further refinement with umbrella sampling molecular dynamics simulations.
2. Methods
2.1. Ligands selection
The DrugBank database [28], [29], [30] was searched for ligands according to two criteria; the candidates should be a small molecule (low molecular weight <900 Daltons) and should be previously approved for commercialization. Ligands were separated into four groups: antiviral drugs, ACE2 inhibitors, drugs for inhalation, and oral administration. Considering safety measures for future application, drugs with action on the central nervous system, antineoplastic agents, and peptides were discarded from the selection. In total, a library was built of 147 candidate ligands with the previously mentioned characteristics (in the case of pharmaceutical salts, the drug structure was used and the ion was discarded), and virtual screening by molecular docking was performed for the selection of five potential drugs to destabilize the SP-ACE2 complex using the best cluster energy score.
2.2. Molecular docking
2.2.1. First screening using molecular docking
The structure of the SP-ACE2 complex was obtained from the public database Protein Data Bank (PDB code: 6M17) [15] using the chain B (ACE2) and E (SP) present in the PDB. Molecular docking was performed with AutoDock 4.2.6 optimized for GPU [31], a grid of 47.25 × 47.25 × 47.25 grid points with a spacing of 0.375 Å, centered on the residues identified as relevant: Q498, T500, N500, K417, Y453, Q474, and F486 in SP, and Y41, Q42, K353, R357, D30, H34, Q24, and M82 in ACE2 [15], was calculated with AutoGrid 4.2.6 [32]. A total of 20 runs and 25,000,000 evaluations were used in a Lamarckian genetic algorithm and Solis-Wets local search [31]. A ligand was selected if it interacted with relevant SP and ACE2 residues and interacted at the interface between SP-ACE2.
2.2.2. Second screening using molecular docking
The second molecular docking was performed using the parameter previously mentioned with a grid of 37.67 × 37.67 × 29.9 grid points with a spacing of 0.299 Å, centered on the residues identified as relevant. A total of 25 runs and 25,000,000 evaluations were used in a Lamarckian genetic algorithm and Solis-Wets local search. Ligands that formed clusters in the region of SP or the SP-ACE2 interface were selected.
2.2.3. Third screening using molecular docking
The third molecular docking was performed using the parameter previously mentioned with a grid of 17.4 × 17.4 × 17.4 grid points with a spacing of 0.290 Å, centered on the ligand with best score in second molecular docking (Table 1 ). A total of 25 runs and 25,000,000 evaluations in a Lamarckian genetic algorithm and Solis-Wets local search were used.
Table 1.
Second grid hits used as the reference for grid calculation in the third screening.
| Grid | Reference ligand | Grid | Reference ligand |
|---|---|---|---|
| 1_1 | Procaterol | 2_1 | Enalaprilat |
| 1_2 | Plerixafor | 2_2 | Cholic acid |
| 1_3 | Tedizolid phosphate | 2_3 | Cerivastatin |
2.3. All atom molecular dynamics simulations (AA-MD)
The protein–ligand complex of the five selected ligands (amprenavir (Amp), brivudine (Bri), cilazapril (Cil), enalaprilat (Ena), and plerixafor (Ple)) from the molecular docking analysis were used for molecular dynamic studies; glycosylations, water, and metal atoms in PDB were retired, and AA-MD simulations were performed in Desmond, using Schrödinger-Maestro software [33], [34] as the graphical interface. The AA-MD systems were built using the System Setup module with an OPLS force-field, adding a POPC lipid membrane, ions, and SPC-water in an NPT assemble at 310.15 K and 1.01325 bar. Once the system was built, the standard relaxation protocol for system relaxation with increasing temperatures and decreasing restrains was used (Berendsen barostat and Langevin thermostat). Then, an MD production simulation of 10 ns was performed in the Molecular Dynamics module (using the Nose-Hoover thermostat and Martyna-Tobias-Klein barostat algorithms) with the trajectory recording interval being every 100 ps (101 frames). The clustering was performed in the Desmond Trajectory Clustering module, to obtain the most representative conformation of the largest cluster (C1). Trajectory analyses were performed in the Simulations Interactions Diagram module in Maestro.
2.4. Umbrella sampling (US) simulations
Using the C1 of each 10 ns AA-MD trajectory, the system for US simulations was built under the previously mentioned conditions. Once the system was obtained and relaxed using the AA-MD relaxation protocol, a 10 ns (101 frames) MD simulation was performed in the Metadynamics module of Desmond using the protein and ligands center of mass distance as the collective variable, and 0.3 kcal/mol height and 0.1 kcal/mol width as gaussian parameters for the umbrella protocol, on an NPT ensemble at 310.15 K and 1.01325 bar. Finally, the analysis was performed in the Metadynamics Analysis module of Desmond. Three types of umbrella sampling were used; umbrella sampling type I (USI), where the center of mass between SP and ACE2 was a collective variable, pulling the SP; umbrella sampling type II (USII), where the center of mass between the SP-ACE2 complex and ligand was a collective variable, pulling the ligand; and umbrella sampling type III (USIII) where the center of mass between specified protein (SP or ACE2) and ligand was a collective variable, pulling the ligand.
