Abstract
Chest X-ray image contains sufficient information that finds wide-spread applications in diverse disease diagnosis and decision making to assist the medical experts. This paper has proposed an intelligent approach to detect Covid-19 from the chest X-ray image using the hybridization of deep convolutional neural network (CNN) and discrete wavelet transform (DWT) features. At first, the X-ray image is enhanced and segmented through preprocessing tasks, and then deep CNN and DWT features are extracted. The optimum features are extracted from these hybridized features through minimum redundancy and maximum relevance (mRMR) along with recursive feature elimination (RFE). Finally, the random forest-based bagging approach is used for doing the detection task. An extensive experiment is performed, and the results confirm that our approach gives satisfactory performance compare to the existing methods with an overall accuracy of more than 98.5%.
Keywords: Covid-19, Convolutional neural network (CNN), Discrete wavelet transform (DWT), Minimum redundancy maximum relevance (mRMR), Recursive feature elimination (RFE), Random forest classifier
1. Introduction
Coronavirus or Covid-19 is a viral disease caused by Severe Acute Respiratory Syndrome (SARS). Covid-19 is 87.99% like the bat-SL-CoVZC45 virus and 96% similar to a bat coronavirus (Li, 2019). That is why it is believed that this virus is transferred to humans via bat species. Coronaviruses are a group of related viruses that cause diseases among mammals and birds. Human coronaviruses were found in 1960 and later 2003 as SARS-CoV, 2004 HCoV NL63, 2005 HKU1, 2012 MERS-CoV, and recently 2019 SERS-CoV-2 (Geller et al., 2012). The outer layer of these viruses is ~80 nm and the spikes are ~20 nm long. This virus was first identified in Wuhan city, the capital of the Hubei province of China. Anyone can get affected through close contact with the affected people. The World Health Organization (WHO) has already declared Covid-19 as a pandemic concerning the threat to public health (Yoo, 2019). The typical Covid-19 clinical syndromes are high fever, cough, sore throat, headache, fatigue, shortness of breath, muscle pain, etc. (Singhal, 2020).
A real-time reverse transcription-polymerase chain reaction (RT-PCR) is the most common standard test technique used in the diagnosis of Covid-19. But radiological image analysis through computed tomography (CT) and X-ray plays a vital role to assist the diagnosis properly due to the low sensitivity of RT-PCR (Kanne et al., 2020, Xie et al., 2020). In RT-PCR diagnosis the target ORF1ab gene is not enough sensitive in case of low viral load and due to SARS-CoV-2 rapid mutation (Pei et al., 2020). This will increase the false-negative result with low sensitivity. For this reason, it is clinically recommended to re-examine the suspicious results with another established method. The fusion of deep features has been proposed in reference (Wang et al., 2020) for the detection of Covid-19 using the CT image dataset. By this time, CT is considered as the established method for screening tools with RT-PCR to detect Covid-19 pneumonia (Gao et al., 2020). However, studies have found that the significant lung diseases had observed about 10 days after the onset of symptoms from the CT scan, though it is normal within 2 days (Bernheim, et al., 2020, Pan, et al., 2019). Moreover, X-ray imaging is the widespread modality for its availability, quick response, and cost-effective nature. The transportation medium of an X-ray digital image is also very flexible from the point of acquisition to analysis. Recently, X-ray imaging using deep learning methods for Covid-19 detection is a widely used mechanism to eliminate the limitations of insufficient test kits, waiting time of test results, and test cost by RT-PCR.
The state-of-the-art technique is persistently focusing on the use of deep learning applications in the detection of chest respiratory abnormalities using radiological imaging. The epidemic outbreak of Covid-19 finds the need for multiclass chest disease detection with higher accuracy. The main challenge of this research is to propose an automatic extraction of optimum features and a classification model for Pneumonia including Covid-19 and normal chest. However, the multiclass classification is more crucial in this experiment as the overlapping features may lead to a confusing result. Exploratory analysis in this work has finally achieved the optimum feature vector with promising classification. Most of the existing works for Covid-19 detection aim to address binary classification task separating Covid and non-Covid cases. Therefore, one of the main focuses of this research is to deal with multiclass scenarios including Covid-19 along with different types of Pneumonia. The experiment has found that the syndromes are likely to be more obvious in chest X-ray than other modalities.
In this study, an automatic feature extraction technique is used without the need for manual feature extraction. This method has proposed a hybrid feature extraction module where early diagnosis is possible. It can remove the limitations of obscuring features in chest X-ray using the combinations of local textural features and deep neural network features. Moreover, the preprocessing techniques play a vital role to dig the potential details. The result shows better accuracy through the utilization of minimum redundancy and maximum relevance (mRMR) features and recursive feature elimination (RFE) techniques. The adaptive boosting technique of ensemble classifiers shows a minimum classification error. The key findings of this research are given below:
-
i)
A preprocessing approach is proposed to improve the early detection of Covid-19 from the chest X-ray.
-
ii)
A hybrid feature extraction mechanism is developed through a pre-trained deep CNN model and the wavelet transform operation.
-
iii)
These features are optimized using the mRMR and RFE techniques to increase accuracy.
-
iv)
An adaptive boosting technique is utilized for final classification in reducing the classification error compared to other methods.
The remaining parts of this article are structured as follows: Section 2 presents related works. Section 3 describes the research materials and methods. Experimental results are presented and discussed in Section 4. Finally, Section 5 concludes the paper.
