Abstract
Antiviral drug discovery continues to be an essential complement to vaccine development for overcoming the global pandemic caused by SARS-CoV-2. The genomic RNA of SARS-CoV-2 contains structural elements important for viral replication and/or pathogenesis making them potential therapeutic targets. Here we report on the stem-loop II motif, a highly conserved noncoding RNA element. Based on our homology model we determined that the G to U transversion in the SARS-CoV-2 stem-loop II motif (S2MG35U) forms a C–U base-pair isosteric to the C-G base-pair in the early 2000’s SARS-CoV (S2M). In addition, chemo-enzymatic probing and molecular dynamics simulations indicate the S2MG35U conformational profile is altered compared to S2M in the apical loop region. We explored S2MG35U as a potential drug target by docking a library of FDA approved drugs. Enzymatic probing of the best docking ligands (aminoglycosides and polymyxins) indicated that polymyxin binding alters the conformational profile and/or secondary structure of the RNA. The SARS-CoV-2 stem-loop II motif conformational differences due to nucleotide transversion and ligand binding are highly significant and provide insight for future drug discovery efforts since the conformation of noncoding RNA elements affects their function.
Keywords: SARS-CoV-2, COVID-19, Stem-loop II motif, S2M, Molecular dynamics, RNA-targeted drug discovery
1. Introduction
The COVID-19 disease and global pandemic caused by the SARS-CoV-2 coronavirus continues to threaten human health and wellbeing [1]. Considerable effort has been focused on developing vaccines and small molecule therapeutics [2] including repurposing of existing drugs [3]. The SARS-CoV-2 coronavirus has genomic RNA structural elements, many in noncoding regions, which play key roles in viral replication and/or pathogenesis [4]. These types of functionally important noncoding RNA structural elements offer an opportunity for small-molecule interactions and the development of RNA-targeted therapeutics [5].
Relatively few small-molecule therapeutics targeting RNA are in the overall drug discovery pipeline [6]. For treating COVID-19, considerable effort is focused on identifying small molecule therapeutics to target SARS-CoV-2 viral proteins, developing vaccines and developing siRNA that target viral RNA to silence viral protein expression [2]. Less focus has been placed on targeting the viral RNA structural elements, although a few studies identified small molecules targeting the frame-shifting RNA element in SARS-CoV [7] and SARS-CoV-2 [8].
One RNA element in SARS-CoV-2 not previously investigated for small molecule targeting is the stem-loop II motif. This element is located in the 3′-UTR close to the poly-A tail [4]. The sequence of the stem-loop II motif is highly conserved across coronaviruses and astroviruses that contain it and its tertiary fold is structurally complex [9]. While the function of this RNA element is unknown [10], it may provide an evolutionary advantage [11] and/or be important for viral pathogenesis [9]. Consistent with this possibility is a recent genomic RNA mapping study which identified secondary structural differences in the apical loop of the SARS-CoV-2 stem-loop II motif in cells compared to that found in vitro indicating the RNA element may be involved in further molecular interactions in the context of an infected cell [12]. Based on the high sequence conservation, complex tertiary structure and cell-dependent structural differences, the stem-loop II motif has the characteristics of an important potential therapeutic target.
In this paper, we investigate the SARS-CoV-2 stem-loop II motif RNA structure and its ligand binding. We developed a computational homology model and used this for both molecular dynamics simulations and ligand docking. We complemented these computational studies with chemo-enzymatic RNA probing experiments. Together, the data indicate that the stem-loop II motif in SARS-CoV-2 has a different conformational profile compared to that of SARS-CoV due to a G to U base transversion and that this conformational profile is altered upon ligand binding. Consequently, since the conformation of noncoding RNA elements often determines their function [13], we suggest that the coronavirus stem-loop II motif may be a druggable target.
2. Materials and methods
2.1. Reagents & computational resources
All reagents were molecular biology grade, RNase/DNase-free or highest purity grade possible. RNases were from ThermoFisher Scientific. Ligands were from Sigma-Aldrich with purity >90%. Custom synthesized HPLC-purified RNAs fluorescently labeled with 5′-DY547 were obtained fully deprotected from Horizon Discovery. RNAs 5′-labeled with DY547 are a substitute for 32P 5′-labeled RNAs in chemo-enzymatic probing experiments [14]. All RNAs were dialyzed in 5 mM MOPS, pH 7.0, 0.01 mM EDTA and renatured prior to use. All computational work was done using the Owens Cluster at the Ohio Supercomputer Center [15] with the exception of the standard precision docking done on a MacPro and the sequence analysis done on a MacBook Pro.
