Abstract
Background
The clinical diagnosis of genetic renal diseases may be limited by the overlapping spectrum of manifestations between diseases or by the advancement of disease where clues to the original process are absent. The objective of this study was to determine whether genetic testing informs diagnosis and facilitates management of kidney disease patients.
Methods
We developed a comprehensive genetic testing panel (KidneySeq) to evaluate patients with various phenotypes including cystic diseases, congenital anomalies of the kidney and urinary tract (CAKUT), tubulointerstitial diseases, transport disorders and glomerular diseases. We evaluated this panel in 127 consecutive patients ranging in age from newborns to 81 years who had samples sent in for genetic testing.
Results
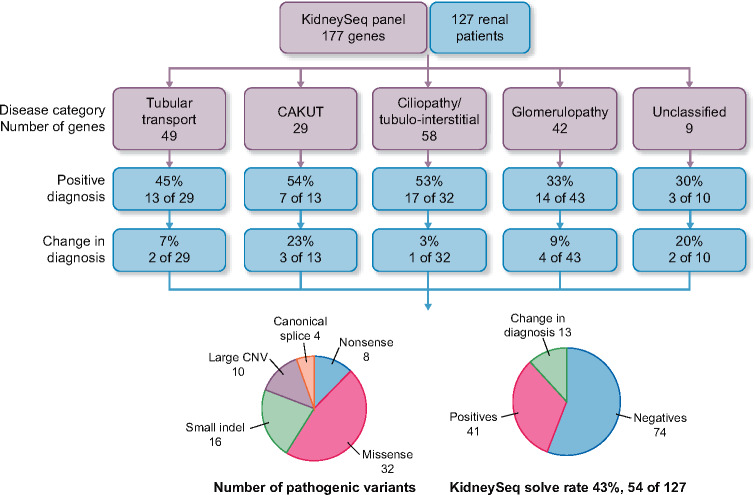
The performance of the sequencing pipeline for single-nucleotide variants was validated using CEPH (Centre de’Etude du Polymorphism) controls and for indels using Genome-in-a-Bottle. To test the reliability of the copy number variant (CNV) analysis, positive samples were re-sequenced and analyzed. For patient samples, a multidisciplinary review board interpreted genetic results in the context of clinical data. A genetic diagnosis was made in 54 (43%) patients and ranged from 54% for CAKUT, 53% for ciliopathies/tubulointerstitial diseases, 45% for transport disorders to 33% for glomerulopathies. Pathogenic and likely pathogenic variants included 46% missense, 11% nonsense, 6% splice site variants, 23% insertion–deletions and 14% CNVs. In 13 cases, the genetic result changed the clinical diagnosis.
Conclusion
Broad genetic testing should be considered in the evaluation of renal patients as it complements other tests and provides insight into the underlying disease and its management.
Keywords: copy number variants, genetic kidney disease, massively parallel sequencing, targeted gene panel
ADDITIONAL CONTENT
An author video to accompany this article is available at: https://academic.oup.com/ndt/pages/author_videos.
INTRODUCTION
The kidney is a complex organ that maintains physiological homeostasis through a myriad of complex processes that include the excretion of excess water, ingested drugs, toxins and metabolic waste products, the regulation of acid–base balance, the reclamation or elimination of various salts, and the synthesis of a variety of endocrine hormones to control blood pressure, erythropoiesis and bone mineralization. Disrupting this function leads to a broad spectrum of disease phenotypes. At one extreme are diseases that manifest as well-recognized Mendelian disorders such as Liddle syndrome, which is characterized by hypertension with hypokalemia from unregulated hyperactivity of the epithelial sodium channel in the connecting tubule and collecting duct [1]. At the other extreme are diseases in which a more global decline in renal function leads to chronic kidney disease (CKD) with a reduction in glomerular filtration rate, retention of urea, phosphorus and potassium, and the development of anemia and bone disease. The development of CKD may blur clues to the inciting insult even with extensive laboratory testing, renal imaging and renal histological examination [2].
Over the past decade, a number of discoveries relevant to renal diseases have improved our understanding of the ciliopathies [3], focal segmental glomerulosclerosis (FSGS) [4], steroid-resistant nephrotic syndrome [5, 6] and congenital anomalies of the kidney and urinary tract (CAKUT) [7, 8]. In recent years, the advancement of next-generation sequencing has facilitated the simultaneous interrogation of multiple genes for molecular diagnosis within many disease categories including those that cause a variety of renal diseases [9, 10]. In addition, exome sequencing (ES) has been used to diagnose monogenic renal diseases [11, 12]. The diagnostic success of disease-focused panels may be limited by difficulty in phenotyping renal diseases into specific categories. Similarly, ES may not be sensitive enough to detect variants in duplicated regions, such as the proximal portion of PKD1. We sought to test the clinical relevance of broad-based genetic testing that targets genes across a wide variety of renal disease phenotypes to inform diagnosis and facilitate management of the renal patient. Using a panel of 177 genes, we tested 127 consecutive renal patients whose samples we received and in this diverse cohort made a genetic diagnosis in 54 patients. Remarkably, in 13 patients, the genetic findings changed the clinical diagnosis.
MATERIALS AND METHODS
Study design
This was a retrospective study of the diagnostic accuracy of comprehensive genetic testing panel used a cohort of 127 consecutive patients where samples were sent to the University of Iowa Institute of Human Genetics for gene screening. There were no exclusion criteria. Patients were classified, based on clinical history provided, into the following broad disease subtypes: ciliopathies/tubulointerstitial diseases, CAKUT, tubular transport disorders and glomerulopathies. American College of Medical Genetics (ACMG) criteria were used to classify genetic variants as pathogenic, likely pathogenic, variant of unknown significance (VUS), likely benign and benign [13].
Gene selection, platform design and validation, and patient recruitment
Genes implicated in a large number of renal diseases were selected for inclusion in the kidney disease panel (KidneySeq v1) and grouped by renal phenotype (e.g. ciliopathy, glomerular diseases and CAKUT). Targeted capture of coding exons and splice sites was optimized using RNA baits selected with Agilent’s SureDesign online software, incorporating 4-fold probe density and 25-base pairs of flanking intronic sequence. Performance metrics were assessed by studying 31 genomic DNA samples from the CEPH consortium (Centre de'Etude du Polymorphism) using results to improve depth-of-coverage (Supplementary data, Table S1). Additional genes were also added to increase the genetically relevant search space. The updated panel (KidneySeq v2) was used in the diagnostic evaluation of sequentially accrued samples from patients with renal disease (Table 1 and Supplementary data, Table S2). There were no exclusionary criteria.
Table 1.
KidneySeq v2 gene list by disease category
| Ciliopathies/tubulointerstitial diseases | |
| Alagille syndrome | NOTCH2 |
| Autosomal recessive polycystic kidney disease | PKHD1 |
| Autosomal dominant polycystic kidney disease | PKD1, PKD2 |
| Autosomal dominant tubulointerstitial kidney disease | HNF1B, REN, UMOD |
| Bardet–Biedl syndrome (BBS) | ARL6, BBS1, BBS2, BBS4, BBS5, BBS7, BBS10, BBS12, CEP290, MKKS, PTHB1, TRIM32, TTC8 |
| COACH syndrome | CC2D2A, RPGRIP1L, TMEM67 |
| HANAC syndrome | COL4A1 |
| Jeune syndrome | IFT80, IFT140, DYNC2H1, NEK1, TTC21B |
| Joubert syndrome | AHI1, ARL13B, ATXN10, CC2D2A, CEP290, CEP41, CSPP1, INPP5E, KIF7, NPHP1, OFD1, RPGRIP1L, TMEM216, TCTN1, TMEM138, TMEM237, TMEM67, TTC21B |
| Juvenile nephronophthisis (JN) | AHI1, ATXN10, IQCB1, CEP290, GLIS2, INVS, NEK8, NPHP1, NPHP3, NPHP4, RPGRIP1L, TMEM67, TTC21B, WDR19, XPNPEP3 |
| Meckel syndrome (MKS)/Meckel–Gruber syndrome | B9D1, B9D2, CC2DA, CEP290, MKS1, NPHP3, RPGRIP1L, TCTN2, TMEM216, TMEM67 |
| Medullary cystic kidney disease 2 | UMOD |
| Oro-facial-digital syndrome 1 | OFD1 |
| Renal cysts and diabetes syndrome | HNF1B |
| Serpentine fibula with polycystic kidney disease (SFPKS)/Hajdu–Cheney syndrome (HJCYS) | NOTCH2 |
| Sensenbrenner syndrome/(CED) | IFT122, IFT43, WDR19, WDR35 |
| Senior–Loken syndrome (JN with retinitis pigmentosa) | CEP290, IQCB1, NPHP1, NPHP3, NPHP4, WDR19 |
| Disorders of tubular ion transport | |
| Apparent mineralocorticoid excess, syndrome of | HSD11B2 |
| APRT deficiency | APRT |
| Autosomal dominant hypocalcemia, ± Bartter syndrome | CASR |
| Bartter syndrome | BSND, CLCNKA, CLCNKB, KCNJ1, SLC12A1 |
| Cystinosis | CTNS |
| Cystinuria | SLC3A1, SLC7A9 |
| Dent disease | CLCN5, OCRL |
| Distal renal tubular acidosis | ATP6V0A4, ATP6V1B1, SLC4A1 |
| Familial hypertension with hyperkalemia (Gordon syndrome), Pseudohypoaldosteronism II | CUL3, KLHL3, WNK1, WNK4 |
| Gitelman syndrome | CLCNKB, SLC12A3 |
| Hypophosphatemic rickets | DMP1, CLCN5, ENPP1, FGF23, PHEX, SLC34A3 |
| Isolated proximal renal tubular acidosis—generalized proximal defect (Fanconi syndrome) | ATP7B, CLCN5, CTNS, EHHADH, FAH, HNF4A, SLC34A1 |
| Liddle syndrome (pseudo hyperaldosteronism) | SCNN1B, SCNN1G |
| Nephrogenic diabetes insipidus (NDI) | AQP2, AVPR2 |
| Nephrogenic syndrome of inappropriate antidiuresis (NSIAD) | AVPR2 |
| Primary hyperoxaluria | AGXT, GRHPR, HOGA1 |
| Pseudohypoaldosteronism I (PHA I) | NR3C2, SCNN1A, SCNN1B, SCNN1G |
| Renal glucosuria | SLC5A2 |
| Renal hypomagnesemia | CLDN16, CLDN19, CNNM2, EGF, HNF1B, TRPM6 |
| Renal tubular acidosis, proximal, with ocular abnormalities | SLC4A4 |
| Glomerular diseases | |
| Alport syndrome | COL4A3, COL4A4, COL4A5 |
| Alstrom syndrome | ALMS1 |
| Congenital nephrotic syndrome (Finnish type) | NPHS1 |
| DDS; Frasier syndrome | WT1 |
| Diffuse mesangial sclerosis | ARHGDIA, PLCE1, WT1 |
| Epstein–Fechtner syndrome (renal disease with macrothrombocytopenia) | MYH9 |
| Fabry disease | GLA |
| FSGS–AD/XL | ACTN4, ANLN, ARHGAP24, CD2AP, COL4A3, COL4A4, COL4A5, INF2, LMX1B, PAX2, TRPC6, WT1 |
| FSGS–AR | APOL1, CRB2, MYO1E, NPHP4, TTC21B |
| FSGS/steroid-resistant nephrotic syndrome (SRNS)–AR | ADCK4, ALG1, ARHGDIA, CUBN, LAMB2, NPHS1, NPHS2, PLCE1, PDSS2, PMM2, PTPRO, SCARB2, ZMPSTE24 |
| Galloway–Mowat syndrome | WDR73 |
| Glomerulopathy with fibronectin deposits | FN1 |
| Hereditary systemic or renal amyloidosis | APOA1, B2M, FGA, LYZ, NLRP3 |
| Muckle–Wells syndrome | NLRP3 |
| Nail patella syndrome | LMX1B |
| Nephrotic syndrome, steroid sensitive | PLCG2 |
| Pierson syndrome (nephrotic syndrome with microcoria) | LAMB2 |
| Thin basement membrane disease (benign familial hematuria) | COL4A3, COL4A4 |
| CAKUT | |
| Branchio-oto-renal syndrome | EYA1, SIX1, SIX5 |
| CAKUT with VACTERL | TRAP1 |
| Cogan oculomotor apraxia | NPHP1 |
| Common CAKUT | AGTR1, AGTR2, CHD1L, DSTYK, EYA1, GATA3, HNF1B, PAX2, RET, ROBO2, SALL1, SIX2, SIX5, TRAP1 |
| Fraser syndrome | FRAS1, FREM2, GRIP1 |
| Hypoparathyroidism, sensorineural deafness and renal dysplasia | GATA3 |
| Isolated renal hypo-dysplasia | BMP4, DSTYK, FGF20, HNF1B, PAX2, RET, SALL1, SIX2 |
| Isolated renal hypoplasia and renal-coloboma syndrome (papillorenal syndrome) | PAX2 |
| Isolated renal hypoplasia | RET, UPK3 |
| Kallmann syndrome | ANOS1 |
| Mayer–Rokitansky–Küster–Hauser syndrome | WNT4 |
| Multicystic dysplastic kidney | CHD1L, HNF1B, ROBO2, SALL1 |
| Posterior urethral valves | CHD1L, HNF1B, ROBO2, SALL1, SIX2 |
| Renal cysts and diabetes syndrome | HNF1B |
| Renal tubular dysgenesis | ACE, AGT, AGTR1, REN |
| Townes–Brocks syndrome | SALL1 |
| Unilateral renal agenesis | DSTYK, HNF1B, RET, SALL1 |
| UPJ obstruction | DSTYK, EYA1, HNF1B, RET, ROBO2, SALL1 |
| UVJ obstruction | CHD1L, PAX2, SIX5 |
| Vesicoureteral reflux | DSTYK, EYA1, GATA3, HNF1B, RET, ROBO2, SALL1, SOX17, TNXB, UPK3A |
| Other | |
| Acrorenoocular syndrome (Okihiro syndrome) | SALL4 |
| Mitochondrial cytopathy | COQ2 |
| Pallister–Hall syndrome | GLI3 |
| Rubinstein–Taybi syndrome | CREBBP |
| Schimke immuno-osseous dysplasia | SMARCAL1 |
| SERKAL syndrome (46XX sex reversal with dysgenesis of kidneys, adrenal and lungs) | WNT4 |
| Simpson–Golabi–Behmel syndrome | GPC3 |
| Smith–Lemli–Opitz syndrome | DHCR7 |
| Tuberous sclerosis | TSC2 |
| Williams syndrome | 7q 11.23 |
Library preparation, targeted genomic enrichment and massively parallel sequencing
After preparing libraries from patient-derived gDNA, library preparation, targeted genomic enrichment and massively parallel sequencing (MPS) were completed as we have described [14]. In brief, libraries were prepared using a modification of the solution-based Agilent SureSelect target enrichment system (Agilent Technologies, Santa Clara, CA, USA) with liquid-handling automation. Hybridization and capture with RNA baits were followed by a second amplification. Before pooling for sequencing, all samples were bar coded and multiplexed. Sequencing was done using Illumina HiSeq (pool of 48 samples) or MiSeq (pools of five samples) instrumentation (Illumina Inc., San Diego, CA, USA). Sanger sequencing was used to amplify and resolve exons 1–34 of PKD1 [15, 16].