2. Related works
Applications using machine learning methods have already gained an adjunct position as a clinical tool. The automatic diagnosis of disease conditions using artificial intelligence is becoming a popular research area for achieving automated, fast, and reliable results. Deep learning technology has already been used in pneumonia detection (Rajpurkar, et al., 1711) using an X-ray image and confirmed a promising result. They had used CheXNet with a 121-dense layer of CNN and pre-trained on the publicly available data repository of ChestX-ray14. Recently, the rapid spread of the epidemic Covid-19 has increased the necessitates for developing expert tools. The machine vision-based automated system can assist the clinicians with a minimum level of radiological expertise. A deep learning model was developed at an earlier stage by Hemdan et al. (Hemdan et al., 2003) to detect Covid-19 from chest X-ray. Their COVID X-Net model used a combination of VGG19 and Dense-Net and obtained a good classification result with an F1-score of 0.91 for Covid-19 cases. Later, Wang and Wong (Wang and Wong, 2003) found a classification accuracy of 92.4% from the limited number of chest X-ray images using their developed COVID-Net. Ioannis et al. (Apostolopoulos and Mpesiana, 2020) obtained an accuracy of 96.78% using the transfer learning model. They had introduced a multiclass (Covid-19, Pneumonia, Normal) approach and find better results using VGG19 and MobileNet V2. Narin et al. (Narin et al., 2003) proposed a three-CNN-based pre-trained model to detect Covid-19 from the X-ray image. They achieved an accuracy of 98% using 5-fold cross-validation. Sethy et al. (Sethy and Behera, 2020) classified Covid-19 from the X-ray with an accuracy of 95.38% using ResNet50 features and a support vector machine classifier. Asif et al. (Asif et al., 2020) obtained a classification accuracy of 98% to detect Covid-19 from chest X-ray using deep CNN. Mangal et al. (Mangal, et al., 2004) proposed a computer-aided system to detect Covid-19 using X-ray and obtained an accuracy of around 90.5%. Going through the literature it is found that redundant features lead to higher false positive (FP) and false-negative (FN) rates. In the multiclass model, it is very crucial to deal with relevant features in the feature vector. Moreover, the hybridization of features from multiple domains will help improve the robustness and accuracy of the system. Besides, preprocessing before feature extraction is absent in most of the existing systems. The current paper tries to fill these gaps.
3. Research methodology
The proposed system utilizes a machine vision approach to detect Covid-19 cases from the Chest X-ray images. It takes the input chest X-ray images of three classes as Covid-19, non-COVID pneumonia, and normal. Non-COVID pneumonia also comprises two subclasses of viral pneumonia and bacterial pneumonia cases. The main contribution of this research is to detect the Covid-19 class from the rest classes using a chest X-ray image. The work-flow diagram of the proposed system is shown in Fig. 1 .
Fig. 1.
Overview of the proposed detection method.
Whenever any input is given to the system, it will be processed into a gray level and then resized the image to a fixed size of 224 × 224 pixels. The next step is to normalize the intensities and remove the noise using the anisotropic diffusion technique. The region of interest is then enhanced by the histogram equalization technique, which is a common technique for enhancing low-contrast images through histogram stretching using cumulative distribution function mapping. A watershed algorithm has been utilized for region segmentation. This segmented image is passed for feature extraction using DWT and CNN techniques. Features from both methods generate a fusion vector, which is optimized through mRMR and RFE techniques. Finally, this optimized vector is used for Covid-19 detection.
3.1. Dataset preparation
The dataset is prepared from several publicly available chest X-ray data sources: Academic torrent repository of confirmed Covid-19 cases of X-ray data by Cohen et al. (Cohen et al., 2003), Kaggle Covid-19 X-ray data (“Covid-19 X rays, 2020), Chest X-ray data of pneumonia (viral and bacterial), and normal cases by Kermany et al. (Kermany, 2018). Fig. 2 exhibits some sample chest X-rays from our experimental dataset.
Fig. 2.
Sample dataset; (a) Covid-19, (b) Viral Pneumonia, (c) Bacterial Pneumonia, (d) Normal chest X-ray.
The reliability of this neural network-based computer-aided diagnosis (CAD) system for Covid-19 depends on a large number of input data for training. The data preparation is explained below.
As it is difficult to get huge labeled data, so, we have used the data augmentation technique, such as zooming, searing, flipping, rotating, changing the brightness, color, and shape in horizontal and vertical positions. After augmentation, this experiment uses a total of 4809 chest X-ray images including 790 confirmed Covid-19 cases, 1215 viral pneumonia cases, 1304 bacterial pneumonia cases, and 1500 normal cases. Our prepared datasets are available at https://github.com/rafid909/Chest-X-ray.
3.2. Data preprocessing
Data preprocessing is one of the vital parts of medical image analysis as it helps in the extraction of effective features. An anisotropic diffusion along with histogram equalization is used for denoising and brightness uniformity for the resized image of 224 × 224 pixels. After that, the watershed technique is applied to segment out the effective region in the X-ray image. The details of these preprocessing steps are given below.