2.2. Sequence analysis
We used the NCBI Viral Database [16] to search for SARS-related coronavirus (taxid: 694009) sequences by collection date for three different time periods: (1) January 1, 1990 to December 1, 2019 (331 retrieved); (2) December 1, 2019 to June 24, 2020 (8171 retrieved); and (3) June 25, 2020 to October 28, 2020 (9302 retrieved). These records were then searched by accession number using BLAST-n [17] for sequences similar to the stem-loop II motif core sequence [9] (nucleotides 10–40, Fig. 1 ). The retrieved sequences were then processed using Jupyter Notebook accessed through Anaconda Navigator and BioPython [18] to parse them according to host and nucleotide identity at the position corresponding to nucleotide 35 in Fig. 1 and complementarity to the core sequence.
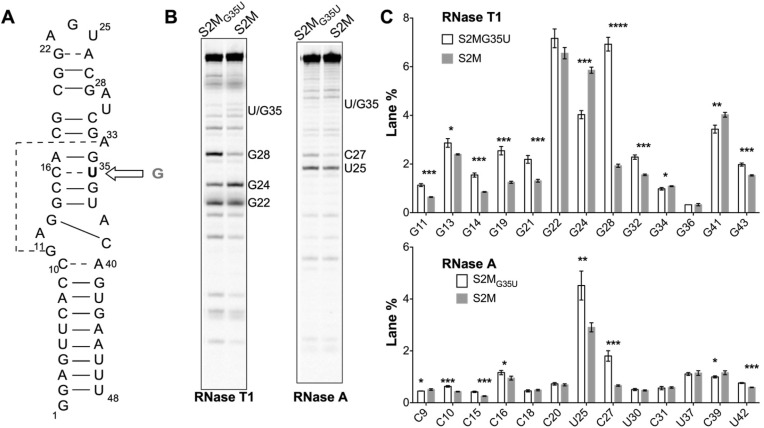
Fig. 1.
SARS-CoV-2 contains G to U mutation in stem-loop II motif that alters secondary structure in apical loop region. (A) SARS-CoV-2 stem-loop II motif model RNA (S2MG35U, core = nucleotides 10–40). Open arrow indicates sequence change from early 2000’s SARS-CoV genomes prior to December 2019 (S2M). Canonical (solid line) and non-canonical (dashed line) base pairs of secondary structure derived from crystal structure PDB ID 1XJR [9]. (B) Representative gel image of RNase A and T1 probing comparing S2MG35U with S2M (gamma = 0.9). (C) Band lane % of RNase probing triplicates. Asterisk indicate two-tailed t-test significance ∗∗∗∗P ≤ 0.0001, ∗∗∗0.001 < P ≤ 0.001, ∗∗0.01 < P ≤ 0.01, ∗0.01 < P ≤ 0.05, no asterisks indicates P > 0.05.
2.3. Computational model RNAs and ligand docking
The computational model RNAs were derived from PDB ID 1XJR, the 2005 crystal structure for a stem-loop II motif (S2M) [9]. In order to facilitate comparison of biochemical and computational results, only the single G35U mutation difference in the core was modeled (i.e., S2M vs. S2MG35U, Fig. 1). Using Protein Preparation Wizard (Schrödinger), we deleted the G1 GTP and refined the structure of S2M still with the hydrated Mg2+. The structure for S2MG35U with hydrated Mg2+ was prepared by deleting the G35 nucleobase, replacing it with a U followed by refinement. For the S2MG35U docking studies, the metal ions and water were deleted and the RNA refined. All of the above refinements used the Protein Preparation Wizard restrained minimization, 0.3 RMSD, OPLS3e forcefield. The S2M and S2MG35U model RNAs were then further minimized using MacroModel (Schrödinger) with OPLS_2005 force field, PRCG minimization method and 0.05 kJ/Å-mol gradient convergence threshold.
