Abstract
The recent outbreak of the coronavirus disease (COVID-19) has left the world clueless. As the WHO declares this new contagion as a pandemic on the 11th of March 2020, the alarming rate of the spawn of the disease in such a short period has disarranged the globe. Standing against this situation researchers are strenuously searching for the key traits responsible for this pandemic. As knowledge regarding the dynamics and host-path interaction of COVID-19 causing Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) is currently unknown, the formulation of strategies concerning antiviral treatment, vaccination, and epidemiological control stands crucial. Before designing adequate therapeutic strategies, it is extremely essential to diagnose the disease at the outset as early detection can have a greater impact on building health system capacity. Hence, a comprehensive review of strategies for COVID-19 diagnosis is essential in this existing global situation. In this review, sequentially, we have provided the clinical details along with genetic and proteomic biomarkers related to COVID-19. The article systematically enlightens a clear overview of the clinically adopted techniques for the detection of COVID-19 including oligonucleotide-based molecular detection, Point-of-Care immunodiagnostics, radiographical analysis/sensing system, and newly developed biosensing prototypes having commercial viability. The commercial kits/analytical methods based-sensing strategies have also been tabulated categorically. The critical insights on the developer, commercial brand name, detection methods, technical operational details, detection time, clinical specimen, status, the limit of detection/detection ability have been discussed comprehensively. We believe that this review may provide scientists, clinicians and healthcare manufacturers valuable information regarding the most recent developments/approaches towards COVID-19 diagnosis.
Keywords: COVID-19, SARS-CoV-2, Clinical diagnosis, RT-PCR, Immunoassay, Serological tests, Point of care
Highlights
-
•
COVID-19 pandemic: Clinical aspects and need of diagnosis are summarized.
-
•
Clinically practiced and nanobio engineered methods for COVID-19 diagnosis are discussed.
-
•
Commercially available diagnostic methodologies are tabulated with their detection strategy and analytical performances.
-
•
Future prospects and the forthcoming diagnostic concepts are elaborated.
1. Introduction
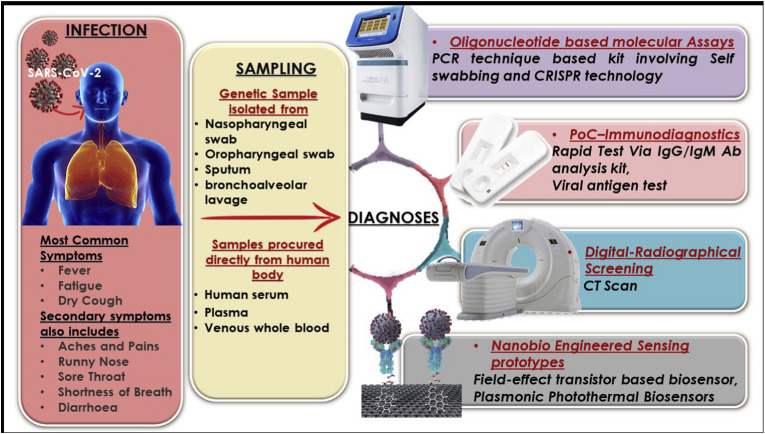
In November 2002, a typical pneumonia called severe acute respiratory syndrome (SARS) began spreading rapidly around the world. The World Health Organization (WHO) declared the ailment as “a worldwide health threat” (World Health Organization, 2014). It had been 17 years since SARS again reappeared in China and within a very short interval, the situation had escalated beyond limits. By the time the global outbreak was confined, the virus had spread to over 8,000 people worldwide and killed almost 800 (Web Reference 1, Jan. 2020). The new SARS-like respiratory illness that emerged in China is caused by a coronavirus, known as SARS-CoV-2 (Coronaviridae Study Group of the International Committee on Taxonomy of Viruses, 2020). Illness caused by this virus is known as COVID-19 (Velavan and Meyer, 2020), which manifests with some routine symptoms such as fever, cough, and shortness of breath. Though presently there are no explicit antiviral treatment or vaccine approved for the disease, clinical trials are currently underway as the spread of the disease becomes uncontrollable. The outbreak of this illness has already surpassed SARS and is expected to grow even more (Web Reference 2, Mar. 2020). As the world races to ramp up testing for this pandemic, various technologies, and tactics are rolling out to make it easier and quicker for people to know whether they have been infected or not. In search of faster and cheaper tests, numerous diagnostics and pharmaceutical companies are working extensively (Web Reference 3, Feb. 2020). Several new detection techniques have been deployed by various organizations and they are ready to ship their products worldwide (Web Reference 4, Feb. 2020). These employ advanced and futuristic strategies that are likely to validate the diagnosis in a shorter period with high precision. The diagnostic spectrum of COVID-19 crucially involves sampling from the suspected patient as the preliminary step. In hospitals, such samples are obtained from the respiratory system specifically nasopharyngeal (NP), oropharyngeal (OP) swab, bronchoalveolar lavage (BAL), and sputum, which are then used to diagnose the virus infection (Holshue et al., 2020; Zhu et al., 2020). This is because, just like the other CoV infections, detection of this virus in the respiratory tract samples is a major key for the clinical diagnosis of COVID-19 (Huang et al., 2020). Examining whole blood, serum, and plasma also yielded positive viral genome via the IgM and IgG antibody profiling in patients with clinical symptoms of SARS-CoV-2 (Guo et al., 2020). In various studies (Li et al., 2020; Normile, 2020; OKBA et al., 2020), it is described that the detection of IgM and IgG antibodies can imply against SARS-CoV-2 in patients by immunodetection methods, as both IgM and IgG antibody levels reported to have surged to a noticeable level after the rapid spread of the infection in the body. Fig. 1 represents various possible procedures adopted for diagnosing the COVID-19 globally in various laboratories and clinical facilities.
Fig. 1.
Schematic representation of SARS-CoV-2 infection eventuated to COVID-19 disease; revealing symptoms, along with clinical sampling and available diagnosis methods.
1.1. Sequencing and molecular data
In an attempt to reveal the nature of the SARS-CoV-2 virus, scientists aimed towards the previously reported human beta-coronaviruses SARS and MERS (Middle East Respiratory Syndrome), which were described as highly pathogenic in humans and can be transmitted between humans causing major outbreaks in healthy individuals and communities. Both SARS and MERS coronaviruses were reported as being zoonotic in origins (Chan et al., 2015). The molecular sequence of SARS-CoV-2 revealed several open reading frames common to the beta-coronaviruses, such as 1ab, which encodes many enzymatic products such as, the spike (S) protein, the small-envelope (E) protein, and the nucleocapsid (N) protein (Wang et al., 2020a, Wang et al., 2020b).
Han et al. sequenced SARS-CoV-2 viral genomes from infected Korean patients during the December 19 outbreak (Han et al., 2020). They performed sequencing studies using upper and lower respiratory tract secretion samples from the presumed patients that were infected with SARS-CoV-2. In this case, the viral genome showed a sequence homology of >99.9% with SARS-CoV-2 which was isolated from patients from other countries. A similar kind of genome sequencing study had been performed by Sah et al. from the OP swab specimen of a 32-year-old Nepalese national tested positive for SARS-CoV-2 (Sah et al., 2020). The sequence homology also revealed >99.99% similarities with two previously sequenced genomes available at GenBank for SARS-CoV-2 from Wuhan, China, with seven additional genome sequences. This varies from the sequence homology compared to SARS and MERS-CoV, where the similarity percentages were 77.5% and 50%, respectively. A study comparing alpha- and beta-coronaviruses based on structural and biochemical experiments, revealed that SARS-CoV-2 may bind to the human receptor Angiotensin-Converting Enzyme 2 (ACE2) gene (Batlle et al., 2020); though the predicted computational evaluations do not idealize the interaction phenomena of the spike proteins (S) and ACE2 gene (Zhou et al., 2020). Moreover, it also shows that the receptor-binding domain sequence is different from those discovered in SARS and MERS-CoV (Suhas et al., 2020). This leads to the conclusion that the high-affinity binding of the SARS-CoV-2 spike protein to human ACE2 is most likely as a result of natural selection and maybe mutation-based modification on a human or other species with high receptor homology to the ACE2 gene (Tang et al., 2020). Yet, it is also suggested that any sort of mutations could affect the virus phenotype and influence the detection of the virus by various biomolecular assays and sensing systems (Zou et al., 2020). This information suggests that SARS-CoV-2 sequencing is likely reasonable for any novel and succeeding outbreaks. Additionally, detection of the location of the mutated region in the viral genome have been successfully attempted predicting the effects on primer and probe's binding sites, which has positively influenced the diagnostic abilities of the presently available marketed kits.
In consideration of such vital molecular and pathophysiological information in terms of molecular probes, primers, genetic biomarkers, and differential levels of antibodies in virus and/or in patients samples/fluids, several analytical methods have been developed recently for the diagnosis of SARS-CoV-2 globally. These tests have been directly applied in the various hospitals worldwide for fast screening and diagnosis of suspects. The subsequent sections will discuss in detail on clinically practiced techniques and recently developed nanobio engineered analytical methods for COVID-19 diagnosis with suitable examples.