Bioinformatics analysis
Data analysis was performed on dedicated computing resources maintained by the Iowa Institute of Human Genetics using a standardized workflow for sequence analysis and variant calling [14]. The freebayes variant caller was used to identify variants in PKD1. Variant annotation was performed with a custom-built reporting tool.
Variant filtering
Library quality was based on the total number of reads per sample and coverage at 30× or greater, excluding low-quality variants [depth <10 or quality by depth (QD) <5] and common variants with a minor allele frequency (MAF) >1% in any population (except for known risk alleles). Nonsynonymous single-nucleotide variants (SNVs), canonical splicing changes and insertion–deletions (indels) were retained. Reference databases routinely queried included the Human Gene Mutation Database, ClinVar, the autosomal dominant polycystic kidney disease (ADPKD) mutation database, the ARUP (COL4A5) database and our in-house Renal Variant Database (RVD). GERP++, PhyloP, MutationTaster, PolyPhen2, SIFT and LRT were used to calculate variant-specific pathogenicity scores as described [14].
Copy number variant analysis
Copy number variant (CNV) analysis was performed using ExomeCopy and ExomeDepth [17]. CNV calls from both programs were manually curated and validated if breakpoints were identified.
Sanger sequencing
Sanger sequencing was performed for platform validation, for PKD1 testing and to confirm pathogenic variants, designing primers using Primer 3 (http://bioinfo.ut.ee/primer3-0.4.0/primer3/) [14].
Variant interpretation
A multidisciplinary board was held semimonthly to discuss all genetic results on a patient-by-patient basis in the context of the available clinical data. Variants were classified following ACMG guidelines. Variants with a MAF >1% were classified as ‘benign’ with a few notable exceptions (APOL1 G1 and G2 alleles). Variants reported as ‘pathogenic’ in the literature with supporting functional evidence were classified as ‘pathogenic’. The ‘likely pathogenic’ classification was assigned to missense variants with pathogenicity scores ≥4 (based on GERP++, PhyloP, MutationTaster, PolyPhen2, SIFT and LRT) if they were also ultra-rare and in a disease-related functional domain. Novel or rare variants that changed protein sequence but had an unknown impact on protein function were classified as VUSs. Based on the clinical phenotype and the genotypic findings, clinical correlation and segregation analysis were recommended.
Institutional Review Board
The study was approved by the Institutional Review Board (IRB No. 201805825) for human subject research and informed consent was waived. The study adheres to the Principles of Medical Research as stated in the Declaration of Helsinki.
RESULTS
Performance metrics
Performance and validation of KidneySeq v1 using 31 CEPH samples showed that >70% of sequence reads overlapped target regions with a mean coverage of ≥400×; >99% of bases were covered by at least 30 reads (30×). This threshold was achievable with at least 5 million reads per sample (Supplementary data, Figure S1). Targeted regions covered at less than 30× were Sanger sequenced; no additional variants were identified (Supplementary data, Table S3). These performance metrics were used to refine the panel by changing probe density.
Variant analysis
Call accuracy in the 31 CEPH controls was determined by Sanger sequencing 29 variants with MAF >1% and 32 variants with QD <10 in all samples (Supplementary data, Table S4); 256 variants that were either heterozygous or homozygous alternate were identified (Supplementary data, Figure S2). All validated variants with a QD <5 were false positives. Between QD >5 and QD <10, there were false-positive calls for both SNVs and indels. Of the 1643 sites, there were 252 true positives, 4 false positives, 1387 true negatives and no false negatives. Specificity (99.71), sensitivity (100), and positive (98.44) and negative (100) predicted values were very high (Supplementary data, Tables S4 and S5).
Validation of sequencing and analysis pipeline
A high-density SNP array was used to interrogate the CEPH sample, NA12287 (1421-14). A comparison of genotype calls from the SNP array and KidneySeq v1 identified only one discordant variant from the 3008 identified (Supplementary data, Figure S3a). Through Sanger confirmation, we verified that the KidneySeq v1 variant call was correct and the SNP array was incorrect. To validate indels, we used Genome-in-a-Bottle (GIAB), which predicts 314 indels in the KidneySeq v1 targeted regions. All predicted indels were identified by KidneySeq v1 in addition to two other indels at QD >5 not reported in the GIAB reference sequence but confirmed by Sanger sequencing (Supplementary data, Figure S3b). To test the reliability and sensitivity of the CNV analysis workflow, positive samples were re-sequenced and re-analyzed. All known CNVs were detected successfully on the repeat samples (Supplementary data, Table S6).
PKD1 gene proximal region
The duplicated region of PKD1 (exons 1–34) was Sanger sequenced to verify variant detection. The panel detected 36 variants in the homologous region of the PKD1 gene in seven patients selected for this validation. Overall, 94.4% (34 of 36) of these variants were verified by Sanger sequence. The variant detected only by MPS (the same variant was detected in two patients) was a false positive in exon 15. No false negatives were detected by Sanger sequencing.
Patients
Genetic testing was completed on 127 patients (77 males). The most common indication was FSGS (17 patients), followed by medullary cystic kidney disease/nephronophthisis (14 patients), Alport or Alport-like syndrome (10 patients), Bartter/Gitelman syndrome (9 patients) and ADPKD (7 patients) (Table 2). Age ranged from newborn to 81 years (0–6 years, 56 patients; 7–14 years, 22 patients; 15–30 years, 26 patients; >30 years, 23 patients) (Table 3).
Table 2.
Indications for testing
| CAKUT | |
| Branchio-oto-renal syndrome | 2 |
| HNF1-β | 1 |
| Multicystic dysplastic kidney | 3 |
| Papillorenal syndrome | 1 |
| Renal hypo/dysplasia | 6 |
| Unspecified | 5 |
| Total | 18 |
| Ciliopathy/tubulointerstitial | |
| ADPKD | 7 |
| ARPKD | 3 |
| Medullary cystic kidney disease/nephronophthisis | 14 |
| Orofacial digital syndrome | 1 |
| Renal cysts | 5 |
| Total | 32 |
| Tubular ion transport | |
| Apparent mineralocorticoid excess | 1 |
| Bartter/Gitelman | 9 |
| Cystinuria | 1 |
| Dent | 5 |
| Fanconi | 2 |
| Hypercalcemia | 3 |
| Hypokalemia | 2 |
| Hypomagnesemia | 3 |
| Hypophosphatemia | 3 |
| Kidney stones | 2 |
| Liddle syndrome | 2 |
| NDI | 3 |
| Pseudohypoaldosteronism I | 2 |
| Renal tubular acidosis | 2 |
| Total | 40 |
| Glomerulopathy | |
| Alport/Alport like | 10 |
| FSGS | 17 |
| Nephrotic proteinuria/nephrotic syndrome | 9 |
| Other glomerular | 2 |
| Total | 40 |
| Other | |
| Nephrogenic rests | 1 |
| Nonrenal | 1 |
| No information | 5 |
| Unclassified kidney disease | 10 |
| Total | 17 |
Some patients had multiple laboratory abnormalities or clinical diagnosis that is listed individually, resulting in larger totals. ARPKD, autosomal recessive polycystic kidney disease.
Table 3.
Clinical renal samples: all patients with indication for testing, family history, disease type and demographics; family history, when known, are shown as positive (Y) or negative (N)
| Case | Indication for testing | Family history | Disease categorya,b | Sex | Age (years) | Ethnicity |
|---|---|---|---|---|---|---|
| 1 | Bilateral multicystic dysplastic kidneys | Y | 1 | F | 6 | Hispanic |
| 2 | Renal dysplasia | Unknown | 1 | M | 1 | Caucasian, non-Hispanic |
| 3 | Stage 5 (CKD), hearing loss | Unknown | 4 | M | 37 | Asian |
| 4 | FSGS at age 40 years | N | 4 | M | 66 | Caucasian, non-Hispanic |
| 5 | Proteinuria, FSGS | Y | 4 | M | 54 | African/African-American, non-Hispanic |
| 6 | Alport syndrome | Y | 4 | M | 34 | White |
| 7 | Dent disease (NDI, failure to thrive) | Unknown | 3 | M | 1 | Caucasian, Hispanic or Latino |
| 8 | Nephronophthisis | Y | 2 | F | 10 | African/African-American |
| 9 | FSGS | Unknown | 4 | M | 54 | African/African-American |
| 10 | Nephrotic syndrome | Unknown | 4 | M | 3 | Hispanic or Latino |
| 11 | Medullary cystic kidney disease | Unknown | 2 | M | 27 | Caucasian, non-Hispanic |
| 12 | Hypomagnesemia | Unknown | 3 | F | 11 | Not provided |
| 13 | FSGS | Unknown | 4 | M | 58 | Caucasian, non-Hispanic |
| 14 | Medullary cystic kidney disease/ nephronophthisis | Unknown | 2 | M | 31 | Caucasian |
| 15 | Hypercalcemia, hypocalciuria | N | 3 | F | 81 | Caucasian |
| 16 | Dilated cardiomyopathy and hypomagnesemia | N | 3 | M | 3 | Caucasian |
| 17 | Fanconi syndrome, hypophosphatemic rickets | Unknown | 3 | M | 2 | Caucasian, aboriginal |
| 18 | ESRD, primary FSGS | Unknown | 4 | M | 55 | Caucasian |
| 19 | Severe CAKUT | Unknown | 1 | M | <1 | Caucasian, Hispanic or Latino |
| 20 | Alport syndrome | Unknown | 4 | M | 5 | Asian (India), non-Hispanic |
| 21 | Hypercalcemia, hypercalciuria, short stature | Unknown | 3 | M | 2 | Caucasian, non-Hispanic |
| 22 | Interstitial nephritis | Unknown | 2 | F | 10 | Caucasian |
| 23 | U/S prenatal echogenic kidneys, postna- tal bilateral cysts, HNF1B disease | Unknown | 1 | M | <1 | Caucasian, non-Hispanic |
| 24 | Bartter syndrome or other | Unknown | 3 | M | 1 | Not provided |
| 25 | ESRD, tubulointerstitial disease | Y | 2 | M | 51 | African/African-American |
| 26 | Bilateral hypoplastic dysplastic kidneys | Unknown | 1 | M | <1 | Caucasian, Hispanic or Latino |
| 27 | Microhematuria, Alport or TBM disease | Unknown | 4 | M | 2 | Caucasian, Hispanic or Latino |
| 28 | FSGS or MCKD | Y | 2, 4 | M | 60 | African/African-American, non-Hispanic |
| 29 | Alport or TBM disease | Unknown | 4 | M | 18 | Caucasian, non-Hispanic |
| 30 | FSGS, SRNS, hypoalbuminemia | Unknown | 4 | M | 17 | Caucasian, non-Hispanic |