3.2.1. Anisotropic diffusion and histogram equalization
Anisotropic diffusion, an iterative process, enhances the quality of X-ray images by removing unnecessary noise and artifacts (Septiana and Lin, 2014), 2014);(Kamalaveni et al., 2015) through a space-variant diffusion filter or coefficient . Here, x and y are the image x-axis and y-axis coordinates, respectively, and t is the iteration step. The diffusion coefficient acts as the function of the local image gradient which is inversely proportional to the magnitude of the gradient. For instance, the gradient magnitude is weak within the inner region and strong close to the boundary. Therefore, it acts as a heat equation to smoothen the inner region and remove noise. It stops the diffusion across the boundary and preserves the edge. For image gradient the partial differential equation (PDE) of anisotropic diffusion can be shown by Equation (1), which is governed by the two edge-stopping functions of the diffusion coefficient given in Equation (2) and Equation (3) according to Perona and Malik (Perona and Malik, 1990).
| (1) |
| (2) |
| (3) |
K is a sensitivity constant, and indicate the two functions of the diffusion coefficient.
For a better perception clinician needs to enhance the visual quality of the image. In an X-ray image, the anisotropic diffusion is used for smoothing fine details and remove the undesirable low contrast and brightness. In addition, the histogram equalization technique can be used to overcome the low contrast problem (Salem et al., 2019). It will work as an efficient contrast enhancement technique in medical imaging operations. In an automatic detection system, more potential features can be attained using the effective presentation of enhanced images. An input Covid-19 chest X-ray image is shown in Fig. 3 (a). Fig. 3(b) and (c) show the images after anisotropic diffusion and histogram equalization, respectively.
Fig. 3.
Image enhancement; (a) Covid-19 chest X-ray, (b) Anisotropic diffusion, (c) Histogram Equalization.
3.2.2. Segmentation using watershed technique
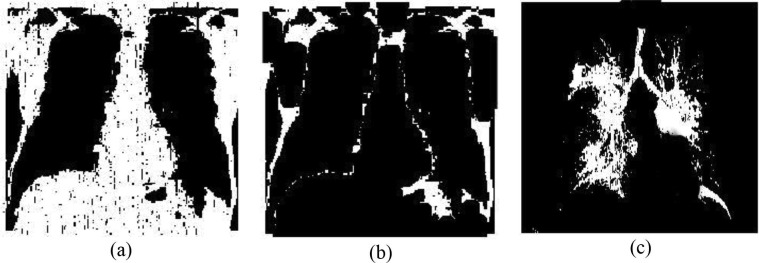
Watershed (Zheng et al., 2008) is a kind of transformation that works on grayscale image for segmenting different regions on the basis of the geological watershed to separate adjacent drainage basins. It performs like a topographic map where the luminance of each point representing its height, and then find out the lines that pass along the tops of ridges. In medical image segmentation, the watershed algorithm provides a complete division that separated meaningful feature regions for diagnosis, hence here this algorithm is used as the non-trivial task of separating the fracture-lung regions. Fig. 4 illustrates the outputs of the watershed segmentation technique of a Covid-19 X-ray image.
Fig. 4.
Outcomes of watershed segmentation technique: (a) Segmented lung, (b) Infected regions, (c) Features in the lung region.
3.3. Feature extraction
This section demonstrates the feature extraction process. Here, two feature extractors namely, discrete wavelet transform (DWT) and convolutional neural network (CNN) are used for feature extraction. As the region of interest varies in different x-ray images, DWT is used in this experiment to find the spatial relationship among pixels for better textural features. These DWT features are combined with deep CNN features for more effective outcomes.
3.3.1. Discrete wavelet transform
Wavelet transform is a powerful tool in medical image decomposition to extract the discriminative features for textural feature analysis. To find the effective DWT features, here we have performed a three-level decomposition task. At each level, it generates a low-resolution image and three detailed images (Wang et al., 2019). So, from three decomposed levels, it produces a total of nine detailed images. After analyzing these images, it is found that the textural information is more obvious in the middle (2nd-level) detail coefficients. So, this experiment has no attempt to decompose more than three-levels as we consider only the coefficients from the second level. Fig. 5 demonstrates the three-level decomposition of the segmented image. The structural and gray-level co-occurrence matrix (GLCM) features are selected from these three decomposed coefficients. Some discriminatory statistical features such as the mean, standard deviation, variance, kurtosis, skewness, and GLCM features namely energy, entropy, correlation, contrast, homogeneity are extracted. These features are calculated from four different orientations 00, 450, 900, 1350 for each of the three detail coefficients. It generates a total of 120 wavelet features.
Fig. 5.
DWT: (a) 1st level decomposition, (b) 2nd level decomposition, (c) 3rd level decomposition.
3.3.2. Convolutional neural networks
For feature extraction, a pre-trained ResNet 50 model is used. This model can handle the gradient disappearance and degradation problem of general CNN by using the residual blocks (Lu et al., 2020). The performance of CNN has improved by the depth of proposed residual blocks. The CNN architecture is a sequence of convoluted layers followed by pooling layers and ending with a fully connected neural network (Mostafiz et al., 2020). The convolutional layers have formed with some learnable filters. Such convolution filters are applied as feature extractors and the features are used to perform classification. Here, we have used the widely used benchmarked ResNet 50 CNN architecture whose self-explanatory schematic diagram is shown in Fig. 6 indicating layers, configurations, and parameters. It contains a total of 49 convolution (conv) layers and a fully connected (FC) layer. There is a max-pooling after the first convolution and an average pooling after the last convolution or before the FC. The FC layer is accounted for the feature extraction.
Fig. 6.
The schematic architecture of the CNN used in this experiment.
ReLU (Rectified Linear Unit) activation function (Nair and Hinton, 2010) is used in each convolutional layer. A dropout layer (Hawkins, 2004) has been added to prevent the overfitting problem. A total of 1024 CNN features are extracted from the FC layer for an input image.