2. Oligonucleotide based molecular detection
To date, the real-time reverse transcription-polymerase chain reaction (RT-PCR) has been employed as the backbone for the diagnosis of SARS-CoV-2 (Lan et al., 2020). RT-PCR is a technique that combines reverse transcription of RNA into DNA (in this context called complementary DNA or cDNA). It also amplifies the specific DNA targets using polymerase chain reaction (PCR) (Freeman et al., 1999). This technique is commonly used to detect the presence of mRNAs, pre-mRNAs, or other types of noncoding RNAs (Rio, 2014). For the detection process of the SARS-CoV-2, initially, the upstream oligonucleotides of the envelope gene (E gene) are screened followed by the confirmation of the nucleocapsid gene (N gene) using RT-PCR approach. Obtaining negative results, further testing is done through the sequencing of the RNA-dependent RNA polymerase (RdRp) gene. In this case, if a positive result is obtained it validates the presence of SARS-CoV-2 in the suspect, and the case is termed corona positive clinically (Chan et al., 2020). The commercially available PCR based SARS-CoV-2 diagnostic kits that are highly specific and sensitive, exploits this algorithm. RT-PCR has its inherent limitations, involving a long turnaround time (considering transportations and queues of viral samples and preparation time), and lack of common measurements and correlations with the Viral Load (VL) which is the numerical expression of the virus quantity in a given volume of a body fluid/blood plasma. Most of the commercialized kits have mentioned using in a specific system in their standard technical/operational manual, hence it may give varying sensitivity in alternate PCR analysis. Moreover, Emergency Usage Laboratories are estimating only cycle threshold values for predicting disease severity, VL, disease progression, and as a cut-off marker to diagnose cases (Gao et al., 2020). Based on the RT-PCR principle several testing kits have been developed worldwide. We have comprehensively discussed such kits with details on the commercial brands, working principle, analytical performance, assay time, etc. in Table 1 (A) .
Table 1.
Table indicating recently developed analytical procedures for detection of COVID-19, (A) Oligonucleotide based molecular detection, (B) Point-of-Care Immunodiagnostics, and (C) Digital Imaging/Sensing system.
| (A) Oligonucleotide based molecular detection: | |||||||||
| Sr. No |
Developer |
Commercial Brand Name |
Analytical Method |
Technical Details |
Detection Time |
Clinical Sample |
Status |
Limit of Detection / Detection Ability |
Source |
| 1 | 1 copy™ | 1 copy™ COVID-19 qPCR Kit | RT-qPCR | Qualitative detection of E gene and RdRP gene of COVID-19 with RT-qPCR via RNA extracted from a specimen of an infected patient | 1 h 50 min | Nasopharyngeal swab, Oropharyngeal swab, Sputum | CE-IVD | 4 copies / reaction | http://www.1drop.co.kr/ |
| 2 | Abbott Molecular Inc. | Abbott RealTime SARS-CoV-2 | RT-PCR | The Abbott RT- SARS-CoV-2 assay is a dual target assay assess for the RdRp and N genes | NR | Nasopharyngeal and oropharyngeal swabs | USA CE-IVD | 1-4 X 102 copies/mL | https://www.molecular.abbott/us/en/products/infectious-disease/RealTime-SARS-CoV-2-Assay |
| 3 | AB ANALITICA | REALQUALITY RQ-2019-nCoV | One-step RT-PCR |
Includes RT-PCR amplification targets to the RdRP gene, E gene and the internal control for qualitative analyses. | 1 h 40 min | Nasopharyngeal swabs, sputum and bronchoalveolar lavage |
Manual; lab-based NAT; CE-IVD |
NR | https://www.abanalitica.com/en/catalogo/product/realquality-rq-2019-ncov/ |
| 4 | A*ccelerate Technology Pte Ltd | A*STAR Fortitude Kit 2.0 | One-step RT-PCR | Runs on the RT-PCR laboratory technique that detects SARS-CoV-2 virus genetic material (E gene and RdRP gene) | 1 h 30 min | Human Clinical Samples | Singapore HSA | 25 copies / reaction | https://www.a-star.edu.sg/News-and-Events/a-star-news/covid-19/news/covid-19/fighting-covid-19-with-fortitude |
| 5 | ADT Biotech | LyteStar 2019-nCoV RT-PCR Kit 1.0 | RT-PCR | Dual target kit for screening and confirmation of SARS-related CoV E-gene and 2019-nCoV specific RdRP-gene | NR | Fresh or frozen human serum and plasma, respiratory specimens, and stool | RUO | NR | https://www.dropbox.com/s/2j8uix6d5dwsvr0/LyteStar%202019-nCoV_Flyer.pdf?dl=0 |
| 6 | Altona Diagnostics | RealStar® SARS-CoV-2 RT-PCR Kit | RT- PCR | Detection and differentiation of lineage ß-betacoronavirus (β-ßCoV) and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) specific RNA | NR | Human Clinical Samples | RUO | NR | https://altona-diagnostics.com/en/products/reagents-140/reagents/realstar-real-time-pcr-reagents/realstar-sars-cov-2-rt-pcr-kit-ruo.html |
| 7 | Beijing Applied Biological Technologies Co., Ltd. | Multiple RT- PCR Kit for Detection of 2019-nCoV | RT- fluorescent PCR detection | Detection kit, screening the specific genes: ORF1ab, N and E gene | 1 h 50 min | Human Clinical Samples | China FDA–EUA; CE-IVD | 1 × 103 copies / mL | http://www.x-abt.com/index.php?m=content&c=index&a=lists&catid=144 |
| 8 | BGI Health (HK) Co. Ltd. | RT- fluorescent RT-PCR kit for detecting 2019 nCoV | RT- Fluorescent RT-PCR kit | No cross-reactivity with the human genome and other pathogens including 54 pathogens, such as human coronavirus OC43, 229E, HKU1 and NL63(HCoV-OC43, HCoV-229E, HCoV-HKU1, HCoV-NL63), etc | 3 h | Throat swab and Bronchoalveolar Lavage Fluid(BALF) samples | China-FDA EUA | 100 copies / mL | https://www.bgi.com/global/molecular-genetics/2019-ncov-detection-kit/ |
| 9 | Caspr Biotech | Phantom 1.0 Dx | Ultrasensitive, rapid, and portable coronavirus SARS-CoV-2 sequence detection | Based on CRISPR-Cas12, Point of Care approach |
> 1 h | Human clinical sample | USA, Validated | 103 copies / mL | https://www.biorxiv.org/content/10.1101/2020.02.29.971127v1.full.pdf |
| 10 | Cancer Rop Co., Ltd. | Q-Sens® 2019-nCoV Detection Kit | RT-PCR based | Detection and amplification of nucleotide sequence of the genes specific to SARS-CoV-2 | 2 h | Sputum, nasopharyngeal smear, and bronchial alveolar lavage fluid | CE-IVD | NR | http://www.cancerrop.com/news_notices/news?tpf=board/view&board_code=6&code=311 |
| 11 | Liming Bio-Products Co., Ltd. | SrongStep® Novel Coronavirus (SARS-CoV-2) Multiplex RT- PCR Kit | Fluorescent probe-based Taqman® RT-qPCR | Designed for the detection of a conserved region of SARS-CoV-2’s ORF1ab gene, E gene, and N gene, avoiding non-specific interference of SARS2003 and Bat-SARS-like virus strains | NR | Human Clinical Samples | CE-IVD | NR | http://www.limingbio.com/index.php?m=content&c=index&a=show&catid=18&id=113 |
| 11 | Novacyt/Primerdesign Ltd. | Genesig RT- PCR COVID-19 | RT- PCR | Rapid detection and exclusive to the COVID-19 strain, high priming efficiency with accurate controls to confirm extraction, and assay validity | NR | Clinical Samples | US-FDA EUA; CE-IVD | NR | https://www.genesig.com/products/10039-coronavirus-covid-19-ce-ivd |
| 12 | PerkinElmer Inc. | PerkinElmer® SARS-CoV-2 RT-PCR Assay | RT-PCR | Detects SARS-CoV-2 ORF1ab and N genes | NR | Human oropharyngeal and nasopharyngeal swab samples | CE-IVD | 20 copies / mL | https://perkinelmer-appliedgenomics.com/home/products/new-coronavirus-2019-ncov-nucleic-acid-detection-kit/ |
| 13 | Pishtaz Teb | COVID-19 Onestep RT-PCR Dual Target Gene | One-Step RT-PCR | The primer probe of this kit adopts the dual-target gene design, which targets the specific conserved sequence encoding the RdRp region and the N region. | 1 h 30 min | Bronchoalveolar lavage fluid, sputum, throat and nasal swab | Iran-FDA | NR | http://assets.pishtazteb.com/uploads/1970806048.covid19-EN-min.pdf |
| 14 | SD BIOSENSOR Inc. | STANDARD M nCoV RT- Detection Kit | One Step RT-PCR | Designed according to WHO interim guidance includes 2019-nCov detects nCoV associated ORF1ab gene, E gene and provides Internal Control | 1 h 30 min | Nasopharyngeal swab, Oropharyngeal swab, Sputum | (Korea-EUA; CE-IVD | NR | http://sdbiosensor.com/xe/product/7653 |
| 15 | Sherlock Biosciences, Cepheid | SHERLOCK™ (Specific High Sensitivity Enzymatic Reporter unlocking) | Rapid CRISPR-based tests for SARS-CoV-2 and other pathogens | Combines SHERLOCK Cas12 and Cas13 enzymes for nucleic acid detection with Cepheid’s GeneXpert test-processing instruments | NR | Genetic material obtained from human clinical samples | US-FDA EUA | NR | https://www.prnewswire.com/news-releases/cepheid-and-sherlock-biosciences-establish-collaboration-on-new-genexpert-tests-for-infectious-diseases-and-oncology-leveraging-crispr-technology-301013198.html |