| 31 | FSGS or Dent disease. Nephroticrange proteinuria, global glomerulosclerosis | Unknown | 3, 4 | M | 18 | African/African-American |
| 32 | ADTKD, tubular proteinuria, no signs of Fanconi | Y | 2 | M | 18 | Unknown |
| 33 | Alport syndrome. Hearing loss, microscopic hematuria, CKD | Unknown | 4 | M | 12 | Caucasian |
| 34 | Renal agenesis/hypoplasia or nephronophthisis | Y | 1, 2 | F | 16 | Hispanic or Latino |
| 35 | Gitelman/Bartter syndrome | Unknown | 3 | F | 17 | Caucasian |
| 36 | Bilateral multicystic dysplastic kidneys, perinatal death | Unknown | 1 | M | 0b | Unknown |
| 37 | Bartter syndrome, NDI or Dent disease. Polyuria, polydipsia, hypercalciuria, medullary nephrocalcinosis | Unknown | 3 | M | 16 | Caucasian, non-Hispanic |
| 38 | Pseudohypoaldosteronism. Hyperkalemia, polyuria | Unknown | 3 | M | 0b | Hispanic or Latino |
| 39 | Multicystic bilateral kidneys | Unknown | 1 | M | 0b | Caucasian, non-Hispanic |
| 40 | Apparent mineral corticoid excess | Unknown | 3 | M | 2 | Not provided |
| 41 | Bartter syndrome. Polyuria, metabolic alkalosis | Unknown | 3 | F | 3 | Caucasian, non-Hispanic |
| 42 | Liddle syndrome. Early onset hypertension and hypokalemia | Y | 3 | F | 19 | Caucasian, Hispanic or Latino |
| 43 | PKD (bilateral renal cysts and hypertension) | Unknown | 2 | M | 15 | Hispanic or Latino |
| 44 | NDI, medullary nephrocalcinosis, vesicoureteral reflux, hypophosphatemia | Unknown | 3 | F | 3 | Caucasian, non-Hispanic |
| 45 | Cystinuria | Y | 3 | F | 19 | Caucasian |
| 46 | FSGS or minimal change disease. Persistent proteinuria | Unknown | 4 | M | 5 | Caucasian, non-Hispanic |
| 47 | Hypokalemia, hypomagnesemia, high urinary Na and K, prior diagnosis of NDI | Unknown | 3 | F | 59 | Caucasian, non-Hispanic |
| 48 | Hypotonia, dysmorphic features, developmental delay, obesity | Unknown | 5 | F | 2 | Caucasian, non-Hispanic |
| 49 | Horseshoe kidney asymptomatic; daughter, son perinatal/fetal demise with CAKUT | Y | 1 | F | 33 | Caucasian, Native American, non-Hispanic |
| 50 | Proximal tubulopathy or Dent or hypophosphatemic rickets, nephrocalcinosis, small stature | Unknown | 3 | F | 13 | Asian, non-Hispanic |
| 51 | FSGS, ESRD, post-kidney transplant | Unknown | 4 | M | 15 | Hispanic or Latino |
| 52 | PKD1, PKD2, HNF1B | Unknown | 2 | M | 6 | Hispanic or Latino |
| 53 | Renal cysts, family history of hereditary nephritis | N | 2 | F | 49 | Asian, non-Hispanic |
| 54 | Polycystic kidney disease, undescended testes, HTN | N | 2 | M | <1 | Caucasian, non-Hispanic |
| 55 | ESRD, FSGS | Y | 4 | M | 64 | African/African-American |
| 56 | HTN, AKI, LVH, congenital nephrotic syndrome or ARPKD | Unknown | 2, 4 | F | <1 | Not provided |
| 57 | Moderate CKD | Unknown | 5 | M | 1 | Not provided |
| 58 | Not provided | Unknown | 5 | F | 16 | Not provided |
| 59 | Bartter/Gitelman syndrome, hypokalemia, hypomagnesemia and metabolic alkalosis | Unknown | 3 | M | 12 | Not provided |
| 60 | Nephronophthisis or MCKD | Y | 2 | M | 58 | Caucasian, non-Hispanic |
| 61 | Polycystic kidney disease | Unknown | 2 | F | 51 | African/African-American |
| 62 | FSGS or MCKD | Y | 2, 4 | M | 56 | African/African-American |
| 63 | FSGS/multicystic dysplastic kidney | Y | 1, 4 | M | 15 | Caucasian, non-Hispanic |
| 64 | Hyperplastic nephrogenic rests, features seen with underlying syndromes such as Beckwith–Wiedemann | Unknown | 5 | F | <1 | Not provided |
| 65 | Hypophosphatemic rickets; distal renal tubular acidosis; isolated proximal renal tubular acidosis, generalized proximal defect | N | 3 | F | 0b | Hispanic or Latino |
| 66 | FSGS | Unknown | 4 | F | 10 | African/African-American, non-Hispanic |
| 67 | Horseshoe kidney, dysmorphic features, VSD | Y | 1 | F | <1 | Egyptian |
| 68 | Kidney stones, paresthesias, hypercalciuria, hypoparathyroidism, ESRD | Y | 3 | M | 58 | Caucasian |
| 69 | Large cystic kidneys | N | 2 | M | 27 | Caucasian, non-Hispanic |
| 70 | Renal cystic dysplasia, ectopic atrial tachycardia, CUA, seizures, LVH; dialysis from birth | Unknown | 2 | F | <1 | Caucasian |
| 71 | Steroid-resistant nephrotic syndrome | N | 4 | F | 8 | Asian, multiracial |
| 72 | MCD, unresponsive to steroids | N | 2 | F | 3 | African/African-American |
| 73 | Glomerulocystic kidneys and hepatoblastoma | N | 2 | F | 3 | Hispanic or Latino |
| 74 | Alport syndrome | 4 | M | 13 | Caucasian | |
| 75 | Steroid-resistant nephrotic syndrome | Y | 4 | M | 4 | Dominican Republic |
| 76 | Gitelman syndrome | N | 3 | F | 23 | Not provided |
| 77 | Not provided | Y | 5 | M | 57 | Not provided |
| 78 | Nephronophthisis | Y | 2 | F | 38 | Caucasian |
| 79 | Premature newborn with severely en larged cystic kidneys noted mid-trimester, severe oligohydramnios, pulmonary hypoplasia | N | 2 | F | 0b | Caucasian, Hispanic or Latino |
| 80 | Alport syndrome | Unknown | 4 | F | 11 | Caucasian |
| 81 | Hyponatremia, hypokalemia, nephrotic- range proteinuria, glucosuria | N | 3 | M | 1 | Caucasian, non-Hispanic |
| 82 | Global glomerulosclerosis | Y | 4 | F | 65 | African/African-American, non-Hispanic |
| 83 | Juvenile nephronophthisis and MCKD | Unknown | 2 | F | 29 | Not provided |
| 84 | Not provided | Unknown | 5 | M | 14 | Not provided |
| 85 | X-linked hypophosphatemic rickets | Unknown | 3 | F | 1 | Caucasian, non-Hispanic |
| 86 | Orofaciodigital syndrome I | Unknown | 2 | F | 21 | Caucasian, non-Hispanic |
| 87 | Bilateral cystic kidneys | Unknown | 2 | M | 0b | Native American, Hispanic or Latino |
| 88 | Renal tubular acidosis | Unknown | 1 | F | 9 | Caucasian, Hispanic |
| 89 | Childhood nephrotic syndrome, possibly collapsing FSGS | Unknown | 4 | F | 9 | African/African-American, non-Hispanic |
| 90 | Alport syndrome | N | 4 | F | 6 | Caucasian |
| 91 | CKD, looking for APOL1 risk variants | N | 4 | F | 18 | African/African-American |
| 92 | Bilateral cystic kidney disease | Unknown | 2 | F | 14 | Caucasian, non-Hispanic |
| 93 | Congenital bilateral echogenic kidneys with small cysts | N | 2 | F | 5 | Not provided |
| 94 | Failure to thrive, presented with HTN and chronic renal failure | N | 5 | F | 6 | Caucasian |
| 95 | FSGS and hypertension | Unknown | 4 | M | 54 | Not provided |
| 96 | Alport syndrome, branchio-oto-renal syndrome (BOR), ESRD, nephronophthisis | Unknown | 2, 4 | M | 16 | Caucasian |
| 97 | Bartter syndrome | Unknown | 3 | F | 2 | Multiracial, Hispanic or Latino |
| 98 | Autosomal recessive polycystic kidney disease | Unknown | 2 | M | 0b | Caucasian |
| 99 | Polycystic kidney disease | Y | 2 | M | 7 | Caucasian |
| 100 | Nephrotic syndrome | N | 4 | M | 2 | Caucasian |
| 101 | Chronic kidney stones and alkaline urine | Unknown | 2 | M | 18 | Not provided |
| 102 | Autosomal recessive polycystic kidney disease | Unknown | 2 | M | 0b | Brazilian/Mexican, Hispanic or Latino |
| 103 | Nephrotic-range proteinuria | N | 4 | M | <1 | Caucasian |
| 104 | Papillorenal syndrome (renal-coloboma syndrome) | N | 1 | M | 2 | Caucasian, Hispanic or Latino |
| 105 | Not provided | N | 5 | M | 14 | Caucasian |
| 106 | ADPKD | N | 2 | M | 12 | Caucasian |
| 107 | Congenital nephrotic syndrome | Unknown | 4 | F | 0b | Hispanic or Latino |
| 108 | Not provided | Unknown | 5 | F | 6 | Not provided |
| 109 | Isolated multicystic dysplastic kidney disease and polycystic kidney disease | Unknown | 2 | M | 7 | Not provided |
| 110 | NDI | N | 3 | M | 1 | Caucasian, non-Hispanic |
| 111 | BOR or isolated CAKUT | Unknown | 1 | F | 2 | Not provided |
| 112 | Dent disease, Bartter or Gitelman syndromes | Unknown | 3 | M | 23 | Caucasian, non-Hispanic |
| 113 | ESRD of unknown etiology | Y | 5 | M | 20 | Hispanic or Latino |
| 114 | IgA nephropathy or FSGS | N | 4 | M | 11 | African/African-American |
| 115 | FSGS or diffuse mesangial sclerosis | Unknown | 4 | M | 4 | Caucasian |
| 116 | Alport syndrome | Y | 4 | M | 13 | Caucasian, non-Hispanic |
| 117 | Liddle syndrome | Unknown | 3 | F | 4 | Not provided |
| 118 | Nephrotic syndrome | Unknown | 4 | M | 8 | African/African-American |
| 119 | CDK Stage 2, FSGS | Unknown | 4 | F | 16 | African/African-American, non-Hispanic |
| 120 | ESRD due to FSGS | Unknown | 4 | F | 20 | Not provided |
| 121 | Juvenile nephronophthisis | Unknown | 2 | M | <1 | Not provided |
| 122 | Zellweger syndrome, Galloway–Mowat syndrome, podocytopathy | Unknown | 4 | M | 1 | Caucasian, non-Hispanic |
| 123 | Steroid-resistant nephrotic syndrome | Unknown | 4 | M | <1 | Caucasian, non-Hispanic |
| 124 | Bartter/Gitelman syndromes, pseudohypoaldosteronism Type 1 | Unknown | 3 | M | <1 | African/African-American |
| 125 | Nephronophthisis | Unknown | 2 | M | 15 | Caucasian |
| 126 | Nephronophthisis | N | 2 | F | 12 | Native Hawaiian or other Pacific Islander, non-Hispanic |
| 127 | Bartter syndrome, Gitelman syndrome or NDI | Y | 3 | M | 2 | Caucasian, non-Hispanic |
Disease category is associated with indication for testing.
Disease categories: 1 = CAKUT; 2 = ciliopathies or tubulointerstitial disease; 3 = disorders of tubular ion transport; 4 = glomerulopathies; 5 = unclassified or other.
ADTKD, autosomal dominant tubulointerstitial disease; ARPKD, autosomal recessive polycystic kidney disease; CUA, calcific uremic arteriolopathy; F, female; HTN, hypertension; LVH, left ventricular hypertrophy; M, male; MCD, minimal change disease; MCKD, medullary cystic kidney disease; TBM, thin basement membrane disease; U/S, ultrasound VSD, ventricular septal defect; Y/N, yes/no.
Variant identification and diagnostic rates in renal patients
A genetic diagnosis was made in 54 patients (43%) (Table 4; 46% solve rate between 0–14 years; 46% from 15–30 years and 22% in those >30 years). By disease group, the solve rate was 54% for CAKUT (7 of 13 patients), 53% for ciliopathies/tubulointerstitial diseases (17 of 32 patients), 45% for disorders of tubular transport (13 of 29 patients) and 33% for glomerulopathies (15 of 43 patients) (Figure 1 and Table 4). A number of identified variants were classified as VUSs as they did not meet ACMG criteria for pathogenicity or likely pathogenicity (Tables 5–7).
Table 4.