3. Results and discussion
3.1. Molecular docking
The three virtual screenings of 147 ligands with the SP-ACE2 complex using molecular docking was performed and resulted in the selection of five ligands with the potential to inhibit or destabilize the interaction between SP and ACE2 in the SARS-CoV-2 infection process.
3.1.1. First screening using molecular docking
The docking score data for the molecular docking of the 147 ligands from the DrugBank database [29] are presented in the Supplementary information (Table S1). After the first screening, 65 ligands that interact at the interface between SP and ACE2 or with only SP were selected. The selection of these ligands was made considering their spatial position in the SP-ACE2 complex; thus, the ligands that were far from the interface region between SP and ACE2 were discarded (Fig. 2 ). Then, the selected ligands were redocked with the respective interaction site (33 with SP and 32 with ACE2).
Fig. 2.
Spatial position of selected ligands in the first screening using molecular docking. a: All ligands in complex with the spike protein-angiotensin-converting enzyme 2 (SP-ACE2) complex, b: ligands in blue selected for proximity to the interface, and ligands in yellow selected for interaction with SP. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.1.2. Second screening using molecular docking
The docking scores of the 65 selected drugs are presented in Table 2, Table 3 , grouped by their binding site. The drugs varied in the mechanism of action and docking score; unexpectedly, antiviral drugs, such as amprenavir, brivudine, and raltegravir, had a good docking score for ACE2. Analyzing the binding site of these 65 drugs by redocking, in the case of the SP-ACE2 interface, 23 of the drugs were positioned in three areas across the interface and eight drugs were positioned in three areas in the SP (Fig. 3 ). Drugs that were positioned at the same site as the reference drugs used for the construction of the second screening grids were selected to continue the molecular dockings.
Table 2.
Docking score of 32 ligands docked within the interface of the spike protein-angiotensin-converting enzyme 2 (SP-ACE2) complex.
| ID | Docking score (kcal/mol) | ID | Docking score (kcal/mol) | ID | Docking score (kcal/mol) | ID | Docking score (kcal/mol) |
|---|---|---|---|---|---|---|---|
| Aceclofenac | −5.36 | Daclatasvir | −7.93 | Idebenone | −7.05 | Plerixafor | −13.25 |
| Allantoin | −4.84 | Docosanol | −7.53 | Idoxuridine | −5.72 | Procaterol | −9.02 |
| Anidulafungin | −7.42 | Dolutegravir | −6.77 | Isavuconazonium | −8.79 | Raltegravir | −8.09 |
| Amprenavir | −10.97 | Elbasvir | −9.67 | Ivacaftor | −6.18 | Ribavirin | −5.12 |
| Asunaprevir | −8.09 | Emtricitabine | −5.88 | Lamivudine | −5.89 | Telbivudine | −5.35 |
| Beclometasone dipropionate | −7.28 | Fosamprenavir | −5.75 | Moexipril | −5.39 | Tedizolid phosphate | −8.96 |
| Bictegravir | −7.28 | Gemifloxacin | −5.09 | Nystatin | −7.46 | Trandolapril | −7.42 |
| Brivudine | −8.08 | Grazoprevir | −7.7 | Pibrentasvir | −8.15 | Zofenopril | −6.55 |
Table 3.
Docking score of 33 ligands docked exclusively with the spike protein.
| ID | Docking score (kcal/mol) | ID | Docking score (kcal/mol) | ID | Docking score (kcal/mol) |
|---|---|---|---|---|---|
| Acipimox | −6.26 | Cidofovir | −3.79 | Fentapril | −4.43 |
| Amlexanox | −6.59 | Cilazapril | −7.23 | Flurbiprofen | −7.1 |
| Benzylpenicillin | −5.36 | Dalsalate | −5.39 | Foscarnet | −3.45 |
| Caffeine | −5.36 | Deferasirox | −6.31 | Foscarnet | −4.36 |
| Glycerophosphate | −2.86 | Deferipone | −4.51 | Fluconazole | −4.5 |
| Captopril | −4.4 | Dexibuprofen | −6.9 | Furosemide | −5.71 |
| Cefoxitin | −7.81 | Elvitegravir | −6.62 | Salsalate | −5.38 |
| Cefpodoxime | −6.06 | Enprofylline | −5.56 | Sofosbuvir | −4.24 |
| Cerivastatin | −5.26 | Enalaprilat | −8.42 | Stavudine | −5.68 |
| Cholic acid | −7.12 | Erdosteine | −4.18 | Vidarabine | −4.00 |
| Salicylate | −5.59 | Favipravir | −5.41 | Zanamivir | −6.27 |
Fig. 3.
