Abstract
Real-time reverse transcription PCR is currently the most sensitive method to detect severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Defining whether a patient could be contagious or not contagious in the presence of residual SARS-CoV-2 RNA is of extreme importance in the context of public health. In this prospective multicenter study, virus isolation was prospectively attempted in 387 nasal swabs from clinically recovered patients showing low viral load (quantification cycle, Cq, value greater than 30). The median Cq value was 36.8 (range 30.0–39.4). Overall, a cytopathic effect was detected in nine samples, corresponding to a culture positivity rate of 2.3% (9/387). The results of this study help to dissect true virus replication and residual viral RNA detection in recovered patients.
Keywords: SARS-CoV-2, COVID-19, Virus isolation, Real-time reverse transcription PCR, Cq value, Infectivity
Introduction
On February 20, 2020, Lombardy, a region in northern Italy, was struck by an outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Soon afterward, the epidemic involved other Italian regions, with a north-south gradient. Several containment measures were adopted, including lockdown of affected areas, social distancing, and quarantining of individuals with laboratory-confirmed COVID-19 as well as their close contacts. Laboratory confirmation of SARS-CoV-2 infection relied on positivity of a nasopharyngeal swab on virus-specific real-time reverse transcription PCR (RT-PCR) targeting several SARS-CoV-2 genes (Centers for Disease Control and Prevention, 2020, WHO, 2020a, Wang et al., 2020).
In the early phases of the epidemic, when containment of the infection was the most urgent goal, prudence required that even minimal amounts of SARS-CoV-2 RNA were considered sufficient for imposing quarantine on a suspected COVID-19 patient. Similarly, quarantine relief rules required two negative nasal swab results from samples taken at least 24 h apart. All these measures were somewhat effective in limiting SARS-CoV-2 circulation, as shown by the marked reduction of new COVID-19 cases by the end of May. On the other hand, while most clinically recovered patients tested negative at the end of the quarantine period and were able to return to their normal working and social life, a substantial proportion (about 16.6%) still tested positive on the required second testing, sometimes forcing them into a never-ending quarantine–positive test loop (Wu et al., 2020). Previous reports indicated that viable virus could not be isolated from samples with low SARS-CoV-2 genome loads (Huang et al., 2020, Atkinson and Petersen, 2020, Wölfel et al., 2020). In the initial phase of the infection (when viable virus is obviously present and the patient is infectious), this finding could be due to sampling bias or culturing inconsistencies (e.g., delayed delivery of the sample to the laboratory). On the other hand, in clinically recovered patients, the presence of residual viral RNA is more likely related to elimination of degrading viral materials.
Our hypothesis is that despite the high sensitivity of SARS-CoV-2-specific molecular assays, in the monitoring of the presence SARS-CoV-2 for release of individuals from quarantine, this approach could be a double-edged sword because of detection of the prolonged presence of a single gene.
Methods
To understand whether residual SARS-CoV-2 RNA load in clinically recovered patients could be associated with ongoing virus replication or is a result of catabolism of the virus or virus-infected cells, we submitted to cell culture isolation 387 nasal swabs from patients resident in the Lombardy, Emilia-Romagna, and Toscana regions showing low SARS-CoV-2 RNA amounts with a quantification cycle (Cq) value greater than 30, according to MIQE guidelines (Bustin et al., 2009). Clinical samples were obtained at the time of discharge from the hospital or during the quarantine period from hospitalized patients, symptomatic healthcare workers, or persons tested as part of the early epidemic response. Samples were anonymized before the analysis, and no clinical information was available.