Patients with a positive genetic diagnosis, showing indication(s) for testing, disease type, genetic variant(s), zygosity, ACMG classification, mean allele frequency and genetic diagnosis
| Case | Indication for testing | Family history | Disease categorya | Sex | Age (years) | Race/ ethnicity | Gene | Variant | Zygosity | ACMG classification [17] | MAF gnomADb | Genetic diagnosis (AD/AR/XLR) | Disease category changea | First reported |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Bilateral multicystic dysplastic kidneys | Y | 1 | F | <1 | H | PKD1 | NM_000296: c.11575delG, p.Ala3859Profs * 85 | het | Pathogenic (PVS1, PM1, PM2) | Not reported | ADPKD | 2 | This manuscript |
| PKD2 | NM_000297: c.2T>A, p.Met1Lys | het | Likely pathogenic (PVS1, PM2, PP3) | 0.02% LAT | http://pkdb.mayo.edu/ | |||||||||
| 2 | Renal dysplasia | Unknown | 1 | M | 2 | 1 | HNF1B | NM_000458: c.516C>G, p.Tyr172* | het | Likely pathogenic (PVS1, PM2, PP3) | Not reported | HNF1B-related nephropathy (AD) | This manuscript | |
| 3 | Stage V (CKD), hearing loss | Unknown | 4 | M | 37 | 4 | COL4A5 | NM_000495: c.529G>C, p.Gly177Arg | hemi | Pathogenic known (PS1, PM1, PM2, PP3) | Not reported | Alport syndrome (XLD) | [18] | |
| 7 | Dent disease (NDI, failure to thrive, anion gap metabolic acidosis) | Unknown | 3 | M | 2 | 1 | AQP2 | NM_000486: c.502G>A, p.Val168Met | het | Pathogenic known PS1, PM2, PP3 | 0.041% LAT | NDI (AR) | [19] | |
| NM_000486: c.656A>G, p.Tyr219Cys | het | Likely pathogenic PM1, PM2, PP3, PP4 | Not reported | This manuscript | ||||||||||
| 8 | Nephronophthisis | Y | 2 | F | 10 | 2 | RPGRIP1L | NM_001127897: c.3118 + 1G>A | het | Pathogenic (PVS1, PM2, PP3) | 0.011% AFR | COACH syndrome (AR) or Joubert syndrome (AR) | This manuscript | |
| NM_001127897: c.1329_1330insA, p.Arg444Thrfs * 10 | het | Pathogenic (PVS1, PM2, PP3) | Not reported | This manuscript | ||||||||||
| 11 | Autosomal dominant polycystic kidney disease | Unknown | 2 | M | 27 | 1 | NPHP1 | NM_000272: c.1756C>T, p.Arg586 * | het | Pathogenic known (PVS1, PS1, PM2, PP3) | 0.0009% NFE | Nephronophthisis 1 (AR) | [20] | |
| Deletion of NPHP1 gene region on chr2 | het | Pathogenic (PVS1, PS1, PM3, PP3) | [21] | |||||||||||
| 20 | Alport syndrome | Unknown | 4 | M | 5 | 4 | COL4A5 | NM_000495: c.1843G>A, p.Gly615Arg | hemi | Likely pathogenic known (PM1, PM2, PM5, PP2, PP3, PP5) | Not reported | Alport syndrome (XLD) | [22] | |
| 23 | U/S prenatal echogenic kidneys, postnatal bilateral cysts, HNF1B disease | Unknown | 1 | M | <1 | 1 | PKD1 | NM_000296: c.8597T>C, p.Leu2866Pro | het | Likely pathogenic known (PS1, PM2, PP3, PP5) | Not reported | ADPKD | 2 | [23] |
| 24 | Bartter syndrome or other | Unknown | 3 | M | 1 | Unknown | KCNJ1 | NM_000220: c.123G>C, p.Arg41Ser | hom | Likely pathogenic (PM1, PM2, PM3, PP2 PP3) | Not reported | Bartter syndrome (AR) | This manuscript | |
| 26 | Bilateral hypoplastic dysplastic kidneys | Unknown | 1 | M | <1 | 1H | EYA1 | NM_000503: c.922C>T, p.Arg308 * | het | Pathogenic known (PVS1, PS3, PM2, PP3) | Not reported | Branchio-oto-renal syndrome (AD) | [24] | |
| 29 | Alport or thin basement membrane disease | Unknown | 4 | M | 18 | 1 | COL4A3 | NM_000091: c.1408 + 2T>C | het | Pathogenic (PVS1, PM2, PP3) | Not reported | Alport syndrome (AD)/thin basement membrane disease (AD) | This manuscript | |
| 33 | Alport syndrome; hearing loss, microscopic hematuria, CKD | Unknown | 4 | M | 12 | 1 | COL4A4 | NM_000092: c.4522G>A, p.Gly1508Ser | het | Likely pathogenic (PS1, PM2, PP3) | 0.00089% NFE | Alport syndrome (AR) | [25] | |
| chr2: 227892566 227974060 del | het | Pathogenic (PVS1, PM2, PM4, PP3) | This manuscript | |||||||||||
| 37 | Bartter syndrome, NDI or Dent disease; polyuria, polydipsia, hypercalciuria, medullary nephrocalcinosis | Unknown | 3 | M | 16 | 1 | SLC12A1 | NM_000338: c.1652C>T, p.Thr551Ile | het | Likely pathogenic (PM1, PM2, PM3, PP3) | 0.0009% NFE | Bartter syndrome (AR) | This manuscript | |
| NM_000338: c.2807G>A, p.Trp936* | het | Pathogenic (PVS1, PM2, PM4) | Not reported | [26] | ||||||||||
| 38 | Pseudohypoaldosteronism; hyperkalemia, polyuria | Unknown | 3 | M | <1 | H | SCNN1B | NM_000336: c.682delG, p.Ala228Hisfs*8 | het | Pathogenic (PVS1, PM2, PM4, PP3) | Not reported | Pseudohypoaldosteronism I (AR) | This manuscript | |
| chr16: 23313555-23315510 del | het | Pathogenic (PVS1, PM2, PP3) | This manuscript | |||||||||||
| 39 | Multicystic bilateral kidneys | Unknown | 1 | M | <1 | 1 | HNF1B | Full gene deletion chr17: 36047234-36104883 del | het | Pathogenic known (PVS1, PM2, PP3) | HNF1B-related nephropathy (AD) | [27] | ||
| 41 | Bartter syndrome; polyuria, metabolic alkalosis | Unknown | 3 | F | 3 | 1 | SLC12A1 | NM_000338: c.2873 + 2_2873 + 3insT | het | Pathogenic (PVS1, PM2, PP3) | 0.0017 % NFE | Bartter syndrome (AR) | This manuscript | |
| NM_000338: c.3164 + 1G>A | het | Pathogenic known (PVS1, PS1 PM2, PP3) | 0.0012% NFE | [28] | ||||||||||
| 42 | Liddle syndrome; early onset hypertension and hypokalemia | Y | 3 | F | 19 | 1H | HSD11B2 | NM_000196: c.623G>A, p.Arg208His | het | Pathogenic (PS1, PS3, PM2, PP3) | 0.006% LAT | Syndrome of apparent mineralocorticoid excess (AR) | [29] | |
| NM_000196: c.667G>A, p.Asp223Asn | het | Pathogenic (PS1, PS3, PM2, PP3) | 0.033% LAT | [30] | ||||||||||
| 45 | Cystinuria | Y | 3 | F | 19 | 1 | SLC7A9 | NM_001126335: c.775G>A, p.Gly259Arg | hom | Pathogenic known (PS1, PM2, PM3, PP2, PP3 | 0.0018% NFE | Cystinuria (AR) | [31] | |
| 52 | PKD1, PKD2, HNF1B | Unknown | 2 | M | 6 | H | PKD1 | NM_000296: c.9395C>T, p.Ser3132Leu | het | Likely pathogenic known (PM1, PM2, PP3, PP5) | Not reported | ADPKD | [32] | |
| 53 | Renal cysts | Y | 2 | F | 49 | 4 | PKD1 | NM_000296: c.10102G>A, p.Asp3368Asn | het | Likely pathogenic known (PS1, PM2, PP3) | 0.3% EA | ADPKD | [33] | |
| UMOD | NM_001008389: c.854C>A, p.Ala285Glu | het | VUS (PM2, PP2, PP3) | Not reported | ||||||||||
| 59 | Bartter/Gitelman syndrome; hypokalemia, hypomagnesemia and metabolic alkalosis. | Unknown | 3 | M | 12 | Unknown | SLC12A3 | NM_000339: c.1836G>T, p.Trp612Cys | hom | Likely pathogenic known (PM1, PM2, PM3, PP3, PP5) | Not reported | Gitelman syndrome (AR) | [34] | |
| CLCNKB | Full gene deletion | het | Likely pathogenic known (PS1, PM2, PM4) | [35] | ||||||||||
| 60 | Nephronophthisis or medullary cystic kidney disease | Y | 2 | M | 58 | 1 | UMOD | UMOD (c.278_289del TCTGCCCCGAAGinsCCGCCTCCT; p.V93_G97del/ins AASC | het | Likely pathogenic known (PS1, PM, PM4) | Not reported | Tubulointerstitial kidney disease (AD) | [36] | |
| 61 | Polycystic kidney disease | Unknown | 2 | F | 51 | 2 | PKD1 | NM_000296: c.6356delA | het | Pathogenic (PVS1, PM2, PP3) | Not reported | ADPKD | This manuscript | |
| 63 | FSGS, multicystic dysplastic kidney | Y | 4/1 | M | 15 | 1 | PAX2 | NM_000278: c.419G>T, p.Arg140Leu | het | Likely pathogenic (PM1, PM2, PP1, PP3) | Not reported | FSGS (AD)/CAKUT | This manuscript | |
| 65 | Hypophosphatemic rickets; distal renal tubular acidosis; isolated proximal renal tubular acidosis, generalized proximal defect | N | 3 | F | <1 | H | ATP6V0A4 | NM_020632: c.154_157 del GTGAp.Val 52 Metfs*25 | het | Pathogenic (PVS1, PM2, PP3) | Not reported | Distal renal tubular acidosis (AR) | This manuscript | |
| NM_020632: c.1231G>T, p.Asp411Tyr | het | Likely pathogenic (PM2, PM3, PP3, PP5) | 0.042% LAT | ClinVar (likely pathogenic) | ||||||||||
| 68 | Kidney stones, paresthesias, hypercalciuria, hypoparathyroidism, ESRD | Y | 3 | M | 58 | 1 | CASR | NM_000388: c.2506G>C, p.Val836Leu | het | Likely pathogenic (PM1, PM2, PP2, PP3) | Not reported | Hypocalcemia (AD) | This manuscript | |
| 69 | Large cystic kidneys | N | 2 | M | 27 | 1 | PKD1 | NM_000296: c.8311G>A, p.Glu2771Lys | het | Likely pathogenic known (PS1, PM1, PM2, PP3) | Not reported | ADPKD | [37] | |
| 70 | Renal cystic dysplasia, ectopic atrial tachycardia, CUA, seizures, LVH; dialysis from birth | Unknown | 2 | F | <1 | 1 | WT1 | NM_000378: c.1249C>T, p.Arg417Cys | het | Likely pathogenic (PS1, PM2, PP3) | Not reported | DDS (AD) | 3 | [38] |
| 79 | Premature newborn with severely enlarged cystic kidneys noted mid-trimester, severe oligohydramnios, pulmonary hypoplasia | N | 2 | F | <1 | 1H | PKHD1 | NM_138694.3: c.9689delA, p.Asp3230Valfs*34 | het | Pathogenic known (PVS1, PM2, PP3, PP4) | 0.039% LAT | ARPKD | [39] | |
| NM_138694.3: c.6297_6300delTG, p.Gln2100Glyfs*7 | het | Pathogenic known (PVS1, PM2, PP3, PP4) | Not reported | [40] | ||||||||||
| 80 | Alport syndrome | Unknown | 4 | F | 11 | 1 | COL4A5 | NM_000495: c.1117C>T, p.Arg373* | het | Pathogenic known (PVS1, PM1, PM2, PP3) | Not reported | Alport syndrome (XLD) | [18] | |
| 84 | Not provided | Unknown | 5 | M | 14 | Unknown | NPHP1 | Whole gene deletion | hom | Pathogenic known (PVS1, PS1, PM2) | Nephronophthisis 1 (AR) | 2 | [41] | |
| 86 | Orofaciodigital syndrome I | Unknown | 2 | F | 21 | 1 | OFD1 | NM_003611: c875_876delAT, p.Met293Glyfs*15 | het | Pathogenic known (PVS1, PM2, PP3) | Not reported | Orofaciodigital syndrome I (AD) | [42] | |
| 87 | Bilateral cystic kidneys | Unknown | 2 | M | <1 | 3H | PKHD1 | NM_138694: c.9559delT, p.Ser3187Leufs*33 | het | Pathogenic (PVS1, PM2, PP3) | Not reported | ARPKD | This manuscript | |
| NM_138694: c.107C>T, p.Thr36Met | het | Likely pathogenic known (PS1, PP3, PP5) | 0.08% NFE | [43] | ||||||||||
| 90 | Alport syndrome | N | 4 | F | 6 | 1 | NPHS2 | NM_014625: c.871C>T, p.Arg291Trp | het | Likely pathogenic known (PS1, PM1, PM2, PP2, PP3, PP5) | 0.029% EA | Steroid-resistant nephrotic syndrome (AR) | [44] | |
| NM_014625: c.686G>A, p.Arg229Gln | het | Likely pathogenic when inherited with a pathogenic known (PS3, PM1, PP2, PP3, PP5) | 6.98% FE | [44] | ||||||||||
| 93 | Congenital bilateral echogenic kidneys with small cysts | N | 2 | F | 5 | Unknown | HNF1B | Whole gene deletion | het | Pathogenic known (PVS1, PM2, PP3) | HNF1B-related nephropathy | [27] | ||
| 94 | Failure to thrive, presented with hypertension and CKD | N | 5 | F | 6 | 1 | TTC21B | NM_024753: c.1516 + 2T>C | het | Pathogenic (PVS1, PM1, PP3) | 0.0009% NFE | Juvenile nephronophthisis (AR), Jeune syndrome (AR), or Joubert syndrome (AR) | 2 | This manuscript |
| NM_024753: c.626C>T, p.Pro209Leu | het | Likely pathogenic known (PS3 PM2, PP3, PP5) | 0.03% LAT | [45] | ||||||||||
| 96 | Alport syndrome, branchio-oto-renal syndrome (BOR), ESRD, nephronophthisis | Unknown | 4 | M | 16 | 1 | COL4A5 | NM_000495: c.796C>T, p.Arg266* | hemi | Pathogenic known (PVS1, PM1, PM2, PM4, PP3, PP5) | Not reported | Alport syndrome (XLD) | [46] | |
| 97 | Bartter syndrome | Unknown | 3 | F | 2 | H | KCNJ1 | NM_000220: c.924C>A, p.Cys308* | het | Pathogenic (PVS1, PM2, PP3, PP5) | Not reported | Bartter syndrome (AR) | This manuscript | |
| NM_000220: c.683G>A, p.Gly228Glu | het | Likely pathogenic known (PS1, PM2, PP3, PP4) | 0.0018% NFE | [47] | ||||||||||
| 98 | Autosomal recessive polycystic kidney disease | Unknown | 2 | M | <1 | 1 | PKHD1 | NM_138694: c.7717C>T, p.Arg2573Cys | het | Likely pathogenic known (PS1, PM2, PP3, PP4, PP5) | 0.0058% EA | ARPKD | [48] | |
| NM_138694: c.3766delC, p.Gln1256Argfs*47 | het | Pathogenic known (PVS1, PS1, PP3) | 0.12% LAT | [48] | ||||||||||
| 99 | Polycystic kidney disease | Y | 2 | M | 7 | 1 | PKD1 | NM_000296: c.12230_12231delAG, p.Glu4077Valfs*78 | het | Pathogenic (PVS1, PM2, PP3) | Not reported | ADPKD | This manuscript | |
| 103 | Nephrotic range proteinuria | N | 4 | M | <1 | 1 | CLCN5 | NM_000084: c.1546C>T, p.Arg516Trp | hemi | Pathogenic known (PS1, PS3, PM2, PP2, PP3, PP5) | Not reported | Dent disease | 3 | [49] |
| 104 | Papillorenal syndrome (renal-coloboma syndrome) | N | 1 | M | 2 | 1H | PAX2 | NM_000278: c.69delC, p.Val26CysfsX2 | het | Pathogenic known (PVS1, PM2, PP3, PP4, PP5) | Not reported | Papillorenal syndrome (AD) | [50] | |
| 106 | Autosomal dominant polycystic kidney disease | N | 2 | M | 12 | 1 | PKD1 | NM_000296: c.776G>A, p.Cys259Tyr | het | Likely pathogenic known (PS1, PM2, PP3, PP5) | 0.027% NFE | ADPKD | [23] | |
| 113 | ESRD of unknown etiology | Y | 5 | M | 20 | H | NPHP1 | Whole gene deletion | hom | Pathogenic known (PVS1, PS1, PM2) | Nephronophthisis 1 (AR) | 2 | [41] | |
| 114 | IgA nephropathy or FSGS | N | 4 | M | 11 | 2 | COL4A4 | NM_000092: c.1856G>A, p.Gly619Asp | het | Likely pathogenic known (PS1, PM2, PP3) | 0.0066% AFR | Alport syndrome (AD) | [51] | |
| 115 | FSGS or diffuse mesangial sclerosis | Unknown | 4 | M | 4 | 1 | WT1 | NM_000378: c.1333C>T, p.Arg445Trp | het | Pathogenic known (PS1, PS3, PM2, PP3) | Not reported | DDS (AD) | [52] | |
| 116 | Alport syndrome | Y | 4 | M | 13 | 1 | COL4A5 | NM_000495: c.1226G>A, p.Gly409Asp | het | Likely pathogenic known (PM1, PM2, PP2, PP3, PP5) | Not reported | Alport syndrome (XLD) | [18] | |
| 118 | Nephrotic syndrome | Unknown | 4 | M | 8 | 2 | APOL1 | NM_001136540: c.1024A>G, p.Ser342Gly | hom | Risk allele | 23% AFR | Dent disease (XLR) and APOL1 G1/G1 | 3 | [53] |
| NM_001136540: c.1152T>G, p.Ile384Met | hom | Risk allele | 22.9% AFR | [53] | ||||||||||
| CLCN5 | NM_000084: c.1909C>T, p.Arg637 * | hemi | Pathogenic known (PS1, PVS1, PM2, PP3, PP5) | Not reported | [54] | |||||||||
| 120 | ESRD due to FSGS | Unknown | 4 | F | 20 | Unknown | PAX2 | NM_000278: c.70_71insG, p.Val26Glyfx*28 | het | Pathogenic known (PS1, PVS1, PM1, PM2, PP3, PP5) | 0.0068% AFR | FSGS (AD); APOL1 G2/G2 | [55] | |
| APOL1 | NM_001136540: c.1160_1165 delATAATT | het | Risk allele | 14.14% AFR | [53] | |||||||||
| 122 | Zellweger syndrome, Galloway–Mowat syndrome, podocytopathy | Unknown | 4 | M | 1 | 1 | OCRL | NM_000276: c.1484C>T, p.Pro495Leu | hemi | Pathogenic known (PS3, PM1, PM2, PP2, PP3, PP5) | Not reported | Lowe syndrome (XLR) | 3 | [56] |
| 124 | Bartter/Gitelman syndromes, pseudohypoaldosteronism type 1 | Unknown | 3 | M | <1 | 2 | NR3C2 | NM_000901: c.1002_1003insGT, p.Ser335Valfs*4 | het | Pathogenic (PVS1, PM2, PP3) | Not reported | Pseudohypoaldosteronism I (AD) | This manuscript | |
| 125 | Nephronophthisis | Unknown | 2 | M | 15 | 1 | NPHP1 | Whole gene deletion | hom | Pathogenic known (PVS1, PS1, PM2) | Nephronophthisis 1 (AR) | [41] | ||
| 126 | Nephronophthisis | N | 2 | F | 12 | 5 | NPHP1 | Whole gene deletion | hom | Pathogenic known (PVS1, PS1, PM2) | Nephronophthisis 1 (AR) | [41] |
Patients in whom the genetic diagnosis changed the clinical diagnosis are shown in bold font.