Binding site of the second molecular docking. a: spike protein-angiotensin-converting enzyme 2 (SP-ACE2) complex, b: interface SP-ACE2 complex, and c: SP.
3.1.3. Third screening using molecular docking
Based on the position and docking score of the drugs presented in Fig. 3, a third screening was performed to select the five best candidates. Therefore, six grids were calculated (three for the SP-ACE2 interface, 1_1 to 1_3, and three for the SP, 2_1 to 2_3) taking the coordinates of the drug with the best docking score from the different sites. This data is shown in Table 4, Table 5, Table 6, Table 7 .
Table 4.
Docking results of ligands preferentially bound at the interface of the spike protein-angiotensin-converting enzyme 2 (SP-ACE2) complex, grid 1_1, 1_2, and 1_3.
| Grid | 1_1 |
1_2 |
1_3 |
||||
|---|---|---|---|---|---|---|---|
| Ligand | Amprenavir | Procaterol | Elbasvir | Pibrentasvir | Plerixafor | Brivudine | Tedizolid phosphate |
| Docking score (kcal/mol) | −10.88 | −8.96 | −8.56 | −7.2 | −13.93 | −5.3 | −7.94 |
| Polar interactions | G354, R393, and N382 | K353, G354, E37, and R393 | V483 and S494 | K31, E484, and Y449 | E484 and E35 | R393 and N394 | F356 |
| Relevant residuesaround 5 Å | K353 | – | H34 | – | H34 | – | K353 |
* = π-π R393.
Table 5.
Docking score of ligands selected with spike protein (SP) grid 2_1.
| Grid | 2_1 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ligand | Acipimox | Caffeine | Cefoxitin* | Salicylate | Cilazapril | Deferipone | Enalaprilat | Foscarnet | Stavudine | Zanamivir |
| Docking score (kcal/mol) | −6.35 | −5.41 | −7.97 | −5.65 | −7.47 | −4.59 | −7.48 | −4.48 | −5.75 | −3.85 |
| Polar interactions | E471 and K458. | E471, R454 and K458. | K458 | – | K458. | E471. | K458 and E471 | I472 and K458. | R454 and K458. | E471, R454, and K458. |
| Relevant residuesaround 5 Å | – | – | Q474 | – | Q474. | – | Q474. | – | Q474. | Q474. |
= π-cation R454.
Table 6.
Docking score of ligands selected with spike protein (SP) grid 2_2.
| Grid | 2_2 |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligand | Glycerophos-phate | Cefpodoxime | Cholic acid | Dalsalate | Erdosteine | Fentapril | Furosemide | Salsalate | Sofosbuvir |
| Docking score (kcal/mol) | −2.45 | −4.45 | −6.36 | −5.87 | −4.91 | −4.53 | −6.75 | −5.93 | −5.79 |
| Polar interactions | C379 | F377 and C379 | S375, Y369, and K378 | K378 and C379 | K378 y C379 | C379 | C379 | C379 | F373, K378, C379, and S383 |
Table 7.
Docking score of ligands selected with spike protein (SP) grid 2_3.
| Grid | 2_3 |
||
|---|---|---|---|
| Ligand | Cerivastatin | Fluconazole* | Vidarabine |
| Docking score (kcal/mol) | −5.61 | −4.09 | −4.64 |
| Polar interactions | R509 and A344 | N343 | S348, N440, L441, and A344. |
= π-π W346 and hydrogen bond with N-acetylglucosamines (NAGs).
3.2. Molecular dynamics
The AA-MD simulations were performed to relax the ligand–protein complex and observe the changes in the interaction between SP and ACE2, and between ligands and the SP-ACE2 complex, comparing the original docking structure (T0) with the most representative conformation of the largest cluster analysis (C1) using the C-alpha root-mean square deviation (RMSD) of the MD trajectory.
3.2.1. Molecular dynamics of the SP-ACE2 complex
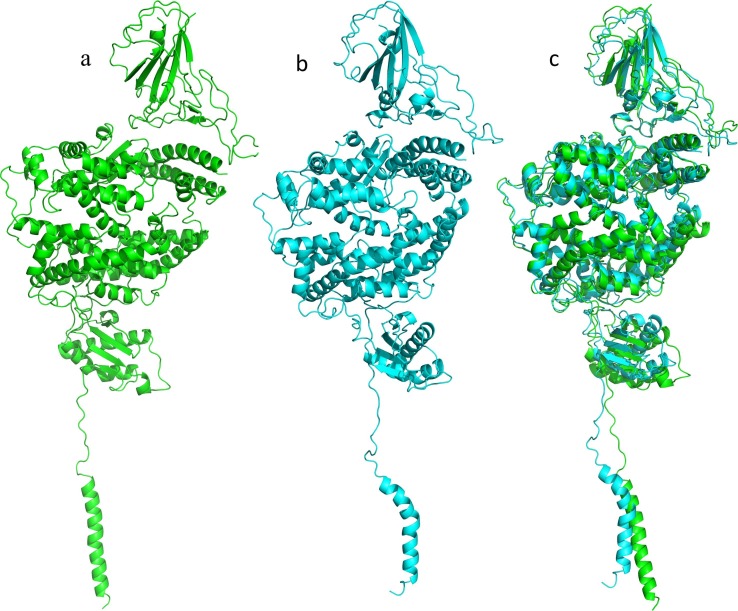
In the C1 of MD of the SP-ACE2 complex, the interaction between SP and ACE2 increased. Among the interactions found, residue K31 in ACE2 interacted with Q493 of SP, which is relevant as K31 is considered important for the SP-ACE2 interaction [10]. The RMSD of the alignment between T0 and C1 was 2.235 Å, and the increased interactions indicating that the system evolves to a more energetically favorable state, which is consistent with the observed good affinity of SP with ACE2 (Fig. 4 and Fig. 5 ).