This study was conduced in collaboration with five different centers: (1) Molecular Virology Unit, Microbiology and Virology Department, Fondazione IRCCS Policlinico San Matteo, Pavia, (2) Istituto Zooprofilattico Sperimentale della Lombardia e dell'Emilia Romagna, (3) Microbiology and Virology Unit, S. Maria delle Scotte University Hospital of Siena, Siena, (4) Virology Unit, Fondazione IRCCS Ca' Granda Ospedale Maggiore Policlinico, Milan, and (5) Microbiology Unit, Clinical Pathology Department, Guglielmo da Saliceto Hospital, Piacenza. Nasal swabs collected in universal transport medium (UTM™, Copan Italia, Brescia, Italy) were prospectively analyzed for the diagnosis of SARS-CoV-2 infection by real-time RT-PCR targeting the E gene according to WHO guidelines (WHO, 2020b) and protocols of Corman et al. (2020) at centers 1 and 2, while at centers 3–5, a commercial multiplex assay including the N gene as a target (Allplex™ 2019-nCoV assay; Seegene, Korea) was used. A series of nasal swabs collected from convalescent patients from April 1 to August 1, 2020, and positive for SARS-CoV-2 RNA with Cq value greater than 30 with the E or N gene were included in the study. To investigate the infectious potential of samples, a 200 μL sample was inoculated into a Vero E6 (VERO C1008 (Vero 76, clone E6, Vero E6); ATCC® CRL-1586™) confluent 24-well microplate for virus isolation. All samples were inoculated between 8 and 24 h after positivity results and kept at 4 °C before processing. After 1 h incubation at 33 °C in 5% CO2 in air, the inoculum was discarded and 1 mL of medium for respiratory viruses (Eagle’s minimum essential medium supplemented with 1% penicillin, streptomycin, and glutamine and trypsin at 5 μg/mL) was added to each well. Cells were incubated at 33 °C in 5% CO2 in air and observed with a light microscope every day for a cytopathic effect. After incubation for 7 days, 200 μL of supernatant from a well showing a cytopathic effect was tested for the presence of SARS-CoV-2 by molecular assay (gene E real-time RT-PCR) or SARS-CoV-2-specific immunofluorescence assays using antibodies to N protein.
In a subset of samples positive for the N gene (with Cq value of 35 or greater), both direct RNA and amplicon sequencing approaches was performed with a MinION instrument (Oxford Nanopore Technologies, UK). The sequencing run was managed by MinKNOW (version 19.12.5), and amplicon reads were mapped to the reference genome Wuhan Wu-1 (GenBank accession no. MN908947).
Results
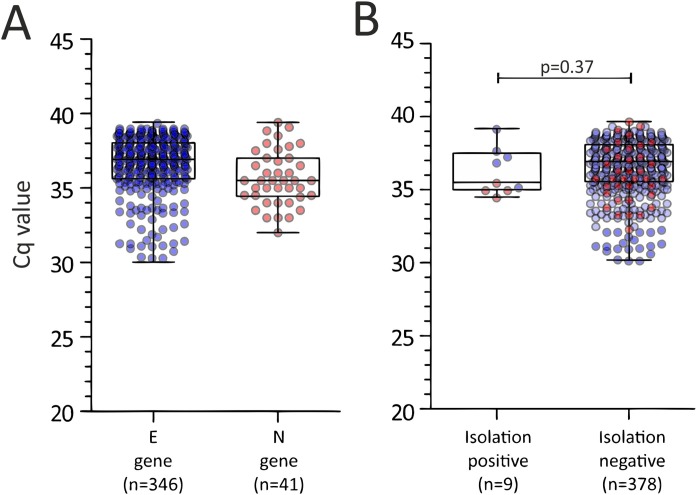
Among the samples tested by real-time RT-PCR, 89.4% (346/387) had Cq values greater than 30 for the E gene and 10.6% (41/387) had Cq values greater for the N gene (Figure 1 A). The median Cq value was 36.8 (range 30.0–39.4). In detail, the median Cq value was 36.9 (range 30.0–39.4) for the E gene and 35.5 (range 32.0–39.4) for the N gene (Figure 1A). A cytopathic effect was observed in only nine samples, corresponding to a culture positivity rate of 2.3% (9/387). Among these samples, five had a high Cq value for the E gene, while the others had a high Cq value for the N gene (Figure 1B). In these samples, the occurrence of active replication of SARS-CoV-2 was determined by real-time RT-PCR in five samples (median Cq value 14.5; range 12.4–16.8) and immunofluorescence assays in the other four samples.