3.4. Feature hybridization and selection
120 wavelet-based feature vectors and 1024 deep CNN feature vectors are fused to generate the hybridized feature vector of size (120 + 1024 = ) 1144. Feature optimization is then used to achieve more interpretable features in the feature vector. After optimization, the feature vector size becomes 100. This has improved the classification accuracy as well as computational cost. The objective is to select the most relevant features and eliminate the irrelevant and redundant features (Mostafiz et al., 2019, Mostafiz et al., 2020, Rashed-Al-Mahfuz et al., 2019). We have investigated two pragmatic feature optimization algorithms: i) Minimal-redundancy-maximal-relevance (mRMR) (Peng et al., 2005) and ii) Double input symmetrical relevance (DISR) (Meyer et al., 2008).
The mRMR algorithm optimizes the mutual dependencies among the selected features. Mutual dependencies of two variable × and y can be calculated using Equation (4) from their probabilistic density function and .
| (4) |
The maximal relevance approximates using Equation (5), where the mean of all mutual dependencies is , c is the class, and S is the feature set. Therefore, minimal redundancies can be added using function denoted by Equation (6).
| (5) |
| (6) |
Equations (5), (6) can be expressed as Eq. (7) to obtain a good subset of features by optimizing the relevance and redundancy.
| (7) |
Recursive feature elimination (RFE) finds the near-optimal feature from Φ of Equation (7).
On the other hand, DISR is a mutual information-based feature selection technique. It selects the most relevant features by utilizing the feature-selection criteria given in Equation (8)
| (8) |
Where T is the desired class, S is a feature set, I is the mutual information and H is the information entropy, and is the feature variable.
3.5. Ensemble (RF) classifier
Random Forest (RF) constructs the classifier using a decision tree (DT) based ensemble technique (Breiman, Oct. 2001). RF is comparatively faster and gives promising accuracy and can handle large data sets through decision trees. The RF classifier in this experiment has used the bagging technique of bootstrap aggregation (Ko et al., 2011). The algorithm works in this experiment as follows:
-
•
Randomly choose a subset of K features from the given data m; where .
-
•
Calculate the node d among K features to find the best split point as a threshold node. At node d the training data is split into the right subset and left subset .
-
•
Recursively splits the nodes for obtaining further best split nodes.
-
•
Continues until a single node is achieved.
-
•
Repeating n times means to create an RF classifier with n number of trees.
The candidate is selected as a threshold node that maximizes the information gain. The entropy estimation is calculated using Equation (9) to measure the information gain. The formation of the RF classifier follows two conditions to complete the training and to stop the iterations.
| (9) |
Analysis of the proposed model has found that the maximum depth 20 and the set of trees 100 will produce the best performance in terms of classification accuracy and computational cost for all cases. The output class of the input test image is obtained by maximizing the value of through Equation (10).
| (10) |
Where T is the total number of trees.
4. Results and analysis
This experiment detects and classifies the Covid-19 chest X-ray in two different scenarios. In the first scenario, only the Covid-19 X-ray is classified from non-COVID classes, which means it is a two-class scenario. The non-COVID class comprises all the chest X-rays of viral pneumonia, bacterial pneumonia, normal chest. Secondly, the experiment also performs the classification of X-ray image based on four classes: Covid-19, viral pneumonia, bacterial pneumonia, normal chest. The total dataset is divided randomly for training and testing at a ratio of 7:3.
For medical image analysis, only accuracy measurement is not enough, so, the method is assessed on recall, precision, F-score also. The performance metrics are shown in Eqs. (11), (12), (13), (14), which are derived from the confusion matrix consisting of True Positive (TP), True Negative (TN), False Positive (FP), and False Negative (FN). In the equations, TP is the prediction of positive class as positive; TN is the prediction of negative class as negative; FP is the prediction of negative class as positive; FN is the prediction of positive class as negative.
| (11) |
| (12) |
| (13) |
| (14) |
Tables 1 and Table 2 show the confusion matrices for the 2-class and the 4-class classification of the test images using the fusion of features of DWT and CNN along with mRMR and RFE optimizations. The classification is performed using the Random Forest (RF) based bagging approach.
Table 1.
Confusion matrix obtained for a 2-class scenario for the test images.
| Actual Class | Predicted Class | |
|---|---|---|
| Covid-19 | Non-COVID | |
| Covid-19 (Total = 237) | (TP) 235 | (FN) 2 |
| Non-COVID (Total = 1206) | (FP) 6 | (TN) 1200 |
Table 2.
Confusion matrix for a 4-class scenario for the test images.
| Actual class | Predict class | |||
|---|---|---|---|---|
| Covid-19 | Viral Pneumonia | Bacterial Pneumonia | Normal | |
| Covid-19(Total = 237) | 233 | 3 | 1 | 0 |
| Viral Pneumonia(Total = 365) | 2 | 357 | 4 | 2 |
| Bacterial Pneumonia(Total = 391) | 1 | 3 | 386 | 1 |
| Normal(Total = 450) | 2 | 0 | 2 | 445 |
Table 1 shows the binary class scenario where only 2 covid positive cases are identified as non-covid among 237 Covid-19 positive samples; i.e. the false positive (FP) rate is almost minimum. For 1206 non-covid cases only 6 are miss classified as covid positive. This little number of non-covid cases overlapped with covid positive cases (FP) because of the highly correlated features of covid and pneumonia chest X-ray. In a multiclass scenario (four-classes), 233 covid positive cases are identified correctly using the same number of samples as binary classification. Viral pneumonia cases are sometimes overlapped with covid cases but no covid positive cases are predicted as the normal chest. The wrong prediction rate has been decreased which is resulted from the efficient optimization of the feature vector with ensemble classification.