| 16 | Thermo Fisher Scientific | TaqManTM SARS-CoV-2 Assay Kit v2 | RT-PCR | 2019-nCoV Assay (Orf1 ab); 2019-nCoV Assay (spike (S) gene); 2019-nCoV Assay (nucleocapsid (N) gene); Rnase P (control) | NR | Human Clinical Samples | US-FDA EUA | NR | https://www.thermofisher.com/jp/ja/home/clinical/clinical-genomics/pathogen-detection-solutions/coronavirus-2019-ncov/genetic-analysis.html#TaqMan-2019-nCoV-Assay-Kit |
| 17 | US CDC | 2019 nCoV RT-PCR Diagnostic Panel | RT-PCR based | The nCoVPC contains non-infectious positive control material supplied in a dried state. nCoVPC consists of in vitro transcribed RNA. nCoVPC will yield a positive result with each assay in the systems diagnostic Panel including RP. | 1 h 20 min | Human sera or pooled leftover negative respiratory specimens | US-FDA EUA | 102.5 Copies / mL | https://www.genscript.com/2019-ncov-qrt-pcr-detection-assay.html?src=topbanner |
| 18 | Wuhan Easydiagnosis Biomedicine Co., Ltd | SARS-CoV-2 nucleic acid test kit | Fluorescent RT-PCR based | Used to qualitatively detect the new coronavirus 2019-nCoV infection in suspects by analysing ORF1ab gene and N gene | NR | Oropharyngeal swabs, nasopharyngeal swabs and sputum samples from patients diagnosed or differentiated | China-FDA EUA; CE-IVD | NR | http://www.mdeasydiagnosis.com/products/InternationalTrade/88.html |
| 19 | Xiamen Zeesan Biotech Co., Ltd. | SARS-CoV-2 Test Kit | RT-PCR based | In vitro qualitative detection of novel coronavirus (SARS-CoV-2) ORF1ab and N gene in samples | NR | Sample includes bronchoalveolar lavage fluid and sputum, and stool sample can be anal swab. |
Manual; lab-based; CE-IVD |
NR | http://www.zeesandx.com/products/sars-cov-2-test-kit-real-time-pcr.html |
| (B) Point-of-Care Immunodiagnostics: | |||||||||
| Sr. No |
Developer |
Commercial Brand Name |
Analytical Method |
Technical Details |
Detection Time |
Clinical Sample |
Status |
Limit of Detection / Detection Ability |
Links |
| 1 | BioMedomics, Inc. | COVID-19 IgM-IgG Dual Antibody Rapid Test | Qualitatively detect IgG and IgM antibodies | This test detects both early and late marker, of corona virus associated IgM/IgG antibodies in human | 10-15 min / test | Whole blood, serum, or plasma | EU (CE Mark), USA (EUA) | NR | https://www.biomedomics.com/products/infectious-disease/covid-19-rt/ |
| 2 | Bioscience (Chongqing) Biotechnology Co., Ltd. | Novel Coronavirus (2019-nCoV) IgM Antibody Detection Kit Novel Coronavirus (2019-nCoV) IgG Antibody Test Kit |
Chemiluminescent immunoassay technology | Identifies IgM/IgG antibodies using magnetic particle chemiluminescence method |
NR | Human clinical blood samples | China (EUA) | NR | https://www.medica-tradefair.com/vis/v1/en/exhibitors/medcom2019.2643477 |
| 3 | Biomerica | 10 Minute Test for COVID-19 Virus | Rapid POC IgM/IgG antibody test | Lateral flow immunoassay | 10 min | Whole blood, sera or plasma | Commenced shipping samples; seeking FDA EUA approval | NR | http://biomerica.com/news/news_view.asp?NewsItem=249 |
| 4 | Guangzhou Wondfo Biotech (Guangzhou, China) | Finecare SARS-CoV-2 Antibody Test | Antibody test | Lateral flow immunoassay that detects IgM and IgG antibodies directed against SARS-CoV-2 | 15 min | Serum or Plasma sample | National Medical Products Administration EUA in China; CE mark in Europe | NR | https://en.wondfo.com.cn/covid-19-3/ |
| 5 | Innovita Biological Technology | 2019-nCoV Ab Test (Colloidal Gold) | IgM/IgG combine detection | Lateral flow immunoassay that detects IgM and IgG antibodies directed against SARS-CoV-2 | 10 min | Human serum, Plasma, venous whole blood | China-FDA EUA; CE-IVD | NR | http://www.innovita.com.cn/index.html |
| 6 | InTec PRODUCTS, INC. | Rapid SARS-CoV-2 Antibody (IgM/IgG) Test | IgM and IgG antibody detection | Rapid immunoassay for the qualitative detection of IgM and IgG antibodies | NR | Single drop of fingerpick whole blood | EU, CE-IVD | NR | https://www.intecasi.com/news/156-intec-products-rapid-sars-cov-2-antibody-test-is-now-available |
| 7 | Snibe Diagnostic | MAGLUMI 2019-nCoV IgM/IgG (CLIA) | MAGLUMI 2019-nCoV IgM/IgG kit | Automated central laboratory rapid test that runs on MAGLUMI chemiluminescence immunoassay system | 30 min | Blood sample | automated IA, CE-IVD | NR | http://www.snibe.com/zh_en/en_newsView.aspx?id=576 |
| 8 | Sugentech | COVID-19 IgM/IgG | SGTi-flex COVID-19 IgM/IgG | Lateral flow immunoassay that detects IgM and IgG antibodies directed against SARS-CoV-2 | 10 min | Human whole blood (finger prick or venous), serum or plasma | CE-IVD | NR | http://sugentech.com/products/products-view.php?ct=7&target=32 |
| 9 | VivaChek Biotech (Hangzhou) Co., Ltd | VivaDiag™ | SARS-CoV-2 IgM/IgG Rapid Test (Colloid Gold) | In vitro qualitative assay to detects IgM and IgG antibodies against SARS-CoV-2 | 15 min | Whole blood (fingertip/venous), serum or plasma | CE-IVD | NR | https://www.vivachek.com/vivachek/English/prods/prod-rapidtest.html |
| 10 | Xiamen AmonMed Biotechnology | COVID-19 IgM/IgG test kit (Colloidal gold) | Antibody detection | Lateral flow immunoassay that detects IgM and IgG antibodies directed against SARS-CoV-2 and Rare Earth Nano Fluorescence Immunochromatography | 10 min | Human blood sample | EU (CE Mark) | NR | http://aode.juejinvr.cn/News/index |
| 11 | Xiamen Biotime Biotechnology Co., Ltd. | SARS-CoV-2 IgG/IgM Rapid Qualitative Test Kit | IgG/IgM combine detection | Rapid Qualitative analysis for IgM and IgG antibodies | 15 min | Serum, plasma, whole blood | EU, CE-IVD | NR | http://www.biotime.cn/En |
| 12 | Zhejiang Orient Gene Biotech | COVID-19 IgG/IgM Rapid Test kit | COVID-19 IgG/IgM Rapid Test | Solid-phase immunochromatographic assay | 2-10 min | Serum/plasma or whole blood specimens | US sublicensed distribution rights from L.B. Resources (Hong Kong) | NR | https://stocknewsnow.com/companynews/5035338834942348/AYTU/101843 |
| 13 | Quidel Corporation | Sofia 2 SARS Antigen FIA | SARS-CoV-2 Antigen detection | Immunofluorescence-based lateral flow technology in a sandwich design for qualitative detection of nucleocapsid protein from SARS-CoV-2 | 15 min | Nasal swab, nasopharyngeal swab | FDA EUA | NR | https://www.quidel.com/immunoassays/rapid-sars-tests/sofia-2-sars-antigen-fia |
| 14 | Avacta Group plc | Affimer® biotherapeutics and reagents | COVID-19 antigen detection | COVID-19 antigen test method using Adeptrix’s proprietary bead-assisted mass spectrometry (BAMS™) platform | NR | Saliva, nasopharyngeal swabs or serum | CE (Europe), FDA (USA) | NR | https://avacta.com/avacta-ships-sars-cov-2-affimer-reagents-to-cytiva-and-adeptrix/?utm_content=129011567&utm_medium=social&utm_source=twitter&hss_channel=tw-2279270671 |
| 15 | Coris Bio Concept | CORIS COVID-19 Ag Respi-Strip | COVID-19 antigen detection | Immunochromatography assay based on a membrane technology with colloidal gold nanoparticles fabricated with monoclonal antibodies directed against the conserved nucleoprotein antigen of SARS-CoV-2 | 15 min | Nasopharyngeal specimens | EU, CE-IVD | 1,25 X104 pfu/ml | https://www.intermed.be/en/professional-products/laboratory-diagnostics/serology-amp-virology/rapid-test/coris-covid-19-ag-respi-strip.html |
| (C) Digital radiographical analysis / Sensing system: | |||||||||
| Sr. No |
Developer |
Commercial Brand Name |
Analytical Method |
Technical Details |
Detection Time |
Clinical Sample |
Status |
Limit of Detection / Detection Ability |
Links |
| 1 | Beijing Infervision Technology Co. Ltd | InferRead CT Pneumonia | AI-based system to detect coronavirus infection via CT scans | Imaging assisted-diagnosis for multiple body areas, but not limited to brain, lung, skeleton, and bone | >20 sec | Physical presence of suspect or patient | Validated | NR | https://global.infervision.com/ |
| 2 | Canary Health Technologies | AiroStotleCV19 | Path Sensors technology which is exclusively licensed from MIT-Lincoln Laboratory, offering cutting-edge pathogen detection capabilities to a variety of industries including food safety, agriculture, and bioterrorism. | Three instrumentation platforms based PathSensors for easy and rapid detection. | > 5 min | Environmental swabs and air monitoring in sensitive spaces such as hospitals, offices, food services, etc | Proof-of-concept | NR | https://news.thomasnet.com/fullstory/new-canary-biosensor-generates-test-results-in-less-than-5-minutes-40034701 |
List of abbreviations: - NR: Not Reported; RT-PCR: Reverse Transcriptase-Polymerase Chain Reaction / qPCR: qualitative Polymerase Chain Reaction; RUO: Research Use Only; CE – IVD: Conformité Européenne (CE certification)-in vitro diagnostics; EUA: Emergency Use Authorization / EU – Emergency Use; FDA: Food and Drug Administration; HSA: Health and Safety Authority / Health and Sciences Authority.