The docking grid for S2MG35U was generated using Glide (Schrödinger) with an OPLS_2005 force field including aromatic hydrogens as hydrogen bond donors and halogens as hydrogen bond acceptors. The centroid of the grid was set to residues 10, 20, 22 and 38 and the midpoint box set at 25, 35, 40 Å X,Y,Z respectively with the bounding box set for docking ligands ≤20 Å. The resulting grid encompassed nucleotides 5 to 45. The FDA-Approved Drug Library Plus (HY-LO22P) was obtained from MedChemExpress as a structure data file. Three-dimensional models of the compounds (including tautomers and stereoisomers for undefined chiral centers) were generated using LigPrep (Schrödinger) with OPLS2005 force field, pH 7.3±0.2. The prepared structures were then docked to the grid using Glide’s high-throughput-virtual-screening mode (one pose per ligand, no post docking minimization). The resulting lowest energy poses were then redocked using the Standard Precision setting with post-docking minimization.
2.4. Molecular dynamics simulations
Molecular dynamics simulations of the model RNAs were performed using Desmond (Schrödinger). Solvated structures of the minimized S2M and S2MG35U model RNAs in the presence of bound Mg2+ were generated in the Desmond system builder. To avoid formation of a ladder-like structure frequently observed in RNA molecular dynamics simulations [19] we applied minimal constraints to the closing base-pairs of the stem: 1.0 (G2, A3, U47); 0.5 (G4,U46); and, 0.2 (U5, A45). A TIP3P explicit water model with a 30 × 30 × 30 orthorhombic boundary box was used, the charges neutralized with 41 Na+ ions and the volume minimized for 250 mM NaCl buffer salt. The TIP3P water model was optimal for other RNA molecular dynamics simulations [20]. The resulting S2M system (95075 atoms, 191476° of freedom) and S2MG35U system (96035 atoms, 193397° of freedom) were relaxed prior to simulation.
A 10.0 ns molecular dynamics simulation was carried out with a 10.0 ps trajectory recording interval and an NPT ensemble class at 300 K, 1.01325 bar. The root mean square fluctuation (RMSF) analyses of each simulation were done using the Desmond Simulation Event Analysis tool and heavy atoms compared to the starting system for S2M. The resulting data were then plotted using Prism (GraphPad). The overall quality of the simulation was verified using the Desmond Simulation Quality Analysis Tool.
2.5. Chemo-enzymatic probing
Enzymatic probing reactions (10 μL) consisted of 10 mM Tris-HCl pH 7.0, 100 mM KCl, 0.1 mM MgCl2 and 4.0 μM 5′-DY547 RNA with RNase T1 (0.004 U/μl) or RNase A (0.0004 μg/ml). For the structure comparison reactions, RNA was incubated for 10 min with RNase T1/A at room temperature prior to separation by denaturing polyacrylamide gel electrophoresis (15% 19:1 acrylamide:bisacrylamide, 7.0 M urea). For the ligand dose-response studies, RNA was incubated with the ligands final concentrations as noted in figure) for 15 min at room temperature, then incubated with RNase A (0.0002 μg/ml) for 10 min at room temperature followed by gel electrophoresis. Gel images were acquired on a BIORAD ChemiDoc™ XRS instrument and data analyzed using the Image Lab™ software. The lane % ((background adjusted volume of a band in a lane)/((background adjusted total volume in that lane)∗100)) of each band was analyzed. Normalized ratios, dose-response slopes and two-tailed t-test statistics were determined using Prism (GraphPad).
Experimental conditions for the Mg2+-facilitated chemical probing were similar to a previous report of low magnesium in-line probing experiments [14]. In-line experiments (10 μL) consisted of 50 mM Tris-HCl (pH 8.3), 100 mM KCl, 1.0 mM MgCl2, 4.0 μM 5′-DY547 RNA and were incubated in the dark at room temperature for 10 days prior to analysis via denaturing polyacrylamide gel electrophoresis (15% 19:1 acrylamide:bisacrylamide, 7.0 M urea). Gels were imaged and analyzed as for the enzymatic probing experiments.
3. Results and discussion
3.1. G to U transversion in SARS-CoV-2 stem-loop II motif alters secondary structure in apical loop
We surveyed genomic databases to investigate the stem-loop II motif RNA element that was previously shown to be rigorously conserved in SARS-related viruses and astroviruses [9,10]. We used the NCBI Viral database to retrieve SARS-CoV related entries (taxid:694009) and BLAST-n to search these for sequences somewhat similar to the core sequence in the stem-loop II motif (S2M) of the 2003 SARS-CoV outbreak (Fig. 1, nucleotides 10–40). All retrieved sequences that were collected prior to December 2019 (163 sequences) had a G at position 35 (S2M, Fig. 1). In contrast, of the human host sequences collected from December 2019 to June 24, 2020 (7641 sequences) and from June 25, 2020 to October 28, 2020 (8711 sequences), all had a U at position 35 (S2MG35U, Fig. 1) with the exception of two sequences having an N or W at this position. Furthermore, >98% of the sequences were fully complementary to the S2MG35U core sequence (nucleotides 10–40, Fig. 1), consistent with an early SARS-CoV-2 genome study [4].