Figure 1.
Cq values observed in samples included in the study according to (A) the target gene used in the diagnostic severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-specific real-time reverse transcription PCR and (B) isolation positive or negative.
The median Cq value of culture-positive samples was not significantly different from that observed in culture-negative samples (35.6 vs. 36.9; p = 0.37, Figure 1B).
In a series of five samples, long-read sequencing data analysis was performed by third-generation sequencing with Nanopore technology. Bioinformatic reconstruction of sequenced RNA showed how samples did not have the whole viral genome, but only few and very short gene fragments (<600 nucleotides). This finding indicates that the residual RNA detected by molecular testing was not functional and represented only pieces of the degraded viral genome.
Discussion
This prospective multicenter observational study demonstrates that residual SARS-CoV-2 RNA load, with a minimum amount of only one gene target, is not substantially associated with ongoing virus replication. Indeed, less than 3% of samples that were positive by real-time RT-PCR with low or very low RNA amounts were still able to transmit infection in cell monolayers. Our data are in agreement with a French study that reported an isolation rate lower than 50.0% in samples with a Cq value between 30 and 35 (only 24 samples) and no isolation for samples with a Cq value greater than 35 (only four samples) (La Scola et al., 2020). Similar results were very recently reported by Singanayagam et al. (2020), where the estimated probability of recovery of virus from samples with a Cq value of 35 or greater was 8.3%. The concept that medium/high viral load is needed for virus isolation was assessed in a recent publication aimed at clarifying the correlation between the culturability of the virus and the RNA copy number (Huang et al., 2020). In that study, the mean Cq value of culturable samples was always less than 30 (Huang et al., 2020). Our data are even more straightforward when one considers that samples were inoculated early after collection, thus minimizing the infectivity loss during transport to the laboratory. In addition, all samples were drawn from clinically recovered patients. The implications of the results of the present study are important at both the individual level and the epidemiologic level. On the individual side, dissection between true virus replication and residual detection of virus genes has the immediate effect of releasing patients from prolonged quarantine, safely allowing them to return to work and social activities. From an epidemiologic standpoint, these data pose the question of whether it is correct to consider these molecular “low-positivity” cases as real virus positives. Indeed, while the analytical positivity of highly sensitive real-time RT PCR methods is correct (a fragment of SARS-CoV-2 RNA is detected), their clinical significance is less than certain (a fragment of viral RNA does not indicate either the presence of the whole virus or that the patient is still contagious). The number of messenger RNAs that originate during SARS-CoV-2 replication is largely different, although they are collinear with the viral genome and are simultaneously targeted by real-time RT PCR methods. Thus, the results of molecular assays have to take into account the virus biology (Huang et al., 2020). Finally, in the infection recovery phase, neutralizing antibodies are generated, and an impact of seroconversion on virus infectivity must be considered. However, specific studies are needed to address this hypothesis.
One limitation of this study is the impossibility to score the quality of sampling and storage before samples are sent to the laboratory and, nevertheless, the sensitivity of cell culture isolation. Comparison of Cq values between E and N gene results could be biased by different sensitivities of the assays; however, WHO and CDC reports showed comparable sensitivity for both assays (WHO, 2020b, Corman et al., 2020, Canters for Disease Control and Prevention, 2020). Indeed, a nationwide external quality assessment conducted for more than 100 Korean laboratories showed overlapping results for Cq values obtained by real-time RT-PCR assays used in the present study (Sung et al., 2020).