Fig. 4.
Alignment of the spike protein-angiotensin-converting enzyme 2 (SP-ACE2) complex. a: original docking structure (T0), b: representative conformation of the largest cluster (C1) with 10 ns all atom molecular dynamics (AA-MD) analysis, c: alignment of T0-C1, RMSD: 2.235 Å.
Fig. 5.
Interactions between spike protein (SP) and angiotensin-converting enzyme 2 (ACE2). a: Interactions at the original docking structure (T0), b: Interactions in the representative conformation of the largest cluster (C1) with 10 ns all atom molecular dynamics (AA-MD) analysis. The B chain corresponds to ACE2 and the E chain corresponds to SP. We can observe an increase in hydrogen bonds and the interaction between K31 (ACE2) and Q493 (SP).
3.2.2. Molecular dynamics of the Amp-SP-ACE2 complex
In this simulation, the RMSD of the alignment between T0 and C1 was 2.387 Å and the interactions between Amp and SP-ACE2 complex were modified.
At T0, are three hydrogen bonds between G354, R382, and R393 (ACE2) and Amp, whereas in C1, residues involved in the polar interactions with Amp are D382, N294 (ACE2), and Y505 (SP). In this conformation, Amp interacts with both ACE2 and SP. Likewise, the interactions between ACE2 and SP are decreased, although interaction between Q493-K31 is present in C1 of SP-ACE2 complex, whereas K31 interacts with E484 in this complex (Fig. 6 ).
Fig. 6.
Amprenavir-spike protein-angiotensin-converting enzyme 2 (Amp-SP-ACE2) complex. a: Alignment of the original docking structure to the representative conformation of the largest cluster (T0-C1) of Amp-SP-ACE2 complex AA-MD; T0 in green and C1 in cyan. b: binding site of amprenavir at T0, c: ligand interaction diagram (LID) of Amp to T0, d: binding site of Amp in C1, e: LID of Amp to C1, f: LID legend, and g: Interactions between SP and ACE2 in C1. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2.3. Molecular dynamics of the Bri-SP-ACE2 complex
In this simulation, the RMSD of alignment T0 and C1 was 2.405 Å and the interactions between Bri and SP-ACE2 decreased. In C1, interactions between R393 and 394 of Bri and ACE2 were lost; however, the conformation of Bri promoted hydrogen bond and π-π-type interactions with F390 in ACE2. Besides, the number of interactions between SP and ACE2 decreased, and the K31-Q493 was modified. In C1, E484 (SP) interacted with K31 (ACE2), and Q493 (SP) interacted with E35 (ACE2); additionally, relevant residues were involved in the interactions between SP-ACE2, T500-N330, N501-K353, K417-D30, and Y489-Q24 by a hydrogen bond (Fig. 7 ).
Fig. 7.
Brivudine-spike protein-angiotensin-converting enzyme 2 (Bri-SP-ACE2) complex. a: Alignment of the original docking structure to the representative conformation of the largest cluster (T0-C1) of Bri-SP-ACE2 complex AA-MD; T0 in green and C1 in cyan. b: binding site of Bri at T0, c: ligand interaction diagram (LID) of Bri to T0, d: binding site of Bri in C1, e: LID of Bri to C1, f: Interactions between SP and ACE2 in C1. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2.4. Molecular dynamics of the Cil-SP-ACE2 complex
In this simulation, the RMSD of the alignment between T0 and C1 was 2.510 Å and the interactions between Cil and SP-ACE2 increased. The conformation of Cil in C1 promotes the polar interactions with R457 and K458 in SP. Moreover, the interactions between SP and ACE2 increased, in which relevant residues involved in polar interactions between SP and ACE2 were T500-Y41 and D355, and K417-D30, G502, and G496 with K353. Finally, the interaction between E484-K31 present in C1 of the AA-MD simulation of Bri and Amp is also present in C1 of the Cil simulation (Fig. 8 ).