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR): The capacity and turnaround statistics are limiting the RT-PCR application in clinical laboratories in this alarming situation. Hence, new approaches for precise and quick detection are important to be developed. CRISPR, in the field of genomic engineering is like a genome programming system that can be programmed to target oligonucleotides having specific genetic code to edit DNA and RNA at precise locations or to use as a diagnostic tool. CRISPR-based diagnosis offers rapid, inexpensive, and sensitive nucleic acid detection that may assist in Point-Of-Care (POC) virus detection, genotyping, and disease monitoring (Gootenberg et al., 2017). Initially, after the COVID-19 outbreak, Sherlock Biosciences Inc. and Mammoth Biosciences have represented their unique CRISPR-Cas13 based proof of concept analytical model for enhanced SARS-CoV-2 diagnostics (Kellner et al., 2019). Recently, the US Food and Drug Administration (FDA) has granted its first Emergency Use Authorization (EUA) certification to Sherlock Biosciences for this CRISPR-based diagnostic kit (Web Reference 5, May 2020). The Specific High-Sensitivity Enzymatic Reporter UnLOCKing (SHERLOCK) platform was first introduced by the team lead by Feng Zhang for highly specific molecular detection of Zika and Dengue viruses, distinguishing pathogenic bacteria, etc. (Gootenberg et al., 2017). The RNA-guided, RNA-targeting CRISPR effector Cas13a enzyme exhibits a “collateral effect” of uninhibited ribonuclease activity upon target recognition. The group has combined the collateral effect of Cas13a with isothermal amplification to establish a CRISPR-based diagnostic (CRISPR-Dx), providing rapid DNA or RNA detection with attomolar sensitivity and single-base mismatch specificity. With a similar approach, a few numbers of laboratories have also developed SARS-CoV-2 diagnostic tests based on CRISPR with less assay time and promising LOD range. Broughton et al. have developed a rapid easy-to-implement and precise CRISPR–Cas12-based lateral flow assay to detect SARS-CoV-2 from respiratory swab RNA extracts, called SARS-CoV-2 DNA Endonuclease-Targeted CRISPR Trans Reporter (DETECTR) (Broughton et al., 2020). This evaluation performs synchronized reverse transcription and isothermal amplification using loop-mediated amplification (RT-LAMP) for RNA extracted from NP or OP swabs in Universal Transport Medium (UTM). This was followed by Cas12 detection of testified coronavirus sequences, after which cleavage of a reporter molecule confirms the presence of the virus in the patient's sample. The assay time as reported is within 30–40 min with LOD of 10 copies/μl input. Their CRISPR DETECTR-based assay claims to provide a visual and faster alternative to the real-time RT-PCR assay, with a 95% positive predictive agreement and 100% negative predictive agreement. Curti et al. also reported a very similar diagnostic technology in their study to detect the SARS-CoV-2 sequence based on CRISPR-Cas 12 (Curti et al., 2020). The detection time and the LOD reported is 30 min and 10 copies/μl, respectively. In another such study, scientists have recently reported a FnCas9 Editor Linked Uniform Detection Assay (FELUDA) CRISPR method, that uses FnCas9, a protein that is highly sensitive to detect the mismatched region, and its position within the reverse transcribed DNA and the positions of these mismatches (Azhar et al., 2020). For the detection of SARS-CoV-2 nucleic acids, the target sequence is amplified by PCR using biotinylated primers, which are then immobilized on streptavidin-coated beads. Further, the fluorescently labeled Cas9 complexes containing single guide RNA interacted with the immobilized target sequences, generating analytical signals. They also improvised this approach to develop a lateral flow strips that contain streptavidin which can bind to the biotinylated targets, offering a LOD of 110 fM for SARS-CoV-2 diagnosis.
3. Point-of-care immunodiagnostics
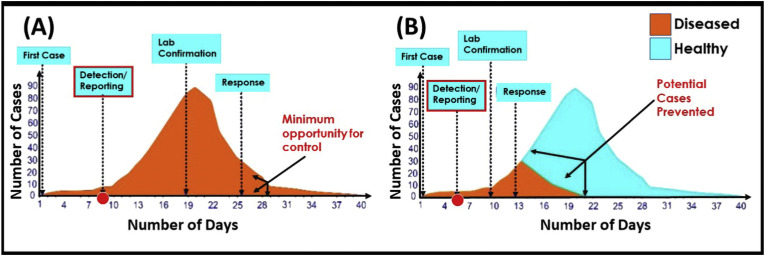
The POC devices have a tremendous role in the diagnosis of various infectious and non-infectious diseases over the years (Chandra, 2016; Mahato et al., 2018a, Mahato et al., 2018b). The basic immunoassays have the potential to provide clinical information on viral exposure along with the diagnostic evidence. According to WHO, the rapid detection method can cut-short the epidemic graph as the early reporting will prevent the outbreak as shown in Fig. 2 .
Fig. 2.
The impact of the rapid detection of infectious diseases in controlling and preventing an outbreak: (A) Impact of delayed diagnosis decreases the opportunity of controlling diseases; (B) the impact of early diagnosis increases the chances of disease prevention. (reproduced with permission from (Nguyen et al., 2020)).
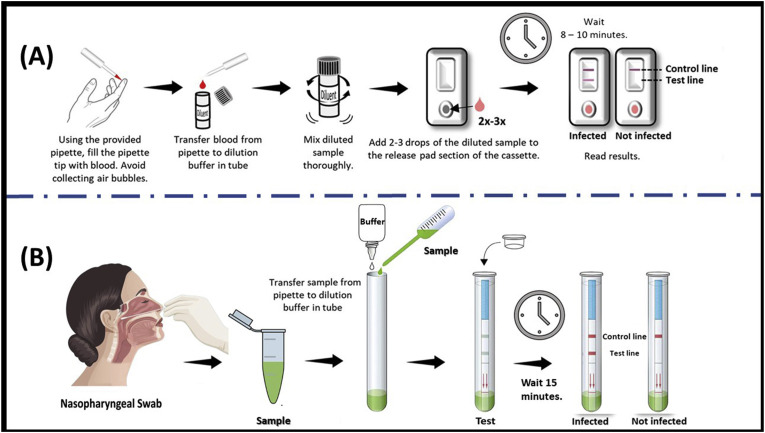
Analysis of the immunocomplexation between monoclonal antibodies (mAbs) i.e. IgG or IgM and SARS-CoV-2 antigens is a very useful method for diagnosing the disease (Li et al., 2020). The lateral-flow assay-based kit involves a dipstick covered in a cartridge containing the capture reagents (Mahato et al., 2020), where either IgM or IgG mAb are immobilized on a nitrocellulose membrane which is directed towards a viral antigen recognized by patients-antibodies separately onto a control line and a test line. In the middle of the strip, the immunoglobulins and SARS-CoV-2 specific protein are coupled, respectively with colloidal gold-nanoparticles, which serve as a detector. At the bottom, there is a well for dropping the samples. While performing the clinical testing, the patient's serum sample and dilution buffer are mixed and added to the sample well. When the sample contains specific antibodies to SARS-CoV-2, they will react with the antigen-gold nanoparticle complex. When the samples cross along the membrane length, a coloured complex of antibodies-antigens-gold nanoparticles will form on the test line containing the specific antigen. The red signal will gradually appear on the test line and become visible by the naked eye. A positive result signifies two parallel lines; where the upper one is the control line, which shows that the device works fine and the lower one is for the test line, which indicates that the serum sample contains SARS-CoV-2 antibodies. In the case of a negative result, the only red color will be seen on the control (upper) line. If red colour is found only on the test line or no lines are visible, the test is invalid. The overall procedure and data interpretation of such immunodiagnostic assay is shown in Fig. 3 (A). In such type of an immunoassay, a few drops of blood after a pinprick is enough to detect a virus in miniaturized settings qualitatively, however it is not possible to know the exact titer using this method.
Fig. 3.
Standard operating procedure of SARS-CoV-2 immunodiagnostics kit, (A) The qualitative detection of IgM or IgG antibodies in human serum, whole blood, or finger prick samples (RayBiotech Life, Georgia); (B) The qualitative detection procedure of viral antigen from nasopharyngeal swab sample (reproduced with permission from Mertens et al., 2020).