Based on the confusion matrices, the performance measures such as accuracy, precision, Recall, F-score of our proposed methodology for the 2-class and 4-class scenarios are shown in Table 3 .
Table 3.
Performance measures of our proposed method.
| Predict classes | Accuracy | Precision | Recall | F1-score |
|---|---|---|---|---|
| 2-class | 0.9945 | 0.9751 | 0.9917 | 0.9833 |
| 4-class | 0.9848 | 0.9789 | 0.9872 | 0.9829 |
To obtain the meaningful feature extraction scheme proposed method fused the textural features and the deep CNN features. The textural feature is formed using both the statistical features and GLCM features. Textural features are extracted from the 2nd level sub-bands of wavelets in different directions. The wavelet transforms on preprocessed chest X-ray shows better evaluation result than normal wavelet features. Table 4 shows the comparative performance of textural feature-based classification for both binary (Covid-19 vs non-COVID) class and multiple class (4-classes: Covid-19, viral pneumonia, bacterial pneumonia, normal chest).
Table 4.
Comparison using textural properties.
| Methods | Predict classes | Accuracy | Precision | Recall | F-score |
|---|---|---|---|---|---|
| DWT Only | 2-class | 0.8426 | 0.8349 | 0.8541 | 0.8445 |
| 4-class | 0.8153 | 0.8096 | 0.8345 | 0.8219 | |
| CNN Only | 2-class | 0.9782 | 0.9715 | 0.9876 | 0.9794 |
| 4-class | 0.9618 | 0.9448 | 0.9809 | 0.9625 | |
| DWT + CNN | 2-class | 0.9945 | 0.9751 | 0.9917 | 0.9833 |
| 4-class | 0.9848 | 0.9789 | 0.9872 | 0.9829 |
Table 4 shows the comparative performance using the features from the single method either CNN or DWT to the fused features (i.e. features from both CNN and DWT). This table confirms the superiority of the fused method.
This experiment attempted different pre-trained CNN models to find the best-suited one. Current studies have used many pre-trained CNN models for the automatic diagnosis of chest X-rays. The exploratory analysis has been performed on those models recommended by various existing literature and find the comparative result tabulated in Table 5 . The performance metrics are obtained using the combination of wavelet features and the CNN features. Working on the training and test data the fine-tuned ResNet50 performs better as a pre-trained model. All the CNN models use a uniform benchmark dataset in training. The test dataset is also identical to examine the performance of all those models. To evaluate each of the fine-tuned models, we have just change the classifier head and train the layers of the desired model. However, this study has also computed the performance of a few scratch models, but the performance peak is not good as fine-tuned models as well.
Table 5.
Comparative performance of the features from different CNN models fused with the DWT features.
| Methods | Predict classes | Accuracy | Precision | Recall | F-score |
|---|---|---|---|---|---|
| ResNet50 | 2-class | 0.9945 | 0.9751 | 0.9917 | 0.9833 |
| 4-class | 0.9848 | 0.9789 | 0.9872 | 0.9829 | |
| VGG19 | 2-class | 0.9627 | 0.9468 | 0.9785 | 0.9623 |
| 4-class | 0.9489 | 0.9347 | 0.9782 | 0.9561 | |
| MobileNet v2 | 2-class | 0.9413 | 0.9334 | 0.9504 | 0.9418 |
| 4-class | 0.9153 | 0.9081 | 0.9244 | 0.9162 | |
| DenseNet201 | 2-class | 0.9241 | 0.9114 | 0.9394 | 0.9252 |
| 4-class | 0.8947 | 0.8837 | 0.9091 | 0.8965 | |
| Inception | 2-class | 0.9084 | 0.8978 | 0.9214 | 0.9095 |
| 4-class | 0.8796 | 0.8693 | 0.8934 | 0.8812 | |
| Xception | 2-class | 0.9046 | 0.8951 | 0.9167 | 0.9058 |
| 4-class | 0.8759 | 0.8639 | 0.8922 | 0.8778 |
The performance result from Table 5 makes it clear that the pre-trained ResNet50 produces a comparatively better result with the combination of the wavelet domain. Using the fine-tuned pre-trained ResNet50 model both the binary classes and multiple classes classification show promising performance working on feature fusion. Two feature selection approaches minimum Redundancy - Maximum Relevance (mRMR) and Double Input Symmetrical Relevance (DISR) are applied separately on the final fused feature vector (wavelet features + CNN features using ResNet50) along with recurrent feature elimination (RFE) technique. The feature selection scheme performs to select the most relevant features and eliminate the redundant features to increase the computation speed and classification performance. By this elimination, the feature dimension becomes more concise with relevant features and obtain better efficiency with less computation required. Table 6 shows the comparative results of mRMR + RFE and DSIR + RFE feature optimizations. This table confirms that mRMR + RFE shows better performance than the DSIR optimization.
Table 6.