Disease category: 1 = CAKUT; 2 = ciliopathies or tubulointerstitial disease; 3 = disorders of tubular ion transport; 4 = glomerulopathies; 5 = unclassified or other. Ethnicity: 1 = Caucasian; 2 = African/African-American; 3 = American Indian or Alaska Native; 4 = Asian; 5 = Native Hawaiian or other Pacific Islander; H = Hispanic or Latino. Zygosity: het, heterozygous; hom, homozygous; hemi, hemizigous.
gnomAD: highest MAF reported.
AFR, African; EA, East Asian; FE, European Finnish; NFE, European (non-Finnish); LAT, Latino; SA, South Asian; AD, autosomal dominant; AR, autosomal recessive; XLR, X-linked recessive; LVH, left ventricular hypertrophy; ARPKD, autosomal recessive polycystic kidney disease; M, male; F, female.
FIGURE 1.
Outcome of KidneySeq panel testing in 127 renal patients. The positive diagnosis rate in each disease category is shown together with the percentage where diagnosis changed. A pie chart shows the number and types of pathogenic variants and the overall solve rate.
Table 5.
VUSs
| Case | Indication for testing | Family history | Disease categorya | Sex | Age (years) | Ethnicity | Gene | Variant | Zygocityb | ACMG classification/ rules [17] | MAF Gnomadc | First reported by | Possibly causald |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | Proteinuria, FSGS | Y | 4 | M | 54 | 2 | FN1 | NM_002026: c.5779C>T, p.Arg1927Cys | het | PM1, PM2, PP3 | 0.007% NFE | Glomerulopathy with fibronectin deposits (AD) | Y |
| 16 | Dilated cardiomyopathy and associated hypomagnesemia | N | 3 | M | 3 | Caucasian | ROBO2 | NM_002942: c.2834T>C, p.Ile945Thr | het | PS3, PM2, PP5 | 0.0027% NFE | [57] | N |
| 17 | Fanconi syndrome, hypophosphatemic rickets | Unknown | 3 | M | 2 | Caucasian, Aboriginal | SLC4A1 | NM_000342: c.2396C>T, p.Ser799Leu | het | PM2, PP3 | 0.0045% NFE | This manuscript | N |
| 18 | ESRD, primary FSGS | Unknown | 4 | M | 55 | Caucasian | ACTN4 | NM_004924: c.2680G>A, p.Gly894Ser | het | PP3 | 0.18% NFE | This manuscript | Y |
| 19 | Severe CAKUT | Unknown | 1 | M | <1 | Caucasian, Hispanic | DSTYK | NM_015375: c.2216G>A, p.Arg739Gln | het | PM2, PP3 | 0.25% LAT | This manuscript | Y |
| 22 | Interstitial nephritis | Unknown | 2 | F | 10 | Caucasian | NPHP4 | NM_015102: c.2849G>A, p.Arg950Gln | het | PM2, PP3 | 0.082% EA | This manuscript | Y |
| NM_015102: c.2542G>A, p.Arg848Trp | het | PM2, PP3, BP6 | 2.56% EF | [58] | Y | ||||||||
| 25 | ESRD, tubulointerstitial disease | Y | 2 | M | 51 | Africa/African-American | CC2D2A | NM_001080522: c.3157A>G, p.Ile1053Val | het | PM2, PP3 | 0.047% AFR | This manuscript | Y |
| NM_001080522: c.3503G>A, p.Arg1168His | het | PM1, PM2, PP3 | 0.035% AFR | This manuscript | |||||||||
| 30 | FSGS, SRNS, hypoalbuminemia | Unknown | 4 | M | 17 | Caucasian non-Hispanic | NPHP3 | NM_153240: c.2881C>G, p.Gln961Glu | het | PP3, PM2 | 0.055% NFE | This manuscript | N |
| 34 | Renal agenesis/hypoplasia or nephronophthisis | Y | 1, 2 | F | 16 | Hispanic | SIX2 | NM_016932: c.126C>G, p.His42Gln | het | PM2, PP3 | Not reported | This manuscript | Y |
| NPHP4 | NM_015102: c.3055G>A, p.Asp1019Asn | het | PM2, PP3 | Not reported | This manuscript | N | |||||||
| 35 | Gitelman/Bartter syndrome; metabolic alkalosis, hypomagnesemia, hypokalemia | Unknown | 3 | F | 17 | Caucasian | KLHL3 | NM_001257194: c.1357G>A, p.Val453Ile | het | PM2, PP2 | 0.002% NFE | This manuscript | N |
| 42 | Liddle syndrome. Early onset hypertension and hypokalemia | Y | 3 | F | 19 | Caucasian, Hispanic | KLHL3 | NM_001257194: c.988C>T, p.Arg330Trp | het | PM2, PP2, PP3, | 0.002% NFE | [59] | N |
| 44 | NDI, medullary nephrocalcinosis, vesicoureteral reflux, hypophosphatemia | Unknown | 3 | F | 3 | Caucasian, non-Hispanic | ANOS1 | NM_000216: c.1759G>T, p.Val587Leu | het | PM1, PM2, PP3, PP5 | Not reported | [60] | N |
| 46 | FSGS or minimal change disease. Persistent proteinuria | Unknown | 4 | M | 5 | Caucasian, non-Hispanic | ANOS1 | NM_000216: c.2015A>G, p.His672Arg | het | PP5 | 0.044% NFE | [61] | N |
| 50 | Proximal tubulopathy or Dent or hypophosphatemic rickets. Nephrocalcinosis, small stature | Unknown | 3 | F | 13 | Asian | FAH | NM_000137: c.181G>T, p.Val61Phe | het | PP3 | 1.907% EA | This manuscript | Y |
| 51 | FSGS. Post deceased kidney transplant | Unknown | 4 | M | 15 | Hispanic | LMX1B | NM_001174146: c.875G>T, p.Arg292Leu | het | PP2, PP3 | 0.21% LAT | This manuscript | Y |
| LAMB2 | NM_002292: c.5234C>A, p.Ala1745Asp | het | PM2, PP3 | Not reported | This manuscript | N | |||||||
| 53 | Renal cysts. Family history of hereditary nephritis | Y | 2 | F | 49 | Asian | UMOD | NM_001008389: c.854C>A, p.Ala285Glu | het | PM2, PP2, PP3 | Not reported | This manuscript | Y |
| 54 | Polycystic kidney disease, undescended testes, HTN | N | 2 | M | <1 | Caucasian, non-Hispanic | NPHS1 | NM_004646: c.563A>T, p.Asn188Ile | het | LB* (PM1, PP5, BP4, BP6) | 0.93% NFE | [62] | N |
| TRAP1 | NM_001272049: c.598A>G, p.Ile200Val | het | PP3 | 2.05% EF | ClinVar | N | |||||||
| 57 | Moderate CKD | Unknown | 5 | M | 1 | Unknown | ACE | NM_000789: c.793C>T, p.Arg265* | het | Pathogenic known (PVS1, PM2, PM4, PP3) | 0.0027% NFE | [63] | Y |
| NM_000789: c.3136G>A, p.Glu1046Ser | het | PM2, PM3, PP3 | Not reported | This manuscript | Y | ||||||||
| 58 | Not provided | Unknown | 5 | F | 16 | Not provided | ACE | NM_000789: c.955G>T, p.Ala319Ser | het | PM2 | Not reported | This manuscript | N |
| GLI3 | NM_000168: c.1616G>A, p.Arg539Lys | het | PM2 | Not reported | This manuscript | U | |||||||
| KLHL3 | NM_001257194: c.203C>T, p.Thr68Met | het | PM2, PP2 | 0.012% NFE | This manuscript | U | |||||||
| SLC3A1 | NM_000341: c.788G>C, p.Ser263Thr | het | PP2, PP3 | 0.27% AFR | ClinVar | U | |||||||
| SMARCAL1 | NM_001127207: c.1271A>T, p.Asp424Val | het | PP3, BP6 | 0.35% NFE | ClinVar | N | |||||||
| 60 | Nephronophthisis or medullary cystic kidney disease | Y | 2 | M | 58 | Caucasian, non-Hispanic | TRAP1 | NM_001272049: c.598A>G, p.Ile200Val | het | PP3, BS1 | 2.05% EF | ClinVar | N |
| 63 | FSGS/multicystic dysplastic kidney | Y | 1, 4 | M | 15 | Caucasian, non-Hispanic | PKD1 | NM_000296: c.971G>T, p.Arg324Leu | het | PM1, PP5 | 0.59% EF | Uniprot | N |
| 64 | Hyperplastic nephrogenic rests, features seen with underlying syndromes such as Beckwith-Wiedemann | Unknown | 5 | F | <1 | Not provided | CHD1L | NM_001256336: c.2179A>G, p.Ile727Val | het | 0.47% NFE | This manuscript | N | |
| IQCB1 | NM_001023570: c.1441G>A, p.Glu481Lys | het | PM1, PP3, BP1 | 0.19% NFE | ClinVar | N | |||||||
| ANOS1 | NM_000216: c.1759G>T, p.Val587Leu | het | PM1, PM2, PP5 | Not reported | [60] | N | |||||||
| 67 | Horseshoe kidney, dysmorphic features, VSD | Y | 1 | F | <1 | Egyptian | PTPRO | NM_002848: c.433G>A, p.Glu145Lys | het | PM2 | 0.001% NFE | This manuscript | N |
| WT1 | NM_000378: c.563C>T, p.Ala188Val | het | 0.007% AFR | This manuscript | N | ||||||||
| 71 | Steroid-resistant nephrotic syndrome | N | 4 | F | 8 | Asian, multiracial | ANLN | NM_018685: c.1741G>C, p.Glu581Gln | het | 0.023% EA | This manuscript | Y | |
| CUBN | NM_001081: c.6095G>A, p.Cys2032Tyr | het | PM2, PP3, BP1 | 0.019% NFE | This manuscript | N | |||||||
| 73 | Glomerulocystic kidneys and hepatoblastoma | N | 2 | F | 3 | Hispanic | CHD1L | NM_001256336: c.1798G>A, p.Gly600Arg | het | PM2, BP4 | Not reported | This manuscript | Y |
| PKD2 | NM_000297: c.2398A>C, p.Met800Leu | het | PM2 | Not reported | Uniprot | N | |||||||
| TMEM67 | NM_001142301: c.272G>A, p.Arg91Gln | het | PM2, PP2, PP3, PP5 | 0.012% LAT | ClinVar | N | |||||||
| 75 | Steroid-resistant nephrotic syndrome | Y | 4 | M | 4 | Dominican Republic | BBS9 | NM_198428: c.1648A>G, p.Ile550Val | het | BP6 | 0.75% AFR | ClinVar | N |
| 76 | Gitelman syndrome | N | 3 | F | 23 | Not provided | EYA1 | NM_000503: c.403G>A, p.Gly135Ser | het | PP3 | 0.064% EA | ClinVar | N |
| 77 | Not provided | Y | 5 | M | 57 | Not provided | TRIM32 | NM_001099679: c.1688G>A, p.Arg563His | het | PM2, PP3 | 0.013% NFE | ClinVar | N |
| 78 | Nephronophthisis | Y | 2 | F | 38 | Caucasian | GLIS2 | NM_032575: c.278A>G, p.Asn93Ser | het | BP1, BP4 | 0.09% EF | This manuscript | N |
| TRPC6 | NM_004621: c.1030G>A, p.Ala344Thr | het | PM2 | Not reported | This manuscript | N | |||||||
| 82 | Global glomerulosclerosis | Y | 4 | F | 65 | African/African-American | COL4A4 | NM_000092: c.3143G>A, p.Gly1048Asp | het | PM2, PP3 | Not reported | This manuscript | Y |
| 83 | Juvenile nephronophthisis and medullary cystic kidney disease | Y | 2 | F | 29 | Not provided | SLC12A3 | NM_000339: c.1967C>T, p.Pro656Leu | het | PP2, PP3 | 0.021% NFE | This manuscript | N |
| 85 | X-linked hypophosphatemic rickets | Unknown | 3 | F | 1 | Caucasian, non-Hispanic | HOGA1 | NM_138413: c.700 + 5G>T | het | PP3, PP5 | 0.208% NFE | ClinVar | N |
| 88 | Renal tubular acidosis | Unknown | 3 | F | 9 | Caucasian, Hispanic | IFT140 | NM_014714: c.1541T>A, p.Leu514His | het | PP3, BP6 | 1.58% EF | ClinVar | N |
| 89 | Childhood nephrotic syndrome, possibly collapsing FSGS | Unknown | 4 | F | 9 | African/African-American | PKD1 | NM_000296: c.5866G>A, p.Val1956Met | het | – | 0.002% NFE | This manuscript | N |
| 90 | Alport syndrome | N | 4 | F | 6 | Caucasian | SLC7A9 | NM_001126335: c.544G>A, p.Ala182Thr | het | PP2, PP3, PP5 | 0.43% NFE | ClinVar | N |
| TMEM67 | NM_001142301: c.803T>C, p.Leu268Ser | het | PM2, PP2, PP3, PP5 | 0.004% NFE | [64] | N | |||||||
| 92 | Bilateral cystic kidneys | Unknown | 2 | F | 14 | 1 | PKD1 | NM_000296: c.8971T>G, p.Tyr2991Asp | het | PM1, PM2, PP3 | Not reported | This manuscript | Y |
| 93 | Congenital bilateral echogenic kidneys with small cysts | N | 2 | F | 5 | Not provided | SLC3A1 | NM_000341: c.647C>T, p.Thr216Met | het | PM2, PP2, PP3, PP5 | 0.018% NFE | [65] | N |
| 102 | Autosomal recessive polycystic kidney disease | Unknown | 2 | M | 0e | Brazilian/Mexican Hispanic | HNF4A | NM_000457: c.1133C>T, p.Ser378Phe | het | PM2 | Not reported | This manuscript | N |
| 107 | Congenital nephrotic syndrome | Unknown | 4 | F | 0e | Hispanic or Latino | COL4A1 | NM_001845: c.1366G>A, p.Glu456Lys | het | PM1, PP2, PP3 | 0.0058% EA | This manuscript | N |
| 108 | Not provided | Unknown | 5 | F | 6 | Not provided | IFT140 | NM_014714: c.886G>A, p.Gly296Arg | het | PM2, PP3 | 0.023% SA | This manuscript | N |