The G35U mutation is notable because it is in a region previously determined to be base-paired and rigorously conserved across coronavirus and astrovirus with minimal structural flexibility [9]. In the stem-loop II motif of the early 2000’s SARS-CoV outbreak, G35 base-pairs with C16 [9]. Of the 16,352 SARS-CoV-2 human host sequences collected December 2019 to Oct 28, 2020 fewer than 2% had an additional mutation or deletion in the core region beyond G35U. Only 6 of the sequences had a mutation at the position corresponding to C16 (a T or Y) indicating that there is no significant occurrence of a corresponding compensatory mutation. The C16-G35 base-pair of S2M of the early 2000’s SARS-CoV is immediately adjacent to G34 and A33, which form key long-range hydrogen bonds that contribute to the core tertiary fold [9]. Consequently, we investigated the structural effect the G35U mutation has on this important anchoring region in S2MG35U of the current SARS-CoV-2.
We probed S2M and S2MG35U model RNAs using single-strand specific RNases T1 and A. We found that both model RNAs have comparable regions of single-stranded character consistent with the 2005 crystal structure of S2M (Fig. 1). The most significant differences in S2MG35U compared to S2M were focused in the apical loop region including a decrease in single-strand character at G24 and an increase in single-stranded character at C27 and G28. Notably, no significant single-strand character was observed at position 35 for either model RNA indicating that position 35 is likely base-paired.
3.2. SARS-CoV-2 stem-loop II motif homology model indicates G to U transversion forms isosteric base-pair
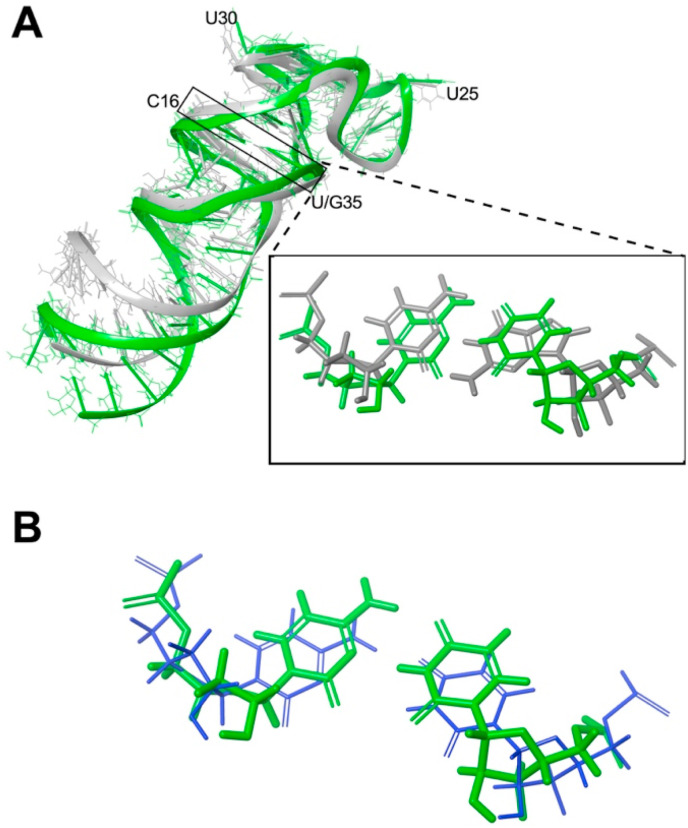
To investigate the implications of the G35U transversion on the tertiary structure of the stem-loop II motif, we prepared a homology model based on the 2005 crystal structure of an S2M (PDBID: 1XJR [9]). The resulting core region of the S2MG35U homology model RNA (nucleotides 10–40) superimposed closely with that of the S2M crystal structure (Fig. 2 , 2.68 RMS). In particular, a C16–U35 base-pair with two hydrogen bonds forms in S2MG35U that is similar to the C16-G35 base-pair in S2M (Fig. 2a inset) and is comparable to an exemplar model [21] for a C–U base-pair (1.65 heavy atom RMS with C764–U900 in PDB ID 1S72 [22], Fig. 2b).