Comparative performance of feature optimization approaches.
| Methods | Predict classes | Accuracy | Precision | Recall | F1-score |
|---|---|---|---|---|---|
| DISR + RFE | 2-class | 0.9879 | 0.9668 | 0.9885 | 0.9776 |
| 4-class | 0.9791 | 0.9647 | 0.9819 | 0.9732 | |
| mRMR + RFE | 2-class | 0.9945 | 0.9751 | 0.9917 | 0.9833 |
| 4-class | 0.9848 | 0.9789 | 0.9872 | 0.9829 |
This experiment has used a Random Forest classifier (RF) as the ensemble of multiple decision trees. RF classifier performs better than the SVM and CNN classifier used in this experiment in terms of performance metrics and computational complexities. Besides, for the overlapping and categorical data, the RF classifier outperforms other models. Table 7 confirms that the Random Forest classifier shows the best performance. The classification has performed after the feature selection process of mRMR and RFE technique.
Table 7.
Comparison of performance of different classifiers used in this experiment.
| Methods | Predict classes | Accuracy | Precision | Recall | F-score |
|---|---|---|---|---|---|
| KNN(K-Nearest Neighbor) | 2-class | 0.9674 | 0.9453 | 0.9696 | 0.9573 |
| 4-class | 0.9518 | 0.9282 | 0.9538 | 0.9408 | |
| CNN(Convolutional Neural Network) | 2-class | 0.9852 | 0.9697 | 0.9826 | 0.9761 |
| 4-class | 0.9746 | 0.9534 | 0.9787 | 0.9658 | |
| SVM(Support Vector Machine) | 2-class | 0.9911 | 0.9634 | 0.9847 | 0.9739 |
| 4-class | 0.9801 | 0.9582 | 0.9821 | 0.9701 | |
| RF(Random Forest) | 2-class | 0.9945 | 0.9751 | 0.9917 | 0.9833 |
| 4-class | 0.9848 | 0.9789 | 0.9872 | 0.9829 |
The visual performance of some tricky cases, as well as one normal case of chest X-ray images, are shown in Fig. 7 . Fig. 7(a) shows a chest X-ray image of a Covid-19 case which is easily detected by both the proposed model and the radiologist as the main critical factor of leverage areas of lung can interpret by auditing opacities, which is the result of a decrease in the ratio of gas to soft tissue in the lung. In Fig. 7(b), the radiologist becomes confused to detect Covid-19 in the chest X-ray, but the proposed model correctly detects it as a Covid-19 class. The transparent design is responsible to gain and interpret the insight infection of Covid-19 is performed by the CAD system but difficult for human experts. However, in Fig. 7(c), both the proposed model and human experts are wrongly predicted as pneumonia but the actual class is Covid-19. The poor quality of X-ray leads to the wrong prediction. In some cases, due to poor-quality X-rays, the proposed system shows misdetection, such as Fig. 7(d). In Fig. 7(d) the normal chest image is detected as bacterial pneumonia.
Fig. 7.
Evaluated by the radiologist and proposed model: (a) Predicted Covid-19 by the proposed model and the radiologist; (b) Predicted Covid-19 by the proposed model but wrongly detected by the radiologist as normal chest; (c) Predict as viral pneumonia by both proposed model and the radiologist but actual class is Covid-19; (d) Predict bacterial pneumonia by the proposed model but actual class is a normal chest.
The 5-fold cross-validation has been performed to obtain a more embodied experimental result. It estimates the model behavior towards the independent data and also useful to minimize the overfitting problem in supervised learning. The performance is measured by averaging the values reported in each fold. In Fig. 8 , the accuracy of each fold is shown for 2-class and 4-class scenarios.
Fig. 8.
Performance graphs of 5-fold cross-validation.
Fig. 8 symbolizes that the peak accuracy is 99.85% for binary classification and 98.91% for multi-class classification. The RF works for classification in each fold based on the selective features of the mRMR technique. As the main motive of this research is to detect Covid-19 from chest X-ray, the performance is measured in two different criteria. The misdetection rate has been minimized by handling the imbalanced data using RFE with mRMR in the ensemble RF classifier. Thus, the proposed method has achieved superior performance than the existing techniques using a comparatively larger dataset. The comparative analysis of the proposed technique is presented in Table 8 with the recently developed techniques.
Table 8.
Comparison of the proposed Covid-19 diagnosis with the existing techniques.