| LAMB2 | NM_002292: c.2974A>G, p.Ile992Val | het | – | 0.413% SA | This manuscript | N | |||||||
| 109 | Isolated multicystic dysplastic kidney disease and polycystic kidney disease | Unknown | 1, 2 | M | 7 | Not provided | ANOS1 | NM_000216: c.98G>C, p.Arg33Pro | het | PP3 | 0.072% LAT | This manuscript | Y |
| 110 | NDI | N | 3 | M | 1 | Caucasian, non-Hispanic | AGTR2 | NM_000686: c.395delT, p.Phe134Leufs*5 | het | PP3, BP6 | 0.102% NFE | ClinVar | N |
| 111 | Branchio-oto-renal syndrome or isolated CAKUT | Unknown | 1 | F | 2 | Not provided | CREBBP | NM_001079846: c.2458C>T, p.Pro820Ser | het | PP3, BP6 | 0.915% AFR | ClinVar | N |
| GRIP1 | NM_001178074: c.2633G>A, p.Arg878His | het | 0.052% AFR | This manuscript | N | ||||||||
| 112 | Dent disease, Bartter or Gitelman syndromes | Unknown | 3 | M | 23 | Caucasian, non-Hispanic | FRAS1 | NM_001166133: c.4648C>T, p.Leu1550Phe | het | BP1 | 0.22% EF | ClinVar | N |
| NLRP3 | NM_001079821: c.128G>A, p.Arg43Lys | het | PP2, BP4 | 0.002% NFE | This manuscript | N | |||||||
| PKD1 | NM_000296: c.7409C>A, p.Pro2470Gln | het | PP3 | 0.0022% NFE | This manuscript | N | |||||||
| 114 | IgA nephropathy or FSGS | N | 4 | M | 11 | African/African-American | PAX2 | NM_000278: c.1178G>C, p.Arg393Pro | het | PM2, PP2 | Not reported | This manuscript | Y |
| 121 | Juvenile nephronophthisis | Unknown | 2 | M | <1 | Not provided | NPHP3 | NM_153240: c.1181T>A, p.Ile394Asn | het | PM2, PP3 | 0.003% LAT | ClinVar | Y |
| NM_153240: c.460G>C, p.Ala154Pro | het | PM2, PP3 | Not reported | ClinVar | Y | ||||||||
| 123 | Steroid-resistant nephrotic syndrome | Unknown | 4 | M | <1 | Caucasian, non-Hispanic | COQ2 | NM_015697: c.854C>G, p.Pro285Arg | het | PM2, PP3, PP5 | 0.001% NFE | ClinVar (likely pathogenic) | Y |
| 125 | Nephronophthisis | Unknown | 2 | M | 15 | Caucasian | NR3C2 | NM_000901: c.731G>A, p.Arg244Gln | het | 0.004% NFE | This manuscript | N | |
| SIX2 | NM_016932: c.722C>T, p.Pro241Leu | het | BP6 | 0.44% FE | [66] | N | |||||||
| 127 | Bartter syndrome, Gitelman syndrome or NDI | Y | 3 | M | 2 | Caucasian, non-Hispanic | TNXB | Full gene deletion | het | LP* (PVS1, PM2) | N | ||
| SLC7A9 | NM_001126335: c.544G>A, p.Ala182Thr | het | PP2, PP3, PP5 | 0.43% NFE | ClinVar | N |
Disease category is associated with the indication for testing. 1 = CAKUT; 2 = Ciliopathies or tubulointerstitial disease; 3 = Disorders of tubular ion transport; 4 = Glomerulopathies; 5 = Unclassified or Other.
Zygosity: het= heterozygous; hom=homozygous; hemi= hemizigous.
gnomAD: highest minor allele frequency reported. AFR= African; EA= East Asian; FE= European Finnish; NFE= European (non-Finnish); LAT= Latino; SA= South Asian.
Yes (Y), no (N) or unknown (U).
Newborn.
Jewish# No gnomAD data.
M, male; F, female. HTN, hypertension; VSD, ventricular septal defect; CUA, calcific uremic arteriolopathy.
Newborn.
Table 6.
Risk alleles
| Case | Indication for testing | Family history | Disease categorya | Sex | Age (year) | Ethnicity | Gene | Variant | Zygocityb | ACMG classification/ rules [17] | MAF gnomADc | Associated disease | First reported by |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 9 | FSGS | Unknown | 4 | M | 54 | African/African-American | APOL1 | NM_001136540: c.1024A>G, p.Ser342Gly | hom | Risk allele | 23% AFR | FSGS, hypertensive nephrosclerosis and HIV associated nephropathy | [53] |
| G1/G1 | NM_001136540: c.1152T>G, p.Ile384Met | Risk allele | 22.9% AFR | [53] | |||||||||
| 15 | Hypercalcemia, hypocalciuria. Suspicion of CaSR inactivating mutation | N | 3 | F | 81 | Caucasian | CaSR | NM_000388: c.2956G>T, p.Ala986Ser | het | PM2, PP2, BP6 | Not reported | Hypercalcemia | [67] |
| 46 | FSGS or minimal change disease. Persistent proteinuria | Unknown | 4 | M | 5 | Caucasian, non-Hispanic | PLCG2 | NM_002661: c.3563C>T, p.Pro1188Leu | het | 0.0067% NFE | Steroid sensitive nephrotic syndrome | This manuscript | |
| 72 | MCD, unresponsive to steroids | N | 2 | F | 3 | African/African-American | APOL1 | NM_001136540: c.1024A>G, p.Ser342Gly | het | Risk allele | 23% AFR | FSGS, hypertensive nephrosclerosis and HIV associated nephropathy | [53] |
| G1/G2 | NM_001136540: c.1152T>G, p.Ile384Met | het | Risk allele | 22.9% AFR | [53] | ||||||||
| NM_001136540: c.1160_1165delATAATT, p.Asn388_Tyr389del | het | Risk allele | 14.14% AFR | [53] | |||||||||
| 101 | Chronic kidney stones and alkaline urine | Unknown | 3 | M | 18 | Not provided | ATP6V1B1 | NM_001692: c.298G>A, p.Asp100Asn | het | PP2, PP3 | 0.16% EA | Kidney stones | This manuscript |
| 118 | Nephrotic syndrome | Unknown | 4 | M | 8 | African/African-American | APOL1 | NM_001136540: c.1024A>G, p.Ser342Gly | hom | Risk allele | 23% AFR | FSGS, hypertensive nephrosclerosis and HIV associated nephropathy | [53] |
| G1/G1 | NM_001136540: c.1152T>G, p.Ile384Met | Risk allele | 22.9% AFR | [53] | |||||||||
| 119 | CDK Stage 2, FSGS | Unknown | 4 | F | 16 | African/African-American | APOL1 | NM_001136540: c.1024A>G, p.Ser342Gly | hom | Risk allele | 23% AFR | FSGS, hypertensive nephrosclerosis and HIV associated nephropathy | [53] |
| G1/G1 | NM_001136540: c.1152T>G, p.Ile384Met | Risk allele | 22.9% AFR | [53] | |||||||||
| 120 | ESRD due to FSGS | Unknown | 4 | F | 20 | Not provided | APOL1G2/G2 | NM_001136540: c.1160_1165delATAATT, p.Asn388_Tyr389del | hom | Risk allele | 14.14% AFR | FSGS, hypertensive nephrosclerosis and HIV associated nephropathy | [53] |
Disease category is associated with the indication for testing. 1 = CAKUT; 2 = Ciliopathies or tubulointerstitial disease; 3 = Disorders of tubular ion transport; 4 = Glomerulopathies; 5 = Unclassified or Other.
Zygosity: het, heterozygous; hom, homozygous; hemi, hemizigous.
gnomAD: highest minor allele frequency reported. AFR, African; EA, East Asian; NFE, European (non-Finnish).
Jewish# No gnomAD data.
N, no; M, male; F, female.
Table 7.
Pathogenic carriers
| Case | Indication for testing | Family history | Disease categorya | Sex | Age (years) | Ethnicity | Gene | Variant | Zygocityb | ACMG classification/rules [17] | MAF gnomADc | Reported in | Associated disease |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 75 | Steroid-resistant nephrotic syndrome | Y | 4 | M | 4 | Dominican Republic | BBS1 | Deletion chr11: 66278119-66301084 | het | This manuscript | BBS carrier | ||
| 83 | Juvenile nephronophthisis and medullary cystic kidney disease | Y | 2 | F | 29 | Not provided | SLC12A3 | NM_000339: c.1967C>T, p.Pro656Leu | het | PP2, PP3 | 0.021% NFE | [68] | Gitelman carrier |
| 85 | X-linked hypophosphatemic rickets | Unknown | 3 | F | 1 | Caucasian, non-Hispanic | HOGA1 | NM_138413: c.700 + 5G>T | het | PP2, PP5 | 0.21% NFE | [69] | Primary hyperoxaluria III carrier |
| 88 | Renal tubular acidosis | Unknown | 1 | F | 9 | Caucasian, Hispanic | IFT140 | NM_014714: c.1541T>A, p.Leu514His | het | PP3, BP6 | 1.58% FE | [70] | Jeune syndrome carrier |
| 108 | Not provided | Unknown | 5 | F | 6 | Not provided | SLC12A1 | NM_000338: c.1872delC | het | Pathogenic (PVS1, PM2, PP3) | 0.032% SA | This manuscript | Bartter syndrome 1 carrier |
| 111 | Branchio-oto-renal syndrome or isolated CAKUT | Unknown | 1 | F | 2 | Not provided | FGF23 | NM_020638: c.59delG, p.Ser20Thrfs*20 | het | LP* (PVS1, PM2) | Not reported | This manuscript | N |
| 112 | Dent disease, Bartter or Gitelman syndromes | Unknown | 3 | M | 23 | Caucasian, non-Hispanic | ATP7B | NM_000053: c.2972C>T, p.Thr991Met | het | Likely pathogenic PS3, PM1, PP2, PP3, PP5 | 0.24% NFE | [71] | Wilson disease carrier |
Disease category is associated with the indication for testing. 1 = CAKUT; 2 = Ciliopathies or tubulointerstitial disease; 3 = Disorders of tubular ion transport; 4 = Glomerulopathies; 5 = Unclassified or Other.
Zygosity: het, heterozygous; hom, homozygous; hemi, hemizigous.
gnomAD: highest minor allele frequency reported. FE, European Finnish; NFE, European (non-Finnish); SA, South Asian.
Jewish# No gnomAD data.
Y, yes; M, male; F, female.
DISCUSSION
We identified a genetic basis for disease in 54 of 127 (44%) patients, demonstrating that broad-based genetic testing can augment current clinical algorithms used to evaluate the renal patient. The solve rate for cases decreased with age from 46% for patients between 0 and 14 years to 22% for patients >30 years old. Among solved cases, 9 were X-linked, 22 were autosomal dominant and 22 were autosomal recessive (6 homozygous and 16 compound heterozygous variants). Family history was positive in six autosomal dominant disorders (13 unknown), four autosomal recessive disorders (14 unknown) and in one X-linked disorder (7 unknown). Pathogenic and likely pathogenic variants included missense (32 of 75), nonsense (9 of 75), canonical splice site variants (4 of 75), small indels (17 of 75) and large CNVs (10 of 75), demonstrating the power to detect all types of genetic variants (Figure 1).
In 41 of 54 patients with a genetic diagnosis, data confirmed the clinical impression (i.e. ADPKD as ADPKD, Bartter as Bartter, etc.) but also provided prognostic information, guided clinical management and/or enabled counseling (Figure 1 and Table 4). For example, the identification of a truncating variant in PKD1 (NM_000296: c.12230_12231delAG) in a 7-year-old child with polycystic kidney disease (Case 99) mandates regular evaluation for increasing kidney volume, since truncating PKD1 variants predict a median onset of end-stage renal disease (ESRD) at 55 years of age, substantially earlier than non-truncating PKD1 variants or any PKD2 variant [72]. In another example, the diagnosis of CKD at age 10 years (Case 8) in two fraternal twins born prematurely led to a clinical suspicion of juvenile nephronophthisis. We identified two null variants in RPGRIP1L, which is reported in the allelic disorders Joubert syndrome, COACH syndrome and Meckel syndrome. Patients with hypomorphic RPGRIP1L variants develop Joubert syndrome or COACH syndrome (Joubert features with congenital hepatic fibrosis), while those with null variants develop Meckel syndrome, which is considered to be at the more severe end of the clinical disease spectrum [73]. While the phenotype can be variable with Joubert and COACH syndrome, awareness of the type of genetic variants should prompt a careful and guided evaluation for extrarenal features, such as liver disease, that may require treatment.