An additional limitation was the unavailability of clinical information because of the anonymization of samples. However, these data, which were consistent among the five different laboratories in northern and central Italy, lead us to propose that the individuals are no longer contagious when the molecular diagnosis is based on a high Cq value (≥35) of one gene target, and this is supported by clinical data. The major caveat of this study is the evidence that most recovered patients (97%) with a high Cq value do not carry infectious virus, and that prolonged quarantine periods are not justified. Thus, revision of current guidelines is highly recommended.
Funding
This study was supported by Ricerca Corrente IRCCS Policlinico San Matteo, Italy (grant no. 80206), by Ricerca Finalizzata from Ministry of Health, Italy (grant no. GR-2013-02358399) and funds from the European Commission Horizon 2020 program (EU project 101003650 - ATAC).
Conflict of interest
The authors declare that they have no competing interests.
Ethics statement
This study was performed according to the guidelines of the Institutional Review Board of our Institute on the use of biological specimens for scientific purposes, in keeping with Italian law (art 13 D. Lgs 196/2003) Samples were anonymized before the analysis and therefore informed consent was not required.
Author contributions
AP collated and analyzed the data and wrote the first draft of the manuscript. MR performed the statistical analysis and constructed the figures. MGC, PP, GL, and RS provided samples for the study. EP and MGC supervised the isolation team. EVN, RD, AF, GS, and CG performed isolation experiments. NV, FB, MT, and FR performed the laboratory PCR work. CG, GA, and CT performed next-geneteration sequencing experiments. MGC and FB supervised and coordinated the samples and testing and critically revised the first version of the manuscript.
Acknowledgment
We thank Daniela Sartori for manuscript editing.
References
- Atkinson B., Petersen E. SARS-CoV-2 shedding and infectivity. Lancet. 2020;395(10233):1339–1340. doi: 10.1016/S0140-6736(20)30868-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Canters for Disease Control and Prevention . 2020. CDC’s diagnostic multiplex assay for Flu and COVID-19 at public health laboratories and supplies.https://www.cdc.gov/coronavirus/2019-ncov/lab/multiplex.html (Accessed 14 September 2020) [Google Scholar]
- Centers for Disease Control and Prevention . 2020. CDC 2019-novel coronavirus (2019-nCoV) real-time RT-PCR diagnostic panel.https://www.fda.gov/media/134922/download [Accessed 18 June 2020] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3):2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.G., Lee K.M., Hsiao M.J., Yang S.L., Huang P.N., Gong Y.N. Culture-based virus isolation to evaluate potential infectivity of clinical specimens tested for COVID-19. J Clin Microbiol. 2020;58(8):e01068–20. doi: 10.1128/JCM.01068-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Scola B., Le Bideau M., Andreani J., Hoang V.T., Grimaldier C., Colson P. Viral RNA load as determined by cell /culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39(6):1059–1061. doi: 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singanayagam A., Patel M., Charlett A., Lopez Bernal J., Saliba V., Ellis J. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. 2020;25(32):2001483. doi: 10.2807/1560-7917.ES.2020.25.32.2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H., Han M.G., Yoo C.K., Lee S.W., Chung Y.S., Park J.S. Nationwide external quality assessment of SARS-CoV-2 molecular testing, South Korea. Emerg Infect Dis. 2020;26(10):2353–2360. doi: 10.3201/eid2610.202551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO.2020a Real-time RT-PCR assays for the detection of SARS-CoV-2. https://www.who.int/docs/default-source/coronaviruse/whoinhouseassays.pdf?sfvrsn=de3a76aa_2. [Accessed 18 June 2020].
- WHO. https://www.who.int/docs/default-source/coronaviruse/protocol-v2-1.pdf. [Accessed 18 June 2020].
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu J., Liu X., Liu J., Liao H., Long S., Zhou N. Coronavirus disease 2019 test results after clinical recovery and hospital discharge among patients in China. JAMA Netw Open. 2020;3(5):e209759. doi: 10.1001/jamanetworkopen.2020.9759. [DOI] [PMC free article] [PubMed] [Google Scholar]