Fig. 8.
Cilazapril-spike protein-angiotensin-converting enzyme 2 (Cil-SP-ACE2) complex. a: Alignment of the original docking structure to the representative conformation of the largest cluster (T0-C1) of Cil-SP-ACE2 complex AA-MD; T0 in green and C1 in cyan. b: binding site of Cil at T0, c: ligand interaction diagram (LID) of Cil to T0, d: binding site of Cil in C1, e: LID of Cil to C1, f: Interactions between SP and ACE2 in C1. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2.5. Molecular dynamics of the Ena-SP-ACE2 complex
The RMSD of alignment between T0 and C1 was 2.723 Å and the polar interactions between Ena and SP-ACE2 complex decreased. The Ena conformation in C1 did not have any polar interactions, the chemical environment corresponded to positively charged residues, and possibly formed an interaction with the aromatic ring in Ena. In contrast, the interactions between SP and ACE2 increased, with relevant residues such as T500-D355 and G502-K353 which are involved in polar interactions between SP and ACE2. In this conformation, K31 interacted with Q493 and F490; the interaction with E484, which was present in Bri and Cil complexes, was lost (Fig. 9 ).
Fig. 9.
Enalaprilat-spike protein-angiotensin-converting enzyme 2 (Ena-SP-ACE2) complex. a: Alignment of the original docking structure to the representative conformation of the largest cluster (T0-C1) of Ena-SP-ACE2 complex AA-MD; T0 in green and C1 in cyan. b: binding site of Ena at T0, c: ligand interaction diagram (LID) of Ena to T0, d: binding site of Ena in C1, e: LID of Ena to C1, f: Interactions between SP and ACE2 in C1. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2.6. Molecular dynamics of the Ple-SP-ACE2 complex
In this simulation, the alignment RMSD of T0 and C1 was 2.753 Å, the polar interactions between Ple and SP-ACE2 and those with the residues around 5 Å of Ple decreased. At T0, Ple exhibited interaction with E35 in ACE2 and E494 in SP; in this conformation, Ple bound with SP and ACE2. In contrast, in the C1 conformation, the interactions between Ple and E35 in ACE2 were lost and the interaction with SP was modified; Ple interacted only with I468. Moreover, the interactions between SP and ACE2 decreased, in which relevant residues involved in polar interactions between SP and ACE2 were K417-D30, Q502-K353, and Q493-K31 (Fig. 10 ).
Fig. 10.
Plerixafor-spike protein-angiotensin-converting enzyme 2 (Ple-SP-ACE2) complex. a: Alignment of the original docking structure to the representative conformation of the largest cluster (T0-C1) of Ple-SP-ACE2 complex AA-MD; T0 in green and C1 in cyan. b: binding site of Ple at T0, c: ligand interaction diagram (LID) of Ple to T0, d: binding site of Ple in C1, e: LID of Ple to C1, f: Interactions between SP and ACE2 in C1. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. Umbrella sampling simulations
Molecular dynamics simulations did not provide enough information to consider that the five selected ligands would destabilize the SP-ACE2 complex. Therefore, three umbrella sampling simulations were performed to calculate the binding energy (ΔG) between SP and ACE2 in the presence of ligands (pulling the SP, USI), ligands and SP-ACE2 complex (pulling the ligands, USII), and ligands and SP or ACE2 protein (pulling the ligands, USIII). The binding energy of the SP-ACE2 complex was considered as a control in USI.
A high negative binding energy value denotes a high binding affinity between protein–ligand and protein–protein complexes. Therefore, if the binding energy (ΔG) is increased owing to the presence of the ligand it would suggest that the complex is destabilized. The distance from the center of mass of SP and ACE2 was used as the collective variable, and the ΔG was calculated for each ligand by taking the difference between maximum and minimum energy calculated from the potential mean force of the US [35].
3.3.1. Umbrella sampling type I (USI)
In USI, the most positive ΔG of the SP-ACE2 complex in the presence of ligands would suggest that the complex would be destabilized. The ΔG of the SP-ACE2 complex and ΔG of SP-ACE2-ligands are presented in Table 8 .
Table 8.
ΔG of the spike protein-angiotensin-converting enzyme 2 (SP-ACE2) complex in umbrella sampling type I.
| ID | ΔG (kcal/mol) | ID | ΔG (kcal/mol) |
|---|---|---|---|
| Amp | −20.13 | Ena | –23.84 |
| Bri | –33.83 | Ple | −19.72 |
| Cil | −26.07 | SP-ACE2 complex | −29.58 |
The ΔG of the SP-ACE2 complex without ligands was used as the control (-29.58 kcal/mol). In the comparison between the ΔG of the complex and ΔG in presence of the ligands, Ple was the ligand that could promote the most destabilization of the SP-ACE2 complex by decreasing the ΔG of the SP-ACE2 complex from −29.58 kcal/mol to −19.72 kcal/mol, a difference of 9.86 kcal/mol. Additionally, Amp had a ΔG decrease of 9.45 kcal/mol, Ena with a decrease of 5.74 kcal/mol, and Cil with a ΔG decrease of 3.51 kcal/mol, also had the potential to destabilize the SP-ACE2 complex.