Such immunodiagnostic kits are portable, direct, easy to use, and can give preliminary results in less than an hour even in the clinical settings (Kumar et al., 2019; Mahato et al., 2018a). The rapid detection potential and adaptability of immunoassays established them as one of the major tests for SARS-CoV-2 diagnosis which is very much evident due to its large manufacturing by various commercial companies across the globe (Web Reference 6, Mar. 2020). Other types of immunoassays, especially the ones based on paper matrix also have several limitations in terms of sensitivity and complications in paper matrix design, optimization, and retaining the biological activity of proteins on the paper matrix (Mahato et al., 2017). Since these immunoassays detect a patient's antibodies to a pathogen, it will unavoidably struggle with the inherent inconsistency of the polyclonal antibody's responses. However, it is reported that tests for the detection of polyclonal antibodies against viral infection (Yurkina et al., 2001) in patients can more rapidly be developed than tests to detect the virus itself, as it only integrates immobilized recombinant antigens which is easier to produce than the mAb. Moreover, it also offers the same levels of accuracy to the mAb-based test kits. During the last few weeks, several companies and institutes have developed various POC immunodiagnostic devices for diagnosing SARS-CoV-2 infection. In Table 1(B) we have discussed in detail the manufacturers of COVID-19 immunodiagnostic kits, their working principle, analytical performance, assay time, etc. The FDA in their latest issue approved the first EUA for a COVID-19 antigen test to the Quidel Corporation for the Sofia 2 SARS Antigen Fluorescent Immunoassay (FIA), a novel testing platform to use in the ongoing pandemic (Web Reference 7, May 2020). This diagnostic platform can detect fragments of proteins found on or within the virus rapidly from the patient's nasal swabs samples. This test is authorized for use in high and moderate complexity laboratories certified by Clinical Laboratory Improvement Amendments (CLIA), as well as for POC testing by facilities operating under a CLIA Certificate of Waiver. The Sofia 2 SARS FIA uses advanced immunofluorescence-based lateral flow technology in a sandwich design for the qualitative detection of N protein from SARS-CoV-2. This immunodiagnostic technology is reported to provide automated as well as unbiased results in merely a few minutes. Another antigen testing technology has been developed by Mertens et al., where they have evaluated the diagnostic efficacy and operational utility of a disposable rapid antigen test to detect SARS-CoV-2 in NP clinical specimens, which is now available worldwide (Mertens et al., 2020). The COVID-19 antigen Respi-Strip follows the same working principle based on lateral flow assays for the detection of viruses in clinical samples as shown in Fig. 3(B). This kit-based test has reported showing a very decent specificity of 99.5%. The detection time was found to be 15 min with the LOD value of 250 pg/mL using this platform. Similarly, Affimer® (Avacta Life Sciences Limited) also developed a POC saliva-based COVID-19 antigen test strip in collaboration with Cytiva, which is pending for the clinical validation (Web Reference 8, May 2020). The role of the COVID-19 antigen strip is complementary to the currently used molecular techniques, though it is an effective and reliable alternative.
Avacta Life Sciences Limited with Adeptrix Corp. also developed a high throughput COVID-19 antigen screening method using Bead-Assisted Mass Spectrometry (BAMS™) platform (Adeptrix's trademarked). The novel BAMS platform provides higher sensitivity to the sample which improves the overall diagnosis of COVID-19. It is predicted that hundreds of samples per day can be analyzed by a single technician using BAMS, beyond the capacity of a single PCR machine, thus contributing significantly to the increase in a global testing capacity. Avacta's newly developed Affimer reagents strongly interact with the SARS-COV-2 spike protein, eventually assisting in enhanced the capturing of the virus particle present in various clinical samples. (Web Reference 8, May 2020).
4. Digital radiographical analysis/sensing system
The initial diagnosis of COVID-19 is based on the indications similar to pneumonia with common indications of dry cough, fever, fatigue, myalgia, and dyspnoea (Cascella et al., 2020). Thus, digitalized chest imaging can play a significant role in the assessment of the disease extent and its consequences as well (Kong and Agarwal, 2020). Digital radiography on patients stereotypically shows opaque cavity-like patch or diffused dissimilar airspace, comparable to the other pre-reported coronavirus types of pneumonia (SARS and MERS-CoV). A test conducted on COVID-19 patients showed that 40 out of 41 patients have bilateral-lung involvement in their chest computerized tomography (CT) scan. Many affected patients diagnosed with the disease also show a typical Ground-Glass Opacity (GGO) appearance of the lungs (Shi et al., 2020). This is in accordance with a more recent study, wherein the digitalized chest-image on an affected family of seven members showed a very similar bilateral patchy GGO pattern of the lungs, the pattern being more pronounced in the elderly members (Qiu et al., 2020a, Qiu et al., 2020b). Similarly, in another study of the initial chest-CT over COVID-19 affected patients, 86% of them were found with abnormal testimony, and 16 out of 18 patients had been reported with bilateral lung involvement (Shi et al., 2020). Furthermore, in a study by Chung et al. 71% of patients had shown the contribution of more than two lobes at chest CT, among them 57% had shown GGO pattern, 33% had shown opacities with rounded morphology, another 33% had shown the peripheral distribution of disease. Additionally, 29% had shown consolidation along with the GGO pattern, and 19% presented with a crazy-paving pattern in the lung (Chung et al., 2020).
Though COVID-19 has indistinguishable similarities compared to the previously reported CT imaging for MERS and SARS-CoV; the initial chest-CT imaging abnormalities in MERS and SARS were more frequent (Hosseiny et al., 2020). In Table 2 the initial observations of the prevalent CT findings are summarized (Wang et al., 2020a, Wang et al., 2020b). Patients with COVID-19 have also reported to have cavitation or lymphadenopathy. Only 1 Out of 99 in confirmed COVID-19 patients was reported having pneumothorax, so it is still uncertain if the pneumothorax is considered a direct impediment of the coronavirus infection (Kong and Agarwal, 2020). In another study, a report of five individuals with confirmed COVID-19 from the swab-test, showed negative results from early CT findings for the diagnosis of the virus (Sun et al., 2020). Though initial reports specified that initial-imaging might confirm normal findings, as low as only 15% of individuals seem to abide by that. However, recent reports have shown that the presence of typical CT-image findings could be helpful for initial screening of the virus in suspected individuals (Li and Xia, 2020). The knowledge acquired from MERS and SARS showed that the follow-up imaging system might be understandable in individuals recovering from COVID-19 for proper indications of long-lasting engrossment of the lungs including interlobular thickening, air-trapping, or fibrosis. In Table 1 (C) highlights the common clinical observations after digital imaging of the lungs of affected patients, furthermore calling attention to the role of radiologists in minimizing the risk of spreading of the pandemic.
Table 2.
Initial CT Findings of patients with Coronavirus Disease (COVID-19) (Wang et al., 2020a,b).
| Features | Observation |
|---|---|
| Common abnormalities | Peripheral multifocal airspace opacities (GGO, consolidation, or both) on chest radiography and CT |
| Rare findings | Pneumothorax |
| Note reported | Cavitation or lymphadenopathy |
| Appearance | Bilateral, multifocal, basal airspace; normal chest radiography findings (15%) |
| Follow-up imaging appearance | Persistent or progressive airspace opacities |
| Indication of poor prognosis | Consolidation (vs GGO) |
| Chronic phase | Unknown, but pleural effusion and interlobar septal thickening have not yet been reported |
| Fibrosis | Not yet reported |
5. Nanobio engineered sensing prototypes
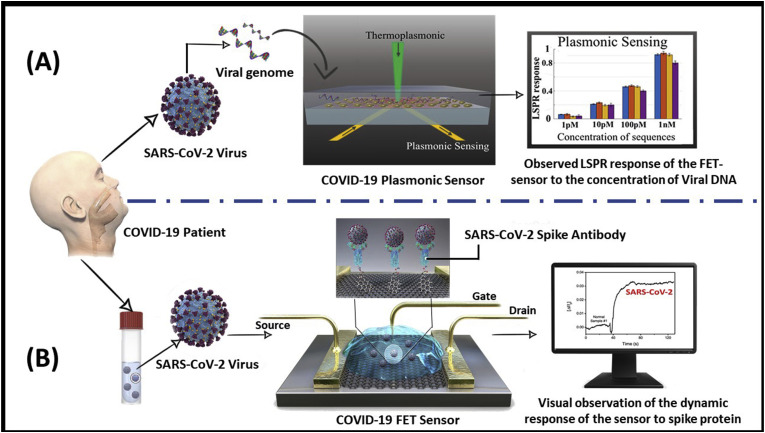
Besides the clinically practiced diagnostic methods which are currently implemented in hospitals, several new nanobio engineered biosensing prototypes have been developed recently for diagnosis of COVID-19. In a most recent work, Qiu et al. have developed a dual-functional plasmonic sensor having high sensitivity, rapidity, and reliable diagnostic capability for the SARS-CoV-2 virus detection (Qiu et al., 2020a, Qiu et al., 2020b). This dual-functional biosensor was integrated with the plasmonic photothermal (PPT) effect and localized surface plasmon resonance (LSPR) sensing transduction on a single cost-effective two-dimensional gold nanoislands (AuNI) chip. Two different angles of incidence have been used to excite the plasmonic resonances of PPT and LSPR at two different wavelengths, which suggestively enhanced the sensing stability, sensitivity, and reliability (Fig. 4 (A)). With this configuration, the LSPR sensing unit attained a real-time and label-free detection of viral sequences including RdRp, ORF1ab, and the E genes from the SARS-CoV-2. Additionally, the in situ PPT enhancement on the AuNI chips suggestively enhanced the hybridization kinetics and the specificity of nucleic acid detection. It predicts the sequences similarity of RdRp genes from SARS-CoV and SARS-CoV-2 and can precisely distinguish between them through the in situ PPT enhancement. The biosensor exhibits a high sensitivity toward the selected SARS-CoV-2 sequences with a LOD value down to the concentration of 0.22 pM and promises a precise detection of the target in a multigene mixture as well. Thus, under the outbreak background of COVID-19, this dual-functional LSPR biosensor may provide a reliable and easy-to-implement diagnosis platform, improving the diagnostic accuracy over the conventional clinical tests and the time-consuming PCR analysis. In another recent study, Seo et al., have developed a graphene-based field-effect transistor (FET) biosensing device, functionalized with the SARS-CoV-2 spike antibody for the detection of a spike protein antigen (Seo et al., 2020). FET-based biosensors are measured to be potentially beneficial in clinical diagnosis, POC testing, and on-site detection (Liu et al., 2019). To develop such a sensing platform, the SARS-CoV-2 spike antibody was immobilized on the fabricated device using 1-pyrenebutyric acid N-hydroxysuccinimide ester bioconjugation system (Fig. 4(B)). The sensor claims to detect the target spike antigen with a LOD of 1 fg/mL with high specificity. The study confirmed its potential against the cultured SARS-CoV-2 virus as well as in patient samples, indicating its tremendous clinical possibilities. Interestingly, it has also been testified that the designed sensor could distinguish the SARS-CoV-2 antigen protein from MERS-CoV. The developed plasmonic biosensing (Qiu et al., 2020a, Qiu et al., 2020b) and FET-based biosensing (Seo et al., 2020) devices have revealed their advantages for rapid, real-time, and label-free probing of biologically relevant analytes, where they have potently detected target molecules at ultralow concentrations and produce compact devices for POC analysis.