Fig. 2.
Computational model of S2MG35Uaccommodates C16–U35base-pair isosteric to S2M C16-G35base pair. (A) Superposition of S2MG35U homology model (green ribbon) compared to S2M crystal structure (grey ribbon, PDB ID 1XJR [9]). Inset: Detail of S2MG35U C16–U35 base-pair compared to S2M C16-G35 base-pair. (B) Superposition of S2MG35U C16–U35 base-pair (green) with exemplar C–U base-pair (blue, C764–U900 from PDB ID 1S72 [22]).
While the in silico studies indicated that the G35U mutation could form a conformationally isosteric base-pair, the hydrogen bonding differences could potentially affect the overall structure/conformational flexibility of the core region. Consequently, we used chemical probing and molecular dynamics simulations to compare S2M and S2MG35U model RNAs.
3.3. S2MG35U single base transversion alters conformational profile compared to S2M
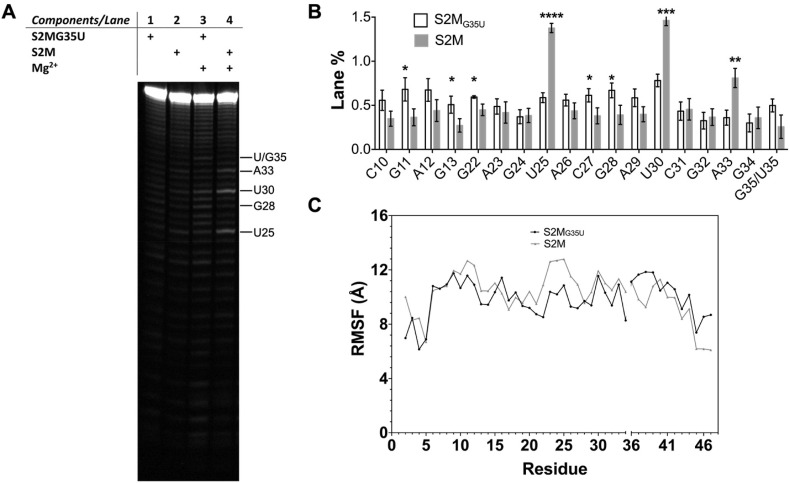
We used magnesium facilitated in-line cleavage to investigate the conformational profile of each model RNA. This in-line cleavage method identifies nucleotides that are conformationally flexible enough to result in in-line attack of the 2′OH to cleave the phosphodiester bond [23]. In-line probing of S2MG35U resulted in a two-fold decrease in cleavage at positions U25, U30 and A33 along with a 50% increase at positions C27 and G28 compared to S2M (Fig. 3 ). Notably, while a slight increase in cleavage was observed at position 35 in S2MG35U, the differences were not statistically significant. These in-line probing results indicate a likely change in the conformational profile of S2MG35U compared to S2M, particularly in the apical loop region.
Fig. 3.
S2MG35Uconformational profile altered compared to S2M. (A) Representative gel image of magnesium-facilitated in-line probing (gamma = 0.9) (B) Triplicate lane % of in-line probing. Asterisks indicate two-tailed t-test statistical significance ∗∗∗∗P ≤ 0.0001, ∗∗∗0.001 < P ≤ 0.001, ∗∗0.01 < P ≤ 0.01, ∗0.01 < P ≤ 0.05, no asterisk indicates P > 0.05. (C) Per residue Root Means Square Fluctuation (RMSF) in 10 ns molecular dynamics simulation of S2MG35U (black line) and S2M (grey line).
We also used molecular dynamics simulations to investigate the effect of the G35U mutation. Molecular dynamics simulations are an effective strategy for exploring RNA conformational sampling and dynamics ranging from bond rotations or sugar puckering occurring on the nanosecond timescale to base-pair opening on the microsecond timescale and more global changes occurring at longer times [19]. The extent of fluctuation during a 10 ns simulation of the hydrated magnesium model RNAs was greater for S2M than it was for S2MG35U particularly in the apical loop region (e.g., U25), consistent with the conformational profile differences observed in the in-line probing experiments (Fig. 3). For U30, however, longer timescale global motions may be why the flexibility differences observed in the in-line probing were not reflected in the molecular dynamics simulations.