In all the ΔG, a decrease in ΔG in the presence of the ligand was observed, except in the presence of Bri in which ΔG did not show a positive difference; thus, Bri was discarded as a candidate for destabilizing the SP-ACE2 complex.
3.3.2. Umbrella sampling type II (USII)
To verify if the ligands can bind with the SP-ACE2 complex once formed (USII), the pulling of the ligand from the SP-ACE2 complex was tested. A ΔG of −28.09 kcal/mol for Amp indicated that this is the ligand with the best affinity to the SP-ACE2 complex, followed by Bri, Ple, and Ena, and Cil was the ligand with the least affinity to the SP-ACE2 complex (Table 9 ).
Table 9.
ΔG of the spike protein-angiotensin-converting enzyme 2 (SP-ACE2) complex in umbrella sampling type II.
| ID | ΔG (kcal/mol) | ID | ΔG (kcal/mol) | ID | ΔG (kcal/mol) |
|---|---|---|---|---|---|
| Amp | −28.09 | Cil | −10.53 | Ple | −14.55 |
| Bri | −14.78 | Ena | −11.64 |
3.3.3. Umbrella sampling type III (USIII)
To calculate the binding energy of the ligands with the respective binding protein calculated in this study (SP and ACE2), USIII or pulling the ligand away from the respective binding protein, was performed. Amp bound with ACE2 and Cil, Ena, and Ple bound with SP; the ΔG of the ligands is presented in Table 10 . Amp had a good affinity with ACE2 (ΔG −28.09 kcal/mol); in contrast, Ena had the best affinity with SP (ΔG of −24.75 kcal/mol), Ple had a ΔG of −23.2 kcal/mol, and Cil had the least affinity with a ΔG of −11.2 kcal/mol.
Table 10.
ΔG of the spike protein-angiotensin-converting enzyme 2 (SP-ACE2) complex in umbrella sampling type III.
| ID | ΔG (kcal/mol) | ID | ΔG (kcal/mol) |
|---|---|---|---|
| Amp | −28.09 | Ena | −24.75 |
| Cil | −11.2 | Ple | −23.2 |
These results indicate that Amp, Ena, and Ple could destabilize the SP-ACE2 complex. Amp is an antiviral drug that can bind with ACE2 either in complex with SP or alone; additionally, Ena is an ACE2 inhibitor drug that can bind with SP, and Ple is an immunostimulatory drug that can bind with SP. These ligands could bind to the SP-ACE2 complex and either SP or ACE2 and could destabilize the SP-ACE2 complex by diminishing the binding energy of SP-ACE2. The energy profiles of all US simulations are presented in Figs. S1, S2, and S3.
The increase in options for the possible treatment of this infection is useful to people without access to one of several vaccines in development, and the people living in countries where the governments have been insensitive to this health problem. The SP in the SARS-CoV-2 is an interesting target to investigate possible drug targeting with antibody neutralizers, fusion inhibitors, and protease inhibitors as options for possible treatment or development of a vaccine [11], [12]; however, the destabilization and inhibition of SP-ACE2 complex formation or membrane fusion is an another approach to possible treatments where the SP is the target, and not only the antibodies and peptides, but also small molecules [36].
4. Concluding remarks
Drug repositioning is a valuable strategy for drug development in sanitary emergencies such as the COVID-19 pandemic; however, to better assess the adequacy of the proposals developed using this strategy, it is not only necessary to choose a relevant biological target, but also to consider the possible mechanism of action as a dynamic process that cannot be studied solely through molecular docking or Quantitative Structure–Activity Relationships (QSAR) studies. These computational analyses of MD and US simulations suggest that Amp, Ena, and Ple are the best candidates for destabilizing the SP-ACE2 complex. Amp interacts with ACE2 according to the docking calculations and in C1 of the AA-MD simulation, interacts with both ACE2 and SP, and decreases interactions between SP and ACE2; however, some interactions between relevant residues such as E484-K31, N487-Q24, and G496 and G502 with K353 remain. However, the ΔG of SP of the Amp-SP-ACE2 complex in the USI approach, was higher than the ΔG of SP without the influence of the ligand; thus, the presence of Amp influences the easy removal of the SP. The ΔG of the Amp-SP-ACE2 complex (USII) indicates that it has a good affinity; therefore, if the SP-ACE2 complex has already formed, Amp could bind with good affinity and possibly destabilize the complex and inhibit membrane fusion.