| Study | Dataset | Method | Result |
|---|---|---|---|
| Hemdan et al. (Pan et al., 2019) | 25 Covid-19 (+);25 Normal X-ray. | VGG19 + MobileNet v2. | Covid-19 and Pneumonia. Accuracy = 90% |
| Wang and Wong et. al. (Rajpurkar et al., 1711) | 358 Covid-19 (+); 5538 Pneumonia (viral + bacterial); 8066 normal chests. | COVID-Net | Covid-19, Pneumonia, and Normal. Accuracy = 93.3% |
| Iaonnis et. al. (Hemdan et al., 2003) | 224 Covid-19 (+); 714 Pneumonia (400 bacterial + 314 viral); 504 normal chests. | MobileNet v2. | Covid-19, Pneumonia, and Normal. Accuracy = 96.78% |
| Narin et. al. (Wang and Wong, 2003) | 50 Covid-19 (+); 50 normal chests. | Deep CNN (ResNet50) | Covid-19 and Normal. Accuracy = 98% |
| Sethy and Behera et. al. (Apostolopoulos and Mpesiana, 2020) | 25 Covid-19 (+); 25 normal chests. | ResNet50 + SVM | Covid-19 and Normal. Accuracy = 95.38% |
| Asif et. al. (Narin et al., 2003) | 309 Covid-19 (+); 2000 Pneumonia; 1000 normal chests. | DenseNet121 | Covid-19, Pneumonia, and Normal. Accuracy = 98% |
| Mangal et. al. (Sethy and Behera, 2020) | 261 Covid-19 (+); 4200 Pneumonia; 2750 normal chests. | COVID-Net | Covid-19, Pneumonia, and Normal. Accuracy = 90.50% |
| Kumar et al (Kumar, 2020) | 62 Covid-19 (+); 4200 Pneumonia; 5610 normal chests. | ResNet152 + RF classifier | Covid-19, Pneumonia, and Normal.Accuracy = 97.3% |
| Ozturk et. al. (Ozturk et al., 2020) | 125 Covid-19 (+); 500 Pneumonia; 500 normal chests. | DarkCovidNet | Covid-19 and Non-Covid Accuracy = 98.08% |
| Sarhan et. al. (Sarhan, 2020) | 88 Covid-19 (+); 88 non– COVID. | Wavelet + SVM | Covid-19 and Non-CovidAccuracy = 94.5% |
| Proposed Method | 790 Covid-19 (+); 1215 viral Pneumonia; 1304 bacterial Pneumonia; 1500 normal chests. | Wavelet + pre-trained ResNet50 + RF classifier | Covid-19 and Non-Covid. Accuracy = 99.45% Covid-19, Viral pneumonia bacterial pneumonia, Normal. Accuracy = 98.48% |
5. Conclusions
This work has designed an intelligent model for Covid-19 detection with high accuracy and low complexity. Proper feature extraction and selection show the satisfactory outcome in Covid-19 classification using the chest X-rays. It resolved some problems in dealing with the large number of feature sets in distinguishing Covid-19 from other similar cases through hybridization of features from CNN and DWT. The mutual information-based feature selection is the strength of this methodology. An ensemble RF classifier shows better performance for the overlapping and unbalanced data like X-ray images. The system has obtained an accuracy of over 98.5% and outperforms the existing techniques. It shows the possibilities of cost-effective, rapid, and automatic diagnosis of coronavirus using the chest X-ray. Scopes are available for investigation of the risk and survival predictions which help treatment strategies and hospitalization management. Though there are many positive sides of the X-ray based detection mechanism, however, there is a limitation of the X-ray based system is the usual radiation hazard compare to the standard RT-PCR technique. In the future, we will investigate the hybridization of other features to improve accuracy by reducing the fewer false-negative and false-positive results.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Peer review under responsibility of King Saud University.
References
- Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020:1. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asif, S., Wenhui, Y., Jin, H., Tao, Y., Jinhai, S. 2020. Classification of COVID-19 from Chest X-ray images using Deep Convolutional Neural Networks. medRxiv.
- Bernheim A., et al. Chest CT findings in coronavirus disease-19 (COVID-19): relationship to duration of infection. Radiology. 2020:200463. doi: 10.1148/radiol.2020200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman Leo. Random forests. Mach. Learn. Oct. 2001;45(1):5–32. [Google Scholar]
- Cohen, J.P., Morrison, P., Dao, L. 2020. COVID-19 image data collection,” arXiv preprint arXiv:2003.11597.
- Covid-19 X rays. https://kaggle.com/andrewmvd/convid19-x-rays (accessed Jul 29, 2020).
- Gao K., Su J., Jiang Z., Zeng L.-L., Feng Z., Shen H., Rong P., Xu X., Qin J., Yang Y. Dual-branch combination network (DCN): Towards accurate diagnosis and lesion segmentation of COVID-19 using CT images. Med. Image Anal. 2020;101836 doi: 10.1016/j.media.2020.101836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller C., Varbanov M., Duval R.E. Human coronaviruses: insights into environmental resistance and its influence on the development of new antiseptic strategies. Viruses. 2012;4(11):3044–3068. doi: 10.3390/v4113044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins D.M. The problem of overfitting. J. Chem. Inf. Comput. Sci. 2004;44(1):1–12. doi: 10.1021/ci0342472. [DOI] [PubMed] [Google Scholar]
- Hemdan, E. E.-D., Shouman, M. A., Karar, M. E., Covidx-net: A framework of deep learning classifiers to diagnose covid-19 in x-ray images. arXiv preprint arXiv:2003.11055, 2020.
- Kamalaveni V., Rajalakshmi R.A., Narayanankutty K.A. Image denoising using variations of Perona-Malik model with different edge stopping functions. Procedia Comput. Sci. 2015;58:673–682. [Google Scholar]
- Kanne J.P., Little B.P., Chung J.H., Elicker B.M., Ketai L.H. Essentials for radiologists on COVID-19: An update—radiology scientific expert panel. Radiol. Soc. NorthAm. 2020 doi: 10.1148/radiol.2020200527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermany D.S., et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172(5):1122–1131. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- Ko B.C., Kim S.H., Nam J.-Y. X-ray image classification using random forests with local wavelet-based CS-local binary patterns. J. Digit. Imaging. 2011;24(6):1141–1151. doi: 10.1007/s10278-011-9380-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, R., et al. 2020. Accurate prediction of Covid-19 using chest X-ray images through deep feature learning model with SMOTE and machine learning classifiers. medRxiv, 2020.
- Li H., et al. Human-animal interactions and bat coronavirus spillover potential among rural residents in Southern China. Biosafety Health. 2019;1(2):84–90. doi: 10.1016/j.bsheal.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Bai Y., Chen Y., Su C., Lu S., Zhan T., Hong X., Wang S. The classification of gliomas based on a pyramid dilated convolution ResNet model. Pattern Recogn. Lett. 2020 [Google Scholar]
- A. Mangal et al., “CovidAID: COVID-19 Detection Using Chest X-Ray,” arXiv preprint arXiv:2004.09803, 2020.