3.4. Computational docking identifies ligands that alter S2MG35U structure
Since the stem-loop II motif is phylogenetically conserved and likely important for viral pathogenesis [9], we investigated the ability of small molecules to target this noncoding RNA element using docking methods previously applied to ligand-RNA complexes [24]. We conducted a high-throughput virtual screening (HTVS) of a library of 2309 FDA approved drugs docking to S2MG35U. Approximately 2% of the library (49 compounds) returned no structures during ligand preparation, however dropped/incorrect structures are not uncommon when working with in silico compound libraries [25]. The resulting set of 3236 prepared structures was docked to the S2MG35U model RNA without the Mg2+ since one key mode by which small molecules bind RNA is via displacement of magnesium ions [26] and since the authors of the original crystal structure suggested a potential site for ligand binding that is available in the RNA without Mg2+ [9]. The resulting ligand-RNA poses obtained (2628 valid poses) were then ranked based on the calculated Emodel value as we have done for other ligand-RNA docking studies [24]. Ligand poses within 10% of the lowest Emodel pose (polymyxin B, −492 kcal/mol) were selected for further analysis: tobramycin (−453 kcal/mol), kanamycin (−425 kcal/mol), colistin (−406 kcal/mol) and capreomycin (−488 kca/mol). In the case of capreomycin, the isomer with the lowest energy docking pose was not a natural stereoisomer due to the original structure data file for this compound not having all the stereocenters specified, consequently, this ligand was not included in further analyses. Compound library structure data files with undefined stereocenters are often encountered [25]. The remaining four compounds were redocked to S2MG35U using a standard precision mode and energy minimized. These compounds docked S2MG35U in the tunnel formed by the long-range G11-A33 base-pair of the complex tertiary fold (Fig. 4 ). This tunnel had previously been proposed as a possible binding site for small molecules [9].
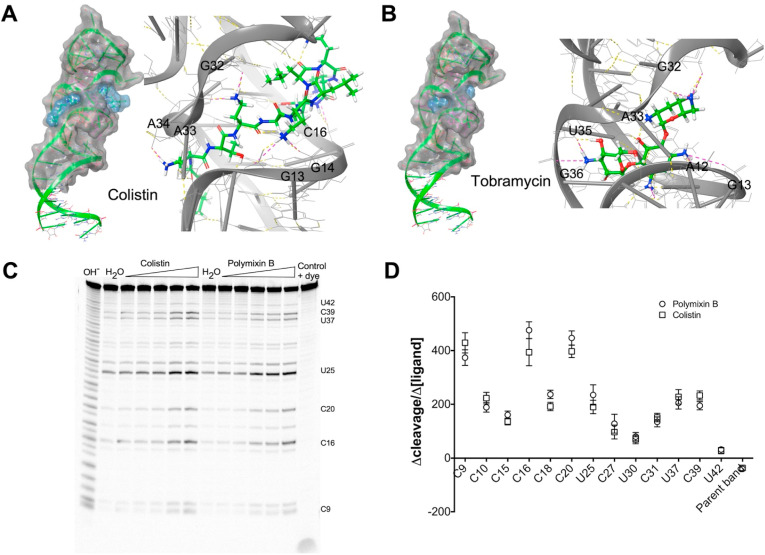
Fig. 4.
Computational docking identifies ligands that alter S2MG35Ustructure (A) & (B) Representative Glide-derived lowest energy ligand docking to S2MG35U (in grey with phosphate backbone ribbon cartoon and grey tubes along lengthwise axis of nucleobase). Ligands shown in thick tube (green = carbon, blue = nitrogen, red = oxygen, grey = hydrogen). Dashed lines indicate hydrogen bonds (yellow) and salt bridges (pink). (C) Gel image of S2MG35U RNase A probing dose-response of polymyxins (0.5 μM–10 μM, gamma = 0.9). (D) Lane % slope of dose-response for individual nucleotide bands.