In the case of Ena, polar interactions with SP were lost in the C1 of the AA-MD, but the ΔG in the USI indicated that the presence of Ena makes it easier for the SP to be removed. However, the calculated affinity with the SP was low compared to the affinity of Amp-ACE2 in USII once the SP-ACE2 complex was formed. In contrast, Ple decreased the interactions between SP-ACE2 in C1 of AA-MD, the ΔG of USI indicated that it facilitates the removal of SP from the complex, having a good affinity with SP alone. Compared to the low affinity to the SP-ACE2 complex, Ple could bind to the SP before the formation of the SP-ACE2 complex and prevent formation of this complex; however, stimulating the immune system could be counterproductive in advanced phases of the disease where the inflammation caused by the cells of the immune system is elevated.
These results present the possibility that Amp, Ena, and Ple could be evaluated for the treatment of COVID-19. However, it is mandatory that these proposals are evaluated in vitro prior to their evaluation in patients. This information was obtained through simulations; therefore, there is the possibility that these drugs may not work. Thus, in vitro evaluation is mandatory to verify the data presented in this paper.
CRediT authorship contribution statement
Ivonne Buitrón-González: Conceptualization, Methodology, Formal analysis, Writing - original draft, Writing - review & editing. Giovanny Aguilera-Durán: Conceptualization, Methodology, Formal analysis, Writing - original draft, Writing - review & editing. Antonio Romo-Mancillas: Conceptualization, Methodology, Formal analysis, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors dedicate this work to the anonymous heroes fighting against the pandemic: to the members of the public health system for their tireless and selfless efforts. It is also dedicated to the deceased, victims of both the pandemic and government incompetence, especially in the region of the “three caballeros”: US, Mexico, and Brazil.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.rechem.2020.100094.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Adhikari S.P., Meng S., Wu Y.-J., Mao Y.-P., Ye R.-X., Wang Q.-Z., Sun C., Sylvia S., Rozelle S., Raat H., Zhou H. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: a scoping review. Infect. Dis. Poverty. 2020;9(1) doi: 10.1186/s40249-020-00646-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.C. BULUT Y. KATO Epidemiology of COVID-19 Turk J Med Sci 50 SI-1 563 570 10.3906/sag-2004-172. [DOI] [PMC free article] [PubMed]
- 3.Ge H., Wang X., Yuan X., Xiao G., Wang C., Deng T., Yuan Q., Xiao X. The epidemiology and clinical information about COVID-19. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39(6):1011–1019. doi: 10.1007/s10096-020-03874-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.OMS, COVID-19 Americas’ Regional Dashboard. Geographic Distribution of Cases and Deaths, (2020). https://who.maps.arcgis.com/apps/dashboards/efb745c3d88647779beccb91c0e715f9.
- 5.Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C., Chen H., Mubareka S., Gubbay J.B., Chan W.C.W. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14(4):3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 6.Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20(6):363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuchun N., Guangwen W., Xuanling S., Hong Z., Yan Q., Zhongping H., Wei W., Gewei L., Xiaolei Y., Liying D., Lili R., Jianwei W., Xiong H., Taisheng L., Hongkui D., Mingxiao D. Neutralizing antibodies in patients with severe acute respiratory syndrome-associated coronavirus infection. J. Infect. Dis. 2004;190:1119–1126. doi: 10.1086/423286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X.i., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin Y., Yang H., Ji W., Wu W., Chen S., Zhang W., Duan G. Virology, epidemiology, pathogenesis, and control of COVID-19. Viruses. 2020;12:372. doi: 10.3390/v12040372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sternberg A., Naujokat C. Structural features of coronavirus SARS-CoV-2 spike protein: Targets for vaccination. Life Sci. 2020;257:118056. doi: 10.1016/j.lfs.2020.118056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Y., Yang C., Xu X.-F., Xu W., Liu S.-W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41(9):1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo Y.-R., Cao Q.-D., Hong Z.-S., Tan Y.-Y., Chen S.-D., Jin H.-J., Tan K.-S., Wang D.-Y., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak – an update on the status. Military Med Res. 2020;7(1) doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science:abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan R., Zhang Y., Li Y., Xia L.u., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science:abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morgenstern B., Michaelis M., Baer P.C., Doerr H.W., Cinatl J., Jr. Ribavirin and interferon-β synergistically inhibit SARS-associated coronavirus replication in animal and human cell lines. Biochem. Biophys. Res. Commun. 2005;326(4):905–908. doi: 10.1016/j.bbrc.2004.11.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim U.J., Won E.-J., Kee S.-J., Jung S.-I., Jang H.-C. Combination therapy with lopinavir/ritonavir, ribavirin and interferon-alpha for Middle East respiratory syndrome: a case report. Antivir Ther. 2015;21(5):455–459. doi: 10.3851/IMP3002. [DOI] [PubMed] [Google Scholar]