Fig. 4.
Schematic representation of the newly developed biosensor prototypes (A) The dual-functional plasmonic biosensor combining the plasmonic photothermal effect and localized surface plasmon resonance sensing transduction for the clinical COVID-19 diagnosis (reproduced with permission from Qiu et al., 2020a, Qiu et al., 2020b); (B) COVID-19 FET sensor operation technique, where graphene is selected as a sensing material, and SARS-CoV-2 spike antibody is conjugated onto the graphene sheet via an interfacing molecule as a probe linker (reproduced with permission from Seo et al., 2020).
6. Additional biochemical monitoring for COVID-19 diagnoses
Apart from the direct diagnosis of COVID-19 in patients, the means of biochemical monitoring is also essential for evaluating the presence of the disease, its severity, and to examine the disease status during therapeutic interventions. Some typical in vitro diagnostic tests (IVD) have been recommended recently for COVID-19 patients where levels of blood cells/various biomarkers have to be tested (Lippi et al., 2020a; 2020b; Ruan et al., 2020). These biochemical tests also provide significant analytical evidence of the COVID-19 disease burden. Table 3 presents a list of these clinical tests recommended based on recent literatures pointing out the major laboratory irregularities related to the adult COVID-19 patients along with the potential clinical observations. Besides these common laboratory tests, some recent studies also suggest that individuals with severe COVID-19 syndrome might be at risk for cytokine storm syndrome as well. Especially, tests for the IL-6 cytokine must be used for patients suffering from the severe viral inflammation (Lippi et al., 2020; Rodriguez-Morales et al., 2020). Significantly, the laboratory profiling of severe COVID-19 in pediatric patients is still not reported clearly because of its inconsistency with SARS. A publication endorses that clinical monitoring of the lymphocyte count, c-reactive protein, and procalcitonin-level may evaluate the severity of the viral infection. Similar to adults, it is suggested that IL-6 should also be inspected as a potent prognostic indicator for children as well (Henry et al., 2020).
Table 3.
List of laboratory test recommended for adult patients with common clinical indications associated with COVID-19 tests (Lippi and Plebani, 2020a, Lippi and Plebani, 2020b, Ruan et al., 2020).
| Sr. No | Recommended Laboratory Tests | Observed abnormalities in patient (associated to COVID-19 progression) | Clinical observations |
|---|---|---|---|
| 1 | Complete blood count | Elevated white blood cell Elevated neutrophil count Lowered lymphocyte count Lowered platelet count |
Bacteraemia Bacteraemia Loss of immune response Abnormal blood coagulation |
| 2 | Serum Albumin | Lowered | ill-fated liver function |
| 3 | Serum LDH (Lactate Dehydrogenase) | Elevated | Pulmonary injury and/or widespread organ damage |
| 4 | Serum glutamic pyruvic transaminase (SGPT) | Elevated | Extensive organ damage/ill-fated liver function |
| 5 | Serum glutamic oxaloacetic transaminase (SGOT) | Elevated | Extensive organ damage/ill-fated liver function |
| 6 | Total bilirubin | Elevated | Liver trauma |
| 7 | Creatinine | Elevated | Kidney trauma |
| 8 | Cardiac troponin | Elevated | Cardiac trauma |
| 9 | D-Dimer | Elevated | Abnormal blood coagulation, distributed coagulopathy |
| 10 | Prothrombin Time | Elevated | Abnormal blood coagulation, distributed coagulopathy |
| Procalcitonin | Elevated | Bacteraemia | |
| 11 | C-reactive protein | Elevated | Severe viral infection |
| 12 | Ferritin | Elevated | Serious inflammation |
| 13 | Cytokines (IL-6) | Elevated | Syndrome associated to “cytokine storm” |
7. Discussions
Following the shadows of the MERS-CoV and SARS-CoV, a recent successor of a similar progeny has recently wrecked the world immensely. What started as causative of ‘pneumonia of unknown etiology’ in the Wuhan province of China in December 2019 was named as “Severe Acute Respiratory Syndrome Coronavirus 2” (SARS-CoV-2) by the WHO (Cascella et al., 2020). Since to date a definite treatment modality or vaccine for SARS-CoV-2 does not exist, only laboratory testing and prompt diagnosis play great roles in containing this highly infectious disease (Fig. 5 ). Naturally, the sensitivity and specificity of these methodologies are very important. RT-PCR is a time-tested method used for COVID-19 detection wherein RNA collected from the swab specimens is amplified. The probe used during this process anneals to a particular sequence. The LOD for these tests shows a positive result of 95% detection even in the lowest concentration of SARS-CoV-2 (Web Reference 9, 2020). The drawback of this testing method, however, is its long processing steps for results and lack of correlation with viral load, especially keeping in mind the rapidity with which the numbers of cases are increasing. Alternative methodologies for prompt testing are hence required, where the immunosensing kits have started to play a significant role. These kits work on the detection of IgM and IgG antibodies from sputum, plasma, or blood specimens and the results are delivered as merely visual interpretations. Consisting of a SARS-CoV-2 antigen and an antibody marker, two detection lines along with a quality control line having fixed anti-IgM and IgG antibody can detect higher levels of coronavirus IgG & IgM antibody. Again, the question looms large on the cent-percent specificity of these tests as compared to the conventional methodology because varied results have been employed with these test kits. A test conducted by collecting the blood from the veins from several Chinese CDC hospitals revealed that out of 397 positive cases (RT-PCR tested) 352 tested positive resulting in a sensitivity of 88.66% (Li et al., 2020). Hence, a correlation of clinical symptoms and other serological data is a must before resorting to this test method as confirmatory (Lee et al., 2020). A need for a more accurate method of testing has led to researchers vouching for CRISPR-based SHERLOCK (Specific High Sensitivity Enzymatic Reporter UnLOCKing) method for the detection of COVID-19. A study by Zhang et al. on the synthetic strain of SARS-CoV-2 shows detection of the COVID-19 target sequence within the range of 20 and 200 aM (10–100 copies per microliter of input) (Zhang et al., 2020). Moreover, A clinical study by Ai et al. on a 56 years old woman who visited Shanghai from Wuhan confirmed the prompt use of CRISPR for the detection of SARS-CoV-2 (Ai et al., 2020). This method of rapid diagnosis can be a good alternative to PCR in laboratories that lack complex resources. Another significant role of diagnosis can be attributed to radiography and digital imaging, as SARS-CoV-2 manifests some typical radiographical findings. These radiographical findings hold clinical correlation in patients who routinely show dyspnoea, cough, and other symptoms of RTI (Respiratory Tract Infections). Just like MERS-CoV and SARS-CoV radiographical findings of opacities and diffused air space in the lungs are observed in the case of COVID-19. Moreover, certain digital radiographs increase the chances of detection of these symptoms to a higher success rate, leading to a quick diagnosis. A recent study by Li et al. showed a success rate of 96.1% in diagnosing COVID-19 via CT imaging (Li et al., 2020). The drawback here comes with the overlapping of some findings being increasingly similar to other RTI and adenovirus pathologies. Hence CT imaging is still limited to differentiating between close pathologies. Further technological advances following the increase in need of sensitive, rapid, and POC diagnostic tools have recently gifted biosensors which can provide flawless information over the conventional clinical tests in miniaturized settings. The studies done so far are highly specific toward SARS-CoV-2 and show a promisingly low LOD, which in turn assures to detect SARS-CoV-2 infection in patients at an early stage. Thus, these upcoming nanobio engineered sensing technologies can be used as one of the major adjuncts for the diagnosis of COVID-19.
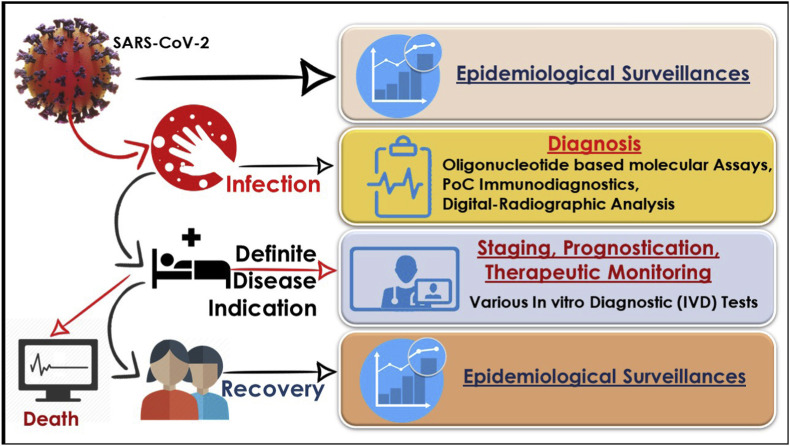
Fig. 5.
The critical role of epidemiological surveillance, diagnosis, therapeutic monitoring in various stages of SARS-CoV-2 infection-causing COVID-19.