It was not surprising that tobramycin and kanamycin were amongst the best docking compounds since they are aminoglycosides, compounds known to bind RNA electrostatically, frequently in divalent metal ion binding sites [26]. In contrast, we were initially surprised that polymixin B and colistin were amongst the best binders since their antibacterial mechanism of action is primarily thought to be via cell wall disruption, however a recent study determined that it might also involve binding to and disrupting ribosomal RNA [27]. Both the aminoglycosides and polymyxins make extensive contacts with S2MG35U primarily with the non-bridging phosphate oxygens lining the tunnel formed by the G11-A33 base-pair and flanking regions. This is the same location where the two hydrated magnesium ions are bound in the crystal structure [9] and is consistent with the electrostatic RNA binding characteristics of these compounds.
To explore the effects of these compounds on the RNA structure, we looked at ligand-induced changes in the RNase A profile of S2MG35U. Tobramycin and kanamycin had no effect at lower concentrations and only showed an overall, nonspecific protection of enzymatic cleavage at high concentrations (>50 μM, Supplementary Material). In contrast, colistin and polymyxin B increased RNase A cleavage of S2MG35U particularly at C16 and C20 starting in the low micromolar range (Fig. 4) indicating that binding of these ligands alters the secondary structure and/or conformational profile of the RNA. At higher concentrations (>0.01 mM) the polymyxins resulted instead in an overall nonspecific protection of enzymatic cleavage (Supplementary Material).
4. Conclusions
RNA conformation and dynamics play a significant role in the functional activity of noncoding RNA elements. This is due to the fact that RNA-ligand binding events take place on a slower time-scale than protein-ligand binding, occurring via a complex interplay of tertiary structure capture and induced-fit [28]. The observed change in the conformational profile of S2MG35U from the current SARS-CoV-2 outbreak may result in a change in its functional activity compared to that of S2M from the early 2000’s SARS-CoV outbreak. Previous studies indicate that the stem-loop II motif may have a host-specific target [11], possibly recruiting the 40S ribosomal subunit to the viral genomic RNA and/or sequestering translation initiation factors [9]. Consistent with these possibilities is a recent report of structural differences concentrated in the apical loop of the stem-loop II motif when in a cellular context [12]. This is in contrast to the global change in the 3′-UTR secondary structure predicted solely based on sequence analysis [29]. Consequently, the stem-loop II motif of the current SARS-CoV-2 outbreak may favor a subset of conformations that facilitate interactions with host or viral factors related to its function and human pathogenesis.
Small molecules binding to noncoding RNA elements can directly block the binding of endogenous factors and also can alter the conformational profile of the RNA, thus disrupting the RNA element’s cellular function [13]. The enzymatic probing changes we observed in the presence of polymyxin B and colistin are consistent with a ligand-induced change in the conformational profile of S2MG35U. Given the possible functional importance of the stem-loop II motif conformational profile discussed above, the identification of ligands which alter the secondary structure and/or conformational profile of the stem-loop II motif in SARS-CoV-2 is significant. These ligand-induced changes indicate the druggability of the stem-loop II motif and may provide an avenue for future drug discovery efforts to treat COVID-19 and other diseases caused by viruses containing the stem-loop II motif.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We wish to thank the National Institutes of Health (grants #1R15GM132841 and 1C06 RR14575-01), the Ohio Supercomputer Center and Ohio University for support of this work and Dr. P. Hines and Prof. S. Bergmeier for helpful discussions.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.bbrc.2021.01.013.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Li H., Liu S.-M., Yu X.-H., Tannkg S.-L., Tang C.-K. Coronavirus disease 2019 (COVID-19): current status and future perspective. Int. J. Antimicrob. Agents. 2020;55:105951. doi: 10.1016/j.ijantimicag.2020.105951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu C., Zhou Q., Li Y., Garner L.V., Watkins S.P., Carter L.J., Smoot J., Gregg A.C., Daniels A.D., Jervey S., et al. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent. Sci. 2020;6:315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Madhusoodanan J. To counter the pandemic, clinicians bank on repurposed drugs. Proc. Natl. Acad. Sci. Unit. States Am. 2020;117:10616–10620. doi: 10.1073/pnas.2007346117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rangan R., Zheludev I.N., Das R. RNA genome conservation and secondary structure in SARS-CoV-2 and SARS-related viruses: a first look. RNA. 2020;26:937–959. doi: 10.1261/rna.076141.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deigan Warner K., Hajdin C.E., Weeks K.M. Principles for targeting RNA with drug-like small molecules. Nat. Rev. Drug Discov. 2018;17:547–558. doi: 10.1038/nrd.2018.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang F., Zuroske T., Watts J. RNA therapeutics on the rise. Nat. Rev. Drug Discov. 2020;19:441–442. doi: 10.1038/d41573-020-00078-0. [DOI] [PubMed] [Google Scholar]