- 18.Yamamoto N., Yang R., Yoshinaka Y., Amari S., Nakano T., Cinatl J., Rabenau H., Doerr H.W., Hunsmann G., Otaka A., Tamamura H., Fujii N., Yamamoto N. HIV protease inhibitor nelfinavir inhibits replication of SARS-associated coronavirus. Biochem. Biophys. Res. Commun. 2004;318(3):719–725. doi: 10.1016/j.bbrc.2004.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W.u., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gimeno A., Mestres-Truyol J., Ojeda-Montes M.J., Macip G., Saldivar-Espinoza B., Cereto-Massagué A., Pujadas G., Garcia-Vallvé S. Prediction of novel inhibitors of the main protease (M-pro) of SARS-CoV-2 through consensus docking and drug reposition. Int. J. Mol. Sci. 2020;21:3793. doi: 10.3390/ijms21113793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shah B., Modi P., Sagar S.R. In silico studies on therapeutic agents for COVID-19: Drug repurposing approach. Life Sci. 2020;252:117652. doi: 10.1016/j.lfs.2020.117652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nutho B., Mahalapbutr P., Hengphasatporn K., Pattaranggoon N.C., Simanon N., Shigeta Y., Hannongbua S., Rungrotmongkol T. Why are lopinavir and ritonavir effective against the newly emerged coronavirus 2019? Atomistic insights into the inhibitory mechanisms. Biochemistry. 2020;59(18):1769–1779. doi: 10.1021/acs.biochem.0c00160.s001. [DOI] [PubMed] [Google Scholar]
- 23.Fantini J., Di Scala C., Chahinian H., Yahi N. Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int. J. Antimicrob. Agents. 2020;55(5):105960. doi: 10.1016/j.ijantimicag.2020.105960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J. Fast identification of possible drug treatment of coronavirus disease-19 (COVID-19) through computational drug repurposing study. J. Chem. Inf. Model. 2020;60(6):3277–3286. doi: 10.1021/acs.jcim.0c00179.s001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao K., Nguyen D.D., Chen J., Wang R., Wei G.-W. Repositioning of 8565 existing drugs for COVID-19. J. Phys. Chem. Lett. 2020;11(13):5373–5382. doi: 10.1021/acs.jpclett.0c01579.s002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elfiky A.A. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020;248:117477. doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin J.H., Bowden N.A. Drug repurposing in the era of COVID- 19: a call for leadership and government investmen. Med. J. Aust. 2020;212:450. doi: 10.5694/mja2.50603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knox C., Law V., Jewison T., Liu P., Ly S., Frolkis A., Pon A., Banco K., Mak C., Neveu V., Djoumbou Y., Eisner R., Guo A.C., Wishart D.S. DrugBank 3.0: a comprehensive resource for 'Omics' research on drugs. Nucleic Acids Res. 2011;39(Database):D1035–D1041. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Law V., Knox C., Djoumbou Y., Jewison T., Guo A.C., Liu Y., Maciejewski A., Arndt D., Wilson M., Neveu V., Tang A., Gabriel G., Ly C., Adamjee S., Dame Z.T., Han B., Zhou Y., Wishart D.S. DrugBank 4.0: shedding new light on drug metabolism. Nucl. Acids Res. 2014;42(D1):D1091–D1097. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z., Assempour N., Iynkkaran I., Liu Y., Maciejewski A., Gale N., Wilson A., Chin L., Cummings R., Le Di, Pon A., Knox C., Wilson M. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santos-martins D., Solis-vasquez L., Koch A., Forli S. Accelerating AUTODOCK 4 with GPUs and gradient-based local search. ChemRxiv. 2019 doi: 10.1021/acs.jctc.0c01006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bowers K.J., Chow D.E., Xu H., Dror R.O., Eastwood M.P., Gregersen B.A., Klepeis J.L., Kolossvary I., Moraes M.A., Sacerdoti F.D., Salmon J.K., Shan Y., Shaw D.E. ACM/IEEE SC 2006 Conf. IEEE; 2006. Scalable algorithms for molecular dynamics simulations on commodity clusters. pp. 43-43 https://doi.org/10.1109/SC.2006.54. [Google Scholar]
- 34.2020. Schrödinger Release 2020-1: Desmond Molecular Dynamics System, D. E. Shaw Research, New York, NY, 2020. Maestro-Desmond Interoperability Tools, Schrödinger, New York, NY, Maestro 2020, (n.d.). https://www.schrodinger.com/desmond.
- 35.Londhe A.M., Gadhe C.G., Lim S.M., Pae A.N. Investigation of molecular details of keap1-Nrf2 inhibitors using molecular dynamics and umbrella sampling techniques. Molecules. 2019;24:4085. doi: 10.3390/molecules24224085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choudhary S., Silakari O.m. Scaffold morphing of arbidol (umifenovir) in search of multi-targeting therapy halting the interaction of SARS-CoV-2 with ACE2 and other proteases involved in COVID-19. Virus Res. 2020;289:198146. doi: 10.1016/j.virusres.2020.198146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.