In the remaining 13 cases (24%), genetic testing changed the clinical diagnosis, helped to direct future care, guided genetic counseling, and/or directed the evaluation process for living donor candidates. For example, the indication for screening in a 1-month-old (Case 26) was bilateral hypoplastic dysplastic kidneys. Upon testing, a null variant was identified in EYA1, consistent with the diagnosis of branchio-oto-renal syndrome 1 (BOR1). BOR1 exhibits variable penetrance and is characterized by hearing loss, branchial defects, preauricular pits and CAKUT [74]. On further evaluation, the child was found to have hearing loss and preauricular pits.
We also identified bilineal autosomal dominant diseases and digenic autosomal recessive disease. As an example of the former, in a 6-year-old female (Case 1) with bilateral multicystic dysplastic kidneys, pathogenic variants were identified in both PKD1 (a single nucleotide deletion) and PKD2 (a nucleotide substitution that converts the start codon to lysine). Each of these variants alone is sufficient to cause ADPKD, and the co-inheritance in this patient is consistent with her severe and atypical phenotype. Bilineal disease is rare in humans, although it has been noted in experimental mice [75–77].
In one case (Case 70), a medically actionable variant in WT1 was incidentally identified in a 6-month-old infant with renal cystic dysplasia, ESRD, ectopic atrial tachycardia, left ventricular hypertrophy and seizures. The variant, p.Arg417Cys, is ultra-rare, predicted pathogenic and previously reported in two patients—one with Denys–Drash syndrome (DDS) and Wilms’ tumor and one child with DDS who died shortly after birth [38, 78]. In light of these reports, the variant was reported to the clinician as likely pathogenic for DDS with the attendant risks of Wilms’ tumor.
In some cases, identified variants had insufficient evidence to be labeled as likely pathogenic or pathogenic and were reported as VUSs (Tables 5–7). In two cases, the genetic variants did not meet strict ACMG criteria for likely pathogenicity and were labeled as VUSs, but in the clinical context, the multidisciplinary group considered these as probably causal (Tables 5–7, Cases 57 and 92). In two other cases, variants classified as likely pathogenic by ACMG criteria were reported as VUSs because the genetic disease appeared irrelevant to the clinical phenotype. One of these was a case with nephrogenic diabetes insipidus (NDI) and nephrocalcinosis with hypophosphatemia (Tables 5–7, Case 44), where an identified variant in KAL1 was classified as likely pathogenic for Kallmann syndrome by ACMG criteria. In the other, a case with hypomagnesemia and dilated cardiomyopathy (Tables 5–7, Case 16), a likely pathogenic variant in ROBO2 for CAKUT was identified but reported as a VUS. In other instances, we identified alleles that increase risk for specific renal diseases (Tables 5–7). Five patients with FSGS, nephrotic syndrome or CKD were homozygous or compound heterozygous for variants in APOL1 that substantially increase the risk for FSGS in Americans of sub-Saharan African descent [79, 80]. Other risk variants were identified in CaSR, PLCG2 and ATP6V1B1, which increase the risk of hypercalcemia, steroid-sensitive nephrotic syndrome and kidney stones, respectively [81–83].
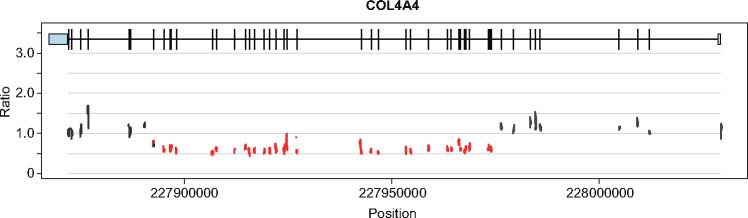
CNVs are significant contributors to genetic renal disease and their detection was an important component of our analysis [84]. We identified pathogenic CNVs in 18% of positive diagnoses, including four cases of autosomal recessive JN1 (NPHP1), two cases of autosomal dominant CAKUT (HNF1B), one case each of autosomal recessive Alport syndrome (COL4A4) and autosomal recessive pseudohypoaldosteronism (SCNN1B) and a possible tri-allelic form of Gitelman syndrome (CLCNKB; Figure 2).
FIGURE 2.
CNV identified in Case 33. The ratio of expected-to-observed sequence reads shows ∼50% reduction in signal, which is consistent with heterozygous deletion of exons 10–40 in COL4A4.
Alternative methods to provide comprehensive unbiased screening for genetic renal disorders include genome sequencing (GS) and/or ES, both of which have been used to diagnose monogenic renal disorders in a research setting and have been used in the clinical setting when locus heterogeneity is extreme, the phenotype is very indistinct, or the renal features are only a minor part of a multisystem disease [85, 86]. Neither GS nor ES is optimized for the renal exome, which includes challenging regions like the first 32 exons of PKD1, which are duplicated as six pseudogenes on chromosome 16. Nevertheless, ES remains an alternative as described by Lata et al., who report a 24% diagnostic rate in a selected population of adults with CKD, excluding ADPKD, where a genetic disease was suspected based on family history or there was early age-of-onset of disease [10]. In another study of a larger cohort of patients with CKD, ES identified diagnostic variants in 9.3% of patients [12].
There are some limitations and caveats to our testing strategy. First, as in ES, some types of genetic variants that occur within tandem oligonucleotide repeats, such as the cytosine insertion within a cytosine repeat sequence in MUC1, are difficult to identify [87]. Second, new genetic causes of kidney disease continue to be identified that may not have been present on the diagnostic gene panel at the time of testing. Included in this category are new and rare causes of kidney disease such as DZIP1L, FAT1 and the NUP and IFT family genes. Third, we were not always able to verify the presence of variants in trans to confirm compound heterozygosity for autosomal recessive disorders due to lack of parent or offspring samples. In addition, we purposefully omitted complement genes on this panel because we have developed a discrete panel for ultra-rare complementopathies, including atypical hemolytic uremic syndrome and C3 glomerulopathy. Finally, it should be noted that our diagnostic yield is high and warrants confirmation with larger studies.
In summary, these data add to the body of literature suggesting that genetic renal diseases are underdiagnosed and underappreciated in both children and adults [10, 88–90]. In this cohort of patients, presumably selected by clinicians based on suspicion of monogenic kidney disease, the genetic diagnostic rate is very high and is likely to be lower if more indiscriminate patient testing becomes the norm. Nevertheless, panels facilitate identification of a broad range of Mendelian diseases, including cystic kidney disease, the CAKUTs, tubulointerstitial disease and glomerular disease, as well as non-Mendelian genetic disease, bilineal and digenic disease, atypical forms of disease and unsuspected disease. As such, comprehensive genetic testing has an important place in the evaluation and care of the renal patient [91].
SUPPLEMENTARY DATA
Supplementary data are available at ndt online.
AUTHORS’ CONTRIBUTIONS
M.A.M., C.P.T. and R.J.S. conceived the study and wrote the manuscript; M.A.M. conducted genetic testing; R.R.S. performed bioinformatic analysis; M.E.F., C.A.C., R.J.S. and C.P.T. interpreted genetic test results with contributions from C.J.N., A.E.K. and M.J.K. All authors approved the final version of the manuscript.
CONFLICT OF INTEREST STATEMENT
None declared.
Supplementary Material
REFERENCES
- 1. Ellison DP, Thomas CP.. Hereditary disorders of connecting tubule and collecting duct sodium and potassium transport In: Mount DB, Pollak MR (eds). Molecular and Genetic Basis of Renal Disease. Philadelphia, PA: Elsevier Saunders, 2007, 251–268 [Google Scholar]
- 2. Snoek R, van Setten J, Keating BJ.. NPHP1 (nephrocystin-1) gene deletions cause adult-onset ESRD. J Am Soc Nephrol 2018; 29: 1772–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Reiter JF, Leroux MR.. Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol 2017; 18: 533–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gbadegesin RA, Hall G, Adeyemo A. et al. Mutations in the gene that encodes the F-actin binding protein anillin cause FSGS. J Am Soc Nephrol 2014; 25: 1991–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Joshi S, Andersen R, Jespersen B. et al. Genetics of steroid-resistant nephrotic syndrome: a review of mutation spectrum and suggested approach for genetic testing. Acta Paediatr 2013; 102: 844–856 [DOI] [PubMed] [Google Scholar]
- 6. Gupta IR, Baldwin C, Auguste D. et al. ARHGDIA: a novel gene implicated in nephrotic syndrome. J Med Genet 2013; 50: 330–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Capone VP, Morello W, Taroni F. et al. Genetics of congenital anomalies of the kidney and urinary tract: the current state of play. Int J Mol Sci 2017; 18: E796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hwang DY, Kohl S, Fan X. et al. Mutations of the SLIT2-ROBO2 pathway genes SLIT2 and SRGAP1 confer risk for congenital anomalies of the kidney and urinary tract. Hum Genet 2015; 134: 905–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bullich G, Domingo-Gallego A, Vargas I. et al. A kidney-disease gene panel allows a comprehensive genetic diagnosis of cystic and glomerular inherited kidney diseases. Kidney Int 2018; 94: 363–371 [DOI] [PubMed] [Google Scholar]
- 10. Lata S, Marasa M, Li Y. et al. Whole-exome sequencing in adults with chronic kidney disease: a pilot study. Ann Intern Med 2018; 168: 100–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mann N, Braun DA, Amann K. et al. Whole-exome sequencing enables a precision medicine approach for kidney transplant recipients. J Am Soc Nephrol 2019; 30: 201–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Groopman EE, Marasa M, Cameron-Christie S. et al. Diagnostic utility of exome sequencing for kidney disease. N Engl J Med 2019; 380: 142–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Richards S, Aziz N, Bale S. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015; 17: 405–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thomas CP, Mansilla MA, Sompallae R. et al. Screening of living kidney donors for genetic diseases using a comprehensive genetic testing strategy. Am J Transplant 2017; 17: 401–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rossetti S, Hopp K, Sikkink RA. et al. Identification of gene mutations in autosomal dominant polycystic kidney disease through targeted resequencing. J Am Soc Nephrol 2012; 23: 915–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tan Y-C, Michaeel A, Blumenfeld J. et al. A novel long-range PCR sequencing method for genetic analysis of the entire PKD1 gene. J Mol Diagn 2012; 14: 305–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Samarakoon PS, Sorte HS, Kristiansen BE. et al. Identification of copy number variants from exome sequence data. BMC Genomics 2014; 15: 661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Renieri A, Bruttini M, Galli L. et al. X-linked Alport syndrome: an SSCP-based mutation survey over all 51 exons of the COL4A5 gene. Am J Hum Genet 1996; 58: 1192–1204 [PMC free article] [PubMed] [Google Scholar]
- 19. Vargas-Poussou R, Forestier L, Dautzenberg MD. et al. Mutations in the vasopressin V2 receptor and aquaporin-2 genes in 12 families with congenital nephrogenic diabetes insipidus. J Am Soc Nephrol 1997; 8: 1855–1862 [DOI] [PubMed] [Google Scholar]
- 20. Caridi G, Dagnino M, Gusmano R. et al. Clinical and molecular heterogeneity of juvenile nephronophthisis in Italy: insights from molecular screening. Am J Kidney Dis 2000; 35: 44–51 [DOI] [PubMed] [Google Scholar]
- 21. Otto EA, Helou J, Allen SJ. et al. Mutation analysis in nephronophthisis using a combined approach of homozygosity mapping, CEL I endonuclease cleavage, and direct sequencing. Hum Mutat 2008; 29: 418–426 [DOI] [PubMed] [Google Scholar]
- 22. Wang F, Zhao D, Ding J. et al. Skin biopsy is a practical approach for the clinical diagnosis and molecular genetic analysis of X-linked Alport’s syndrome. J Mol Diagn 2012; 14: 586–593 [DOI] [PubMed] [Google Scholar]
- 23. Rossetti S, Consugar MB, Chapman AB. et al. Comprehensive molecular diagnostics in autosomal dominant polycystic kidney disease. J Am Soc Nephrol 2007; 18: 2143–2160 [DOI] [PubMed] [Google Scholar]
- 24. Abdelhak S, Kalatzis V, Heilig R. et al. A human homologue of the Drosophila eyes absent gene underlies branchio-oto-renal (BOR) syndrome and identifies a novel gene family. Nat Genet 1997; 15: 157–164 [DOI] [PubMed] [Google Scholar]
- 25. Storey H, Savige J, Sivakumar V. et al. COL4A3/COL4A4 mutations and features in individuals with autosomal recessive Alport syndrome. J Am Soc Nephrol 2013; 24: 1945–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nozu K, Iijima K, Kanda K. et al. The pharmacological characteristics of molecular-based inherited salt-losing tubulopathies. J Clin Endocrinol Metab 2010; 95: E511–E518 [DOI] [PubMed] [Google Scholar]
- 27. Duval H, Michel-Calemard L, Gonzales M. et al. Fetal anomalies associated with HNF1B mutations: report of 20 autopsy cases. Prenat Diagn 2016; 36: 744–751 [DOI] [PubMed] [Google Scholar]
- 28. Brochard K, Boyer O, Blanchard A. et al. Phenotype-genotype correlation in antenatal and neonatal variants of Bartter syndrome. Nephrol Dial Transplant 2009; 24: 1455–1464 [DOI] [PubMed] [Google Scholar]
- 29. Kitanaka S, Katsumata N, Tanae A. et al. A new compound heterozygous mutation in the 11 beta-hydroxysteroid dehydrogenase type 2 gene in a case of apparent mineralocorticoid excess. J Clin Endocrinol Metab 1997; 82: 4054–4058 [DOI] [PubMed] [Google Scholar]