8. Conclusions
This is the first report which describes the clinically practiced and commercially viable nanobio engineered approaches for COVID-19 diagnosis to the best of our knowledge. It covers minute details on RT-PCR, immunodiagnostic assays, radiographical analysis/sensing systems, and novel biosensing technologies. This review not only focuses on the diagnostics presently available for SARS-CoV-2 but also highlights the upcoming products/technologies as well as the challenges in rapid diagnostics concerning the outbreak. The COVID-19 disease is reported to be instigated by coronavirus originating from animals. Though the biological, as well as the clinical presentations of the disease, is under extensive review. The characterizing gene reservoir of SARS-CoV-2 is still not confirmed, but few of them have undoubtedly been exploited as a potential genetic marker for the diagnosis of the disease. The emphasis on early diagnosis is now primarily focused on conventional RT-PCR based assays. Moreover, advances in biotechnology also established CRISPR based diagnosis concept which not only assures to increase and accelerate the detection of SARS-CoV-2 but also ensures to provide prompt treatment to the patients. Besides, the POC lateral-flow assay based on immobilized IgM/IgG antibodies, antigen-based detection kits have also established their potent application for serological diagnosis of SARS-CoV-2 in suspects and patients under treatments. The method due to its rapid detection property is now under the focus of all clinical trials. The digitalized visual quantitative analysis based on CT scanning has also shown higher consistency and diagnostic capability in the clinical classification. Astonishingly, the novel nanobiosensor prototypes have certainly delivered some promising results for COVID-19 detection with much lower LOD range in considerably less time. Thus, in upcoming days it is expected that these technologies could precisely determine the clinical severity of COVID-19 and will provide further improvised clinical information.
9. Future prospects
Concerning the time-consuming process of the PCR technique, new approaches are being reported in the urge of rapid and POC diagnosis. It is desired that SARS-CoV-2 patients will be seroconverted eventually in the upcoming phases; thus, serological tests may also not remain a practical approach. Several molecular tests focusing on non-PCR-based methods are thus proposed recently. This includes the Nucleic Acid sequence-based Amplification Technique (NAAT), loop-mediated isothermal amplification for the detection of SARS-CoV-2 RNA. The recombinant DNA technology may also offer the possible rapid POC screening for SARS-CoV-2. The POC or near-POC-NAAT platform-based techniques are reporting to provide prospects for the execution of automated self-contained trials in research laboratories with limited expertise, especially for endemic-prone zones. Moreover, the expansion of the test menu regarding the proposed platforms will promisingly support the in-country way of dealing with the endemic disease. There is also a necessity for more subtle and upgradable serological assays with minor costing and least cross-reactivity that can be used as observation tools. A more thorough understanding of SARS-CoV-2 viral and antibody kinetics is also desirable for a wide range of various sample types to optimize the usage of present assays and to focus-on ongoing technical challenges in the recognition of minor and asymptomatic contagions. In the future, integration of redox-cyclic approaches and signal amplification due to nanoengineered materials with COVID-19 immunosensing kit can also increase the sensitivity, selectivity, probability of the existing system. Furthermore, support for trial biobanks with well-characterized specimens-reference standards will simplify diagnostic progress and quality assurance for worldwide SARS-CoV-2 diagnostics.
CRediT authorship contribution statement
Supratim Mahapatra: Writing - original draft, Writing - review & editing. Pranjal Chandra: Conceptualization, Investigation, Writing - original draft, Writing - review & editing, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Dr. Pranjal Chandra thanks Prof. Pramod Kumar Jain, Director IIT(BHU) for encouragement and providing the necessary facility for completion of this work.
References
- Ai J.-W., Zhang Y., Zhang H.-C., Xu T., Zhang W.-H. Era of molecular diagnosis for pathogen identification of unexplained pneumonia, lessons to be learned. Emerg. Microb. Infect. 2020 doi: 10.1080/22221751.2020.1738905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azhar M., Phutela R., Ansari A.H., Sinha D., Sharma N., Kumar M., Aich M., Sharma S., Singhal K., Lad H., Patra P.K., Makharia G., Chandak G.R., Chakraborty D., Maiti S. Rapid, field-deployable nucleobase detection and identification using FnCas9. bioRxiv. 2020 doi: 10.1101/2020.04.07.028167. [DOI] [Google Scholar]
- Batlle D., Wysocki J., Satchell K. Soluble angiotensin-converting enzyme 2: a potential approach for coronavirus infection therapy? Clin. Sci. (Lond.) 2020 doi: 10.1042/CS20200163. [DOI] [PubMed] [Google Scholar]
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020 doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascella M., Rajnik M., Cuomo A., Dulebohn S.C., Di Napoli R. StatPearls; 2020. Features, Evaluation and Treatment Coronavirus (COVID-19) [PubMed] [Google Scholar]
- Chan J.F.-W., Yip C.C.-Y., To K.K.-W., Tang T.H.-C., Wong S.C.-Y., Leung K.-H., Fung A.Y.-F., Ng A.C.-K., Zou Z., Tsoi H.-W., Choi G.K.-Y., Tam A.R., Cheng V.C.-C., Chan K.-H., Tsang O.T.-Y., Yuen K.-Y. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-polymerase chain reaction assay validated in vitro and with clinical specimens. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00310-20. JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.W., Lau S.K.P., To K.K.W., Cheng V.C.C., Woo P.C.Y., Yue K.Y. Middle East Respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 2015 doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra P. 2016. Nanobiosensors for Personalized and Onsite Biomedical Diagnosis, Nanobiosensors for Personalized and Onsite Biomedical Diagnosis. [DOI] [Google Scholar]
- Chung M., Bernheim A., Mei X., Zhang N., Huang M., Zeng X., Cui J., Xu W., Yang Y., Fayad Z.A., Jacobi A., Li K., Li S., Shan H. CT imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2020 doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020 doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curti L., Pereyra-Bonnet F., Gimenez C. bioRxiv; 2020. An Ultrasensitive, Rapid, and Portable Coronavirus SARS-CoV-2 Sequence Detection Method Based on CRISPR-Cas12. [DOI] [Google Scholar]
- Freeman W.M., Walker S.J., Vrana K.E. Quantitative RT-PCR: pitfalls and potential. Biotechniques. 1999;26(1):112‐125. doi: 10.2144/99261rv01. [DOI] [PubMed] [Google Scholar]
- Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., Verdine V., Donghia N., Daringer N.M., Freije C.A., Myhrvold C., Bhattacharyya R.P., Livny J., Regev A., Koonin E.V., Hung D.T., Sabeti P.C., Collins J.J., Zhang F. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;80 doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Li T., Han M., Li X., Wu D., Xu Y. Diagnostic utility of clinical laboratory data determinations for patients with the severe COVID-19. J. Med. Virol. 2020 doi: 10.1002/jmv.25770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Ren L., Yang S., Xiao M., Chang D., Yang F., Dela Cruz C.S., Wang Y., Wu C., Xiao Y., Zhang L., Han L., Dang S., Xu Yan, Yang Q., Xu S., Zhu H., Xu Yingchun, Jin Q., Sharma L., Wang L., Wang J. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J.-M.K., Yoon-Seok C., Hye Jun J., Nam-Joo L., Mi Seon K., Sang Hee W., Sehee P., Jee Woong K., Heui Man K., Myung G. Identification of coronavirus isolated from a patient in korea with COVID-19. Osong Publ. Health Res. Perspect. 2020 doi: 10.24171/j.phrp.2020.11.1.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry B.M., Lippi G., Plebani M. Laboratory abnormalities in children with novel coronavirus disease 2019. Clin. Chem. Lab. Med. 2020 doi: 10.1515/cclm-2020-0272. [DOI] [PubMed] [Google Scholar]
- Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L.A., Patel A., Gerber S.I., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M.A., Weldon W.C., Biggs H.M., Uyeki T.M., Pillai S.K. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseiny M., Kooraki S., Gholamrezanezhad A., Reddy S., Myers L. Radiology perspective of coronavirus disease 2019 (COVID-19): lessons from severe acute respiratory syndrome and Middle East respiratory syndrome. AJR Am. J. Roentgenol. 2020 doi: 10.2214/AJR.20.22969. [DOI] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019 doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W., Agarwal P.P. Chest imaging appearance of COVID-19 infection. Radiol. Cardiothorac. Imag. 2020 doi: 10.1148/ryct.2020200028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Purohit B., Mahato K., Chandra P. RSC Detection Science. 2019. Chapter 11: advance engineered nanomaterials in point-of-care immunosensing for biomedical diagnostics. [DOI] [Google Scholar]
- Lan L., Xu D., Ye G., Xia C., Wang S., Li Y., Xu H. Positive RT-PCR test results in patients recovered from COVID-19. J. Am. Med. Assoc. 2020 doi: 10.1001/jama.2020.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N.-Y., Li C.-W., Tsai H.-P., Chen P.-L., Syue L.-S., Li M.-C., Tsai C.-S., Lo C.-L., Hsueh P.-R., Ko W.-C. A case of COVID-19 and pneumonia returning from Macau in Taiwan: clinical course and anti-SARS-CoV-2 IgG dynamic. J. Microbiol. Immunol. Infect. 2020 doi: 10.1016/j.jmii.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Xia L. Coronavirus disease 2019 (COVID-19): role of chest CT in diagnosis and management. AJR Am. J. Roentgenol. 2020 doi: 10.2214/AJR.20.22954. [DOI] [PubMed] [Google Scholar]
- Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., Sun R., Wang Y., Hu B., Chen W., Zhang Y., Wang J., Huang B., Lin Y., Yang J., Cai W., Wang X., Cheng J., Chen Z., Sun K., Pan W., Zhan Z., Chen L., Ye F. Development and clinical application of A rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020 doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippi G., Lavie C.J., Sanchis-Gomar F. Cardiac troponin I in patients with coronavirus disease 2019 (COVID-19): evidence from a meta-analysis. Prog. Cardiovasc. Dis. 2020 doi: 10.1016/j.pcad.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippi G., Plebani M. The critical role of laboratory medicine during coronavirus disease 2019 (COVID-19) and other viral outbreaks. Clin. Chem. Lab. Med. 2020 doi: 10.1515/cclm-2020-0240. [DOI] [PubMed] [Google Scholar]
- Lippi G., Plebani M. Laboratory abnormalities in patients with COVID-2019 infection. Clin. Chem. Lab. Med. 2020 doi: 10.1515/cclm-2020-0198. [DOI] [PubMed] [Google Scholar]
- Liu J., Chen X., Wang Q., Xiao M., Zhong D., Sun W., Zhang G., Zhang Z. Ultrasensitive monolayer MoS 2 field-effect transistor based DNA sensors for screening of down syndrome. Nano Lett. 2019 doi: 10.1021/acs.nanolett.8b03818. [DOI] [PubMed] [Google Scholar]
- Mahato K., Kumar S., Srivastava A., Maurya P.K., Singh R., Chandra P. Handbook of Immunoassay Technologies. 2018. Electrochemical immunosensors: fundamentals and applications in clinical diagnostics. [DOI] [Google Scholar]
- Mahato K., Maurya P.K., Chandra P. Fundamentals and commercial aspects of nanobiosensors in point-of-care clinical diagnostics. Biotechnology. 2018;3 doi: 10.1007/s13205-018-1148-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahato K., Purohit B., Kumar A., Chandra P. Comprehensive Analytical Chemistry. 2020. Paper-based biosensors for clinical and biomedical applications: emerging engineering concepts and challenges. [DOI] [Google Scholar]
- Mahato K., Srivastava A., Chandra P. Paper based diagnostics for personalized health care: emerging technologies and commercial aspects. Biosens. Bioelectron. 2017 doi: 10.1016/j.bios.2017.05.001. [DOI] [PubMed] [Google Scholar]
- Mertens Pascal, De Vos Nathalie, Delphine Martiny, Christian Jassoy, Ali Mirazimi, Lize Cuypers, Van den Wijngaert Sigi, Vanessa Monteil, Pierrette Melin, Karolien Stoffels, Yin Nicolas, Davide Mileto, Sabrina Delaunoy, Henri Magein, Katrien Lagrou, Bouze V.J. Development and potential usefulness of the COVID-19 Ag Respi-Strip diagnostic assay in a pandemic context. Front. Med. 2020;7:225. doi: 10.3389/fmed.2020.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T., Duong Bang D., Wolff A. 2019 Novel Coronavirus Disease (COVID-19): Paving the Road for Rapid Detection and Point-of-Care Diagnostics. Micromachines. 2020 doi: 10.3390/mi11030306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normile D. Singapore claims first use of antibody test to track coronavirus infections. Science (80) 2020 doi: 10.1126/science.abb4942. [DOI] [Google Scholar]
- OKBA N.M.A., Muller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M., Lamers M.M., Sikkema R.S., Bruin E. de, Chandler F.D., Yazdanpanah Y., Hingrat Q. Le, Descamps D., Houhou-Fidouh N., Reusken C.B.E.M., Bosch B.-J., Drosten C., Koopmans M.P.G., Haagmans B.L. medRxiv; 2020. SARS-CoV-2 Specific Antibody Responses in COVID-19 Patients. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020 doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- Qiu H., Wu J., Hong L., Luo Y., Song Q., Chen D. Clinical and epidemiological features of 36 children with coronavirus disease 2019 (COVID-19) in Zhejiang, China: an observational cohort study. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30198-5. [Published Online First: 2020/03/25] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio D.C. Reverse transcription–polymerase chain reaction. Cold Spring Harb. Protoc. 2014 doi: 10.1101/pdb.prot080887. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Morales A.J., Cardona-Ospina J.A., Gutiérrez-Ocampo E., Villamizar-Peña R., Holguin-Rivera Y., Escalera-Antezana J.P., Alvarado-Arnez L.E., Bonilla-Aldana D.K., Franco-Paredes C., Henao-Martinez A.F., Paniz-Mondolfi A., Lagos-Grisales G.J., Ramírez-Vallejo E., Suárez J.A., Zambrano L.I., Villamil-Gómez W.E., Balbin-Ramon G.J., Rabaan A.A., Harapan H., Dhama K., Nishiura H., Kataoka H., Ahmad T., Sah R. Clinical, laboratory and imaging features of COVID-19: a systematic review and meta-analysis. Trav. Med. Infect. Dis. 2020 doi: 10.1016/j.tmaid.2020.101623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Q., Yang K., Wang W., Jiang L., Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020 doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sah R., Rodriguez-Morales A.J., Jha R., Chu D.K.W., Gu H., Peiris M., Bastola A., Lal B.K., Ojha H.C., Rabaan A.A., Zambrano L.I., Costello A., Morita K., Pandey B.D., Poon L.L.M. Complete genome sequence of a 2019 novel coronavirus (SARS-CoV-2) strain isolated in Nepal. Microbiol. Resour. Announc. 2020 doi: 10.1128/mra.00169-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G., Kim S.-J., Lee J.-O., Kim B.T., Park E.C., Kim S. Il. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020 doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- Shi H., Han X., Jiang N., Cao Y., Alwalid O., Gu J., Fan Y., Zheng C. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhas Srinivasan, Cui Hongzhu, Gao Ziyang, Liu Ming, Lu Senbao, Mkandawire Winnie, Narykov Oleksandr, Mo Sun D.K. Structural genomics of SARS-CoV-2 indicates. Viruses. 2020;12 doi: 10.3390/v12040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Xu X., Xie J., Li J., Huang X. Evolution of computed tomography manifestations in five patients who recovered from coronavirus disease 2019 (COVID-19) pneumonia. Korean J. Radiol. 2020 doi: 10.3348/kjr.2020.0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Wu C., Li X., Song Y., Yao X., Wu X., Duan Y., Zhang H., Wang Y., Qian Z., Cui J., Lu J. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 2020 doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velavan T.P., Meyer C.G. The COVID-19 epidemic. Trop. Med. Int. Health. 2020 doi: 10.1111/tmi.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Liu Z., Chen Z., Huang X., Xu M., He T., Zhang Z. The establishment of reference sequence for SARS‐CoV‐2 and variation analysis. J. Med. Virol. 2020 doi: 10.1002/jmv.25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Dong C., Hu Y., Li C., Ren Q., Zhang X., Shi H., Zhou M. Temporal changes of CT findings in 90 patients with COVID-19 pneumonia: a longitudinal study. Radiology. 2020 doi: 10.1148/radiol.2020200843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Web Reference 1 COVID-19 coronavirus pandemic. https://www.worldometers.info/coronavirus/
- Web Reference 2 Estimating the global growth of coronavirus. https://towardsdatascience.com/estimating-the-global-growth-of-coronavirus-33ca3c28f066
- Web Reference 3 Coronavirus and the race to distribute reliable diagnostics. https://www.nature.com/articles/d41587-020-00002-2 [DOI] [PubMed]
- Web Reference 4 Shipping of CDC 2019 novel coronavirus diagnostic test kits begins. https://www.cdc.gov/media/releases/2020/p0206-coronavirus-diagnostic-test-kits.html
- Web Reference 5 First CRISPR test for the coronavirus approved in the United States. https://www.nature.com/articles/d41586-020-01402-9?fbclid=IwAR1gTF4e5L9tZ0_lFS1uyAbGngbVsrmkIfR49CKf2GdeEJ8l-chJkiKS_f0#ref-CR1 [DOI] [PubMed]
- Web Reference 6: Fast, portable tests come online to curb coronavirus pandemic, https://www.nature.com/articles/d41587-020-00010-2. [DOI] [PubMed]
- Web Reference 7 Coronavirus (COVID-19) update: FDA authorizes first antigen test to help in the rapid detection of the virus that causes COVID-19 in patients. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-first-antigen-test-help-rapid-detection-virus-causes
- Web Reference 8 Avacta ships SARS-COV-2 affimer reagents to Cytiva and Adeptrix. https://www.businesswire.com/news/home/20200511005754/en/Avacta-Ships-SARS-COV-2-Affimer-Reagents-Cytiva-Adeptrix
- Web Reference 9 . 2020. Accelerated Emergency Use Authorization (EUA) Summary COVID-19 RT-PCR Test (Laboratory Corporation of America) FDA.https://www.fda.gov/media/136151/download [Google Scholar]
- World Health Organization . WHO Publ; 2014. The Evolving Threat of Antimicrobial Resistance: Options for Action. [Google Scholar]
- Yurkina E.A., Ofitserov V.I., Samukov V.V., Sabirov A.N., Mizenko G.A. In: Antibodies to Distinct Protein Epitopes from a Single Polyclonal Serum BT - Peptides: the Wave of the Future: Proceedings of the Second International and the Seventeenth American Peptide Symposium, June 9–14, 2001, San Diego, California, U.S.A. Lebl M., Houghten R.A., editors. Springer Netherlands; Dordrecht: 2001. pp. 994–995. [DOI] [Google Scholar]
- Zhang F., Abudayyeh O.O., Gootenberg J.S., Sciences C., Mathers L. 2020. A Protocol for Detection of COVID-19 Using CRISPR Diagnostics 1–8. [Google Scholar]
- Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020 doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J., Yin J., Fang L., Yang M., Wang T., Wu W., Zhang P. 2020. Computational Prediction of Mutational Effects on the SARS-CoV-2 Binding by Relative Free Energy Calculations. [DOI] [PubMed] [Google Scholar]