- Meyer P.E., Schretter C., Bontempi G. Information-theoretic feature selection in microarray data using variable complementarity. IEEE J. Sel. Top. Signal Process. 2008;2(3):261–274. [Google Scholar]
- Mostafiz Rafid, R, Hasan Mosaddik, M, Mosaddik Imran, I, Rahman Mohammad Motiur., M An intelligent system for gastrointestinal polyp detectionin endoscopic video using fusion of bidimensional empirical mode decomposition and convolutional neural network features. Int. J. Imag. Sys. Tech. 2019;30(1):224–233. doi: 10.1002/ima.22350. [DOI] [Google Scholar]
- Mostafiz R., Rahman M.M., Islam A.K.M., Belkasim S. Focal liver lesion detection in ultrasound image using deep feature fusions and super resolution. Mach. Learn. Knowl. Extrac. 2020;2(3):172–191. [Google Scholar]
- Nair, V., Hinton, G.E. 2010 Rectified linear units improve restricted boltzmann machines. Proceedings of the 27th international conference on machine learning (ICML-10), pp. 807–814.
- Narin, A., Kaya, C., Pamuk, Z. 2020. Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks. arXiv preprint arXiv:2003.10849, 2020. [DOI] [PMC free article] [PubMed]
- Mostafiz, Rafid, R, Uddin, Mohammad Shorif, MS, Alam, Nur-A-, NA, Hasan, Md. Mahmodul, M., Rahman, Mohammad Motiur, MM, et al., 2020. MRI-Based Brain Tumor Detection using the Fusion of Histogram Oriented Gradients and Neural Features. Evolutionary Intelligence 13, 550. 10.1007/s12065-020-00550-1. Submitted for publication. [DOI]
- Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of Covid-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F., et al. Time course of lung changes on chest CT during recovery from 2019 novel coronavirus (COVID-19) pneumonia. Radiology. 2020:200370. doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei, F., Wang, L., Zhao, H., Li, H., Ji, M., Yang, W., Wang, Q., Zhao, Q., Wang, Y. 2020. Sensitivity evaluation of 2019 novel coronavirus (SARS-CoV-2) RT-PCR detection kits and strategy to reduce false negative. medRxiv. [DOI] [PMC free article] [PubMed]
- Peng H., Long F., Ding C. Feature selection based on mutual information criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005;27(8):1226–1238. doi: 10.1109/TPAMI.2005.159. [DOI] [PubMed] [Google Scholar]
- Perona P., Malik J. Scale-space and edge detection using anisotropic diffusion. IEEE Trans. Pattern Anal. Mach. Intell. 1990;12(7):629–639. [Google Scholar]
- Rajpurkar, P., et al. 2017. Chexnet: Radiologist-level pneumonia detection on chest x-rays with deep learning. arXiv preprint arXiv:1711.05225, 2017.
- Rashed-Al-Mahfuz, M., Hoque, M.R., Pramanik, B.K., Hamid, M.E., Moni, M.A. 2019. SVM model for feature selection to increase accuracy and reduce false positive rate in falls detection. 2019 International Conference on Computer, Communication, Chemical, Materials and Electronic Engineering (IC4ME2), 2019, pp. 1–5.
- Salem N., Malik H., Shams A. Medical image enhancement based on histogram algorithms. Procedia Comput. Sci. 2019;163:300–311. [Google Scholar]
- Sarhan, A.M. 2020. Detection of Covid-19 Cases in chest X-ray images using wavelets and support vector machines.
- Septiana, L., Lin, K.-P. 2014. X-ray image enhancement using a modified anisotropic diffusion. 2014 IEEE International Symposium on Bioelectronics and Bioinformatics (IEEE ISBB 2014), pp. 1–4.
- Sethy K., Behera S.K. Detection of coronavirus disease (covid-19) based on deep features. Preprints. 2020;2020030300:2020. [Google Scholar]
- Singhal T. A review of coronavirus disease-2019 (COVID-19) Indian J. Pediatrics. 2020:1–6. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L. Wang and A. Wong, “COVID-Net: A Tailored Deep Convolutional Neural Network Design for Detection of COVID-19 Cases from Chest X-Ray Images,” arXiv preprint arXiv:2003.09871, 2020. [DOI] [PMC free article] [PubMed]
- Wang S.-H., Zhang Y.-D., Yang M., Liu B., Ramirez J., Gorriz J.M. Unilateral sensorineural hearing loss identification based on double-density dual-tree complex wavelet transform and multinomial logistic regression. Integr. Comput.-Aided Eng. 2019;26:411–426. [Google Scholar]
- Wang S.-H., Govindaraj V.V., Górriz J.M., Zhang X., Zhang Y.-D. Covid-19 classification by FGCNet with deep feature fusion from graph convolutional network and convolutional neural network. Inf. Fusion. 2020 doi: 10.1016/j.inffus.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical 2019-nCoV pneumonia: Relationship to negative RT-PCR testing. Radiology. 2020:200343. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo J.-H. The fight against the 2019-nCoV outbreak: an arduous march has just begun. J. Korean Med. Sci. 2019;35(4):2019. doi: 10.3346/jkms.2020.35.e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W., Yang H., Sun H.-S., Fan H.-Q. 2008 2nd International Conference on Bioinformatics and Biomedical Engineering. 2008. Multiscale reconstruction and gradient algorithm for X-ray image segmentation using watershed; pp. 2519–2522. [Google Scholar]