- 30. Carvajal CA, Gonzalez AA, Romero DG. et al. Two homozygous mutations in the 11 beta-hydroxysteroid dehydrogenase type 2 gene in a case of apparent mineralocorticoid excess. J Clin Endocrinol Metab 2003; 88: 2501–2507 [DOI] [PubMed] [Google Scholar]
- 31. Feliubadalo L, Font M, Purroy J. et al. Non-type I cystinuria caused by mutations in SLC7A9, encoding a subunit (bo,+AT) of rBAT. Nat Genet 1999; 23: 52–57 [DOI] [PubMed] [Google Scholar]
- 32. Cornec-Le Gall E, Audrezet MP, Chen JM. et al. Type of PKD1 mutation influences renal outcome in ADPKD. J Am Soc Nephrol 2013; 24: 1006–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang S, Mei C, Zhang D. et al. Mutation analysis of autosomal dominant polycystic kidney disease genes in Han Chinese. Nephron Exp Nephrol 2005; 100: e63–e76 [DOI] [PubMed] [Google Scholar]
- 34. Lee JW, Lee J, Heo NJ. et al. Mutations in SLC12A3 and CLCNKB and their correlation with clinical phenotype in patients with Gitelman and Gitelman-like syndrome. J Korean Med Sci 2016; 31: 47–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nozu K, Fu XJ, Nakanishi K. et al. Molecular analysis of patients with type III Bartter syndrome: picking up large heterozygous deletions with semiquantitative PCR. Pediatr Res 2007; 62: 364–369 [DOI] [PubMed] [Google Scholar]
- 36. Smith GD, Robinson C, Stewart AP. et al. Characterization of a recurrent in-frame UMOD indel mutation causing late-onset autosomal dominant end-stage renal failure. Clin J Am Soc Nephrol 2011; 6: 2766–2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rossetti S, Strmecki L, Gamble V. et al. Mutation analysis of the entire PKD1 gene: genetic and diagnostic implications. Am J Hum Genet 2001; 68: 46–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Royer-Pokora B, Beier M, Henzler M. et al. Twenty-four new cases of WT1 germline mutations and review of the literature: genotype/phenotype correlations for Wilms tumor development. Am J Med Genet A 2004; 127A: 249–257 [DOI] [PubMed] [Google Scholar]
- 39. Bean LJ, Tinker SW, da Silva C et al.. Free the data: one laboratory’s approach to knowledge-based genomic variant classification and preparation for EMR integration of genomic data. Hum Mutat 2013; 34: 1183–1188 [DOI] [PubMed] [Google Scholar]
- 40. Denamur E, Delezoide AL, Alberti C. et al. Genotype-phenotype correlations in fetuses and neonates with autosomal recessive polycystic kidney disease. Kidney Int 2010; 77: 350–358 [DOI] [PubMed] [Google Scholar]
- 41. Saunier S, Calado J, Benessy F. et al. Characterization of the NPHP1 locus: mutational mechanism involved in deletions in familial juvenile nephronophthisis. Am J Hum Genet 2000; 66: 778–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Prattichizzo C, Macca M, Novelli V. et al. Mutational spectrum of the oral-facial-digital type I syndrome: a study on a large collection of patients. Hum Mutat 2008; 29: 1237–1246 [DOI] [PubMed] [Google Scholar]
- 43. Ward CJ, Hogan MC, Rossetti S. et al. The gene mutated in autosomal recessive polycystic kidney disease encodes a large, receptor-like protein. Nat Genet 2002; 30: 259–269 [DOI] [PubMed] [Google Scholar]
- 44. Tory K, Menyhard DK, Woerner S. et al. Mutation-dependent recessive inheritance of NPHS2-associated steroid-resistant nephrotic syndrome. Nat Genet 2014; 46: 299–304 [DOI] [PubMed] [Google Scholar]
- 45. Davis EE, Zhang Q, Liu Q. et al. TTC21B contributes both causal and modifying alleles across the ciliopathy spectrum. Nat Genet 2011; 43: 189–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang F, Wang Y, Ding J. et al. Detection of mutations in the COL4A5 gene by analyzing cDNA of skin fibroblasts. Kidney Int 2005; 67: 1268–1274 [DOI] [PubMed] [Google Scholar]
- 47. Ji W, Foo JN, O’Roak BJ. et al. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat Genet 2008; 40: 592–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gunay-Aygun M, Tuchman M, Font-Montgomery E. et al. PKHD1 sequence variations in 78 children and adults with autosomal recessive polycystic kidney disease and congenital hepatic fibrosis. Mol Genet Metab 2010; 99: 160–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Smith AJ, Reed AA, Loh NY. et al. Characterization of Dent’s disease mutations of CLC-5 reveals a correlation between functional and cell biological consequences and protein structure. Am J Physiol Renal Physiol 2009; 296: F390–F397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Negrisolo S, Benetti E, Centi S. et al. PAX2 gene mutations in pediatric and young adult transplant recipients: kidney and urinary tract malformations without ocular anomalies. Clin Genet 2011; 80: 581–585 [DOI] [PubMed] [Google Scholar]
- 51. Wu Y, Hu P, Xu H. et al. A novel heterozygous COL4A4 missense mutation in a Chinese family with focal segmental glomerulosclerosis. J Cell Mol Med 2016; 20: 2328–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pelletier J, Bruening W, Kashtan CE. et al. Germline mutations in the Wilms’ tumor suppressor gene are associated with abnormal urogenital development in Denys-Drash syndrome. Cell 1991; 67: 437–447 [DOI] [PubMed] [Google Scholar]
- 53. Kopp JB, Nelson GW, Sampath K. et al. APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol 2011; 22: 2129–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Takemura T, Hino S, Ikeda M. et al. Identification of two novel mutations in the CLCN5 gene in Japanese patients with familial idiopathic low molecular weight proteinuria (Japanese Dent’s disease). Am J Kidney Dis 2001; 37: 138–143 [DOI] [PubMed] [Google Scholar]
- 55. Ford B, Rupps R, Lirenman D. et al. Renal-coloboma syndrome: prenatal detection and clinical spectrum in a large family. Am J Med Genet 2001; 99: 137–141 [DOI] [PubMed] [Google Scholar]
- 56. Hichri H, Rendu J, Monnier N. et al. From Lowe syndrome to Dent disease: correlations between mutations of the OCRL1 gene and clinical and biochemical phenotypes. Hum Mutat 2011; 32: 379–388 [DOI] [PubMed] [Google Scholar]
- 57. Lu W, van Eerde AM, Fan X. et al. Disruption of ROBO2 is associated with urinary tract anomalies and confers risk of vesicoureteral reflux. Am J Hum Genet 2007; 80: 616–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Otto E, Hoefele J, Ruf R. et al. A gene mutated in nephronophthisis and retinitis pigmentosa encodes a novel protein, nephroretinin, conserved in evolution. Am J Hum Genet 2002; 71: 1161–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Louis-Dit-Picard H, Barc J, Trujillano D. et al. KLHL3 mutations cause familial hyperkalemic hypertension by impairing ion transport in the distal nephron. Nat Genet 2012; 44: 456–460. [DOI] [PubMed] [Google Scholar]
- 60. Miraoui H, Dwyer AA, Sykiotis GP. et al. Mutations in FGF17, IL17RD, DUSP6, SPRY4, and FLRT3 are identified in individuals with congenital hypogonadotropic hypogonadism. Am J Hum Genet 2013; 92: 725–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Marcos S, Sarfati J, Leroy C. et al. The prevalence of CHD7 missense versus truncating mutations is higher in patients with Kallmann syndrome than in typical CHARGE patients. J Clin Endocrinol Metab 2014; 99: E2138–E2143 [DOI] [PubMed] [Google Scholar]
- 62. Koziell A, Grech V, Hussain S. et al. Genotype/phenotype correlations of NPHS1 and NPHS2 mutations in nephrotic syndrome advocate a functional inter-relationship in glomerular filtration. Hum Mol Genet 2002; 11: 379–388 [DOI] [PubMed] [Google Scholar]
- 63. Gribouval O, Moriniere V, Pawtowski A. et al. Spectrum of mutations in the renin-angiotensin system genes in autosomal recessive renal tubular dysgenesis. Hum Mutat 2012; 33: 316–326 [DOI] [PubMed] [Google Scholar]
- 64. Doherty D, Parisi MA, Finn LS. et al. Mutations in 3 genes (MKS3, CC2D2A and RPGRIP1L) cause COACH syndrome (Joubert syndrome with congenital hepatic fibrosis). J Med Genet 2010; 47: 8–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Botzenhart E, Vester U, Schmidt C. et al. Cystinuria in children: distribution and frequencies of mutations in the SLC3A1 and SLC7A9 genes. Kidney Int 2002; 62: 1136–1142 [DOI] [PubMed] [Google Scholar]
- 66. Weber S, Taylor JC, Winyard P. et al. SIX2 and BMP4 mutations associate with anomalous kidney development. J Am Soc Nephrol 2008; 19: 891–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cole DE, Vieth R, Trang HM. et al. Association between total serum calcium and the A986S polymorphism of the calcium-sensing receptor gene. Mol Genet Metab 2001; 72: 168–174 [DOI] [PubMed] [Google Scholar]
- 68. Vargas-Poussou R, Dahan K, Kahila D. et al. Spectrum of mutations in Gitelman syndrome. J Am Soc Nephrol 2011; 22: 693–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Beck BB, Baasner A, Buescher A. et al. Novel findings in patients with primary hyperoxaluria type III and implications for advanced molecular testing strategies. Eur J Hum Genet 2013; 21: 162–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Schmidts M, Frank V, Eisenberger T. et al. Combined NGS approaches identify mutations in the intraflagellar transport gene IFT140 in skeletal ciliopathies with early progressive kidney disease. Hum Mutat 2013; 34: 714–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Luoma LM, Deeb TM, Macintyre G. et al. Functional analysis of mutations in the ATP loop of the Wilson disease copper transporter, ATP7B. Hum Mutat 2010; 31: 569–577 [DOI] [PubMed] [Google Scholar]
- 72. Cornec-Le Gall E, Audrezet MP, Rousseau A. et al. The PROPKD score: a new algorithm to predict renal survival in autosomal dominant polycystic kidney disease. J Am Soc Nephrol 2016; 27: 942–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Stokman M, Lilien M, Knoers N.. Nephronophthisis In: Pagon RA, Adam MP, Ardinger HH (eds). GeneReviews. Seattle, WA: University of Washington, 2016 [PubMed] [Google Scholar]
- 74. Chang EH, Menezes M, Meyer NC. et al. Branchio-oto-renal syndrome: the mutation spectrum in EYA1 and its phenotypic consequences. Hum Mutat 2004; 23: 582–589 [DOI] [PubMed] [Google Scholar]
- 75. Pei Y, Paterson AD, Wang KR. et al. Bilineal disease and trans-heterozygotes in autosomal dominant polycystic kidney disease. Am J Hum Genet 2001; 68: 355–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Dedoussis GVZ, Luo Y, Starremans P. et al. Co-inheritance of a PKD1 mutation and homozygous PKD2 variant: a potential modifier in autosomal dominant polycystic kidney disease. Eur J Clin Invest 2008; 38: 180–190 [DOI] [PubMed] [Google Scholar]
- 77. Wu G, Tian X, Nishimura S. et al. Trans-heterozygous Pkd1 and Pkd2 mutations modify expression of polycystic kidney disease. Hum Mol Genet 2002; 11: 1845–1854 [DOI] [PubMed] [Google Scholar]
- 78. Jeanpierre C, Denamur E, Henry I. et al. Identification of constitutional WT1 mutations, in patients with isolated diffuse mesangial sclerosis, and analysis of genotype/phenotype correlations by use of a computerized mutation database. Am J Hum Genet 1998; 62: 824–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Genovese G, Friedman DJ, Ross MD. et al. Association of trypanolytic ApoL1 variants with kidney disease in African-Americans. Science 2010; 329: 841–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kruzel-Davila E, Wasser WG, Aviram S. et al. APOL1 nephropathy: from gene to mechanisms of kidney injury. Nephrol Dial Transplant 2016; 31: 349–358 [DOI] [PubMed] [Google Scholar]
- 81. Zhang J, Fuster DG, Cameron MA. et al. Incomplete distal renal tubular acidosis from a heterozygous mutation of the V-ATPase B1 subunit. Am J Physiol Renal Physiol 2014; 307: F1063–F1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Gbadegesin RA, Adeyemo A, Webb NJ. et al. HLA-DQA1 and PLCG2 are candidate risk loci for childhood-onset steroid-sensitive nephrotic syndrome. J Am Soc Nephrol 2015; 26: 1701–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lorentzon M, Lorentzon R, Lerner UH. et al. Calcium sensing receptor gene polymorphism, circulating calcium concentrations and bone mineral density in healthy adolescent girls. Eur J Endocrinol 2001; 144: 257–261 [DOI] [PubMed] [Google Scholar]
- 84. Sanna-Cherchi S, Kiryluk K, Burgess KE. et al. Copy-number disorders are a common cause of congenital kidney malformations. Am J Hum Genet 2012; 91: 987–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yang Y, Muzny DM, Reid JG. et al. Clinical whole-exome sequencing for the diagnosis of Mendelian disorders. N Engl J Med 2013; 369: 1502–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Xue Y, Ankala A, Wilcox WR et al.. Solving the molecular diagnostic testing conundrum for Mendelian disorders in the era of next-generation sequencing: single-gene, gene panel, or exome/genome sequencing. Genet Med 2015; 17: 444–451 [DOI] [PubMed] [Google Scholar]
- 87. Kirby A, Gnirke A, Jaffe DB. et al. Mutations causing medullary cystic kidney disease type 1 lie in a large VNTR in MUC1 missed by massively parallel sequencing. Nat Genet 2013; 45: 299–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Daga A, Majmundar AJ, Braun DA. et al. Whole exome sequencing frequently detects a monogenic cause in early onset nephrolithiasis and nephrocalcinosis. Kidney Int 2018; 93: 204–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Cornec-Le Gall E, Harris PC.. The underestimated burden of monogenic diseases in adult-onset ESRD. J Am Soc Nephrol 2018; 29: 1583–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Mallett AJ, McCarthy HJ, Ho G. et al. Massively parallel sequencing and targeted exomes in familial kidney disease can diagnose underlying genetic disorders. Kidney Int 2017; 92: 1493–1506 [DOI] [PubMed] [Google Scholar]
- 91. Posey JE, Harel T, Liu P. et al. Resolution of disease phenotypes resulting from multilocus genomic variation. N Engl J Med 2017; 376: 21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.