- 7.Hermann T. Small molecules targeting viral RNA. WIREs RNA. 2016;7:726–743. doi: 10.1002/wrna.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haniff H.S., Tong T., Liu X., Chen J.L., Suresh B.M., Andrews R.J., Peterson J.M., O’Leary C.A., Benhamou R.I., Moss W.N., et al. Targeting the SARS-CoV-2 RNA genome with small molecule binders and ribonuclease targeting chimera (RIBOTAC) degraders. ACS Cent. Sci. 2020;6:1713–1721. doi: 10.1021/acscentsci.0c00984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robertson M.P., Igel H., Baertsch R., Haussler D., Ares M., Jr., Scott W.G. The structure of a rigorously conserved RNA element within the SARS virus genome. PLoS Biol. 2005;3:86–94. doi: 10.1371/journal.pbio.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tengs T., Jonassen C.M. Distribution and evolutionary history of the mobile genetic element s2m in coronaviruses. Diseases. 2016;4:27. doi: 10.3390/diseases4030027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tengs T., Kristoffersen A.B., Bachvaroff T.R., Jonassen C.M. A mobile genetic element with unknown function found in distantly related viruses. Virol. J. 2013;10:132. doi: 10.1186/1743-422X-10-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huston N.C., Wan H., de Cesaris Araujo Tavares R., Wilen C., Pyle A.M. Comprehensive in-vivo secondary structure of the SARS-CoV-2 genome reveals novel regulatory motifs and mechanisms. Molecular Cell. 2021;(January 1) doi: 10.1016/j.molcel.2020.12.041. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Umuhire J.A., Patwardhan N.N., Hargrove A.E. Understanding the contributions of conformational changes, thermodynamics and kinetics of RNA-small molecule interactions. ACS Chem. Biol. 2019;17:824–838. doi: 10.1021/acschembio.8b00945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diaz J.P., Chirayil R., Chirayil S., Tom M., Head K.J., Luebke K.J. Association of a peptoid ligand with the apical loop of pri-miR-21 inhibits cleavage by Drosha. RNA. 2014;20:528–539. doi: 10.1261/rna.042911.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohio Supercomputer Center . Ohio Supercomputer Center. Ohio Supercomputer Center; Columbus OH: 1987. http://osc.edu/ark:/19495/f5s1ph73 [Google Scholar]
- 16.NCBI. NCBI Virus Database. National Library of Medicine.
- 17.Johnson M., Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., Madden T.L. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cock P.A., Antao T., Chang J.T., Chapman B.A., Cox C.J., Dalke A., Friedberg I., Hamelryck T., Kauff F., et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sponer J., Bussi G., Kreple M., Banãs P., Bottaro S., Cunha R.A., Gil-Ley A., Pinamonti G., Poblete S., Jurečka P., et al. RNA structural dynamics as captured by molecular simulations: a comprehensive overview. Chem. Rev. 2018;118:4171–4338. doi: 10.1021/acs.chemrev.7b00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Musiani F., Rosetti G., Capece L., Gerger T.M., Micheletti C., Varani G., Carloni P. Molecular dynamics simulations identify time scale of conformational changes responsible for conformational selection in molecular recognition of HIV-1 transactivation responsive RNA. J. Am. Chem. Soc. 2014;136:15631–15637. doi: 10.1021/ja507812v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.NDB Nucleic acid database RNA basepair catalog. http://ndbserver.rutgers.edu/ndbmodule/services/BPCatalog/bpCatalog.html
- 22.Klein D.J., Moore P.B., Steitz T.A. The roles of ribosomal proteins in the structure, assembly and evolution of the large ribosomal subunit. J. Mol. Biol. 2004;340:141–177. doi: 10.1016/j.jmb.2004.03.076. [DOI] [PubMed] [Google Scholar]
- 23.Soukup G.A., Breaker R.R. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orac C.M., Zhou S., Means J.A., Boehme D., Bergmeier S.C., Hines J.V. Synthesis and stereospecificity of 4,5-disubstituted oxazolidinone ligands binding to T-box riboswitch RNA. J. Med. Chem. 2011;54:6786–6795. doi: 10.1021/jm2006904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brooks W.H., Guida W.C., Daniel K.G. The significance of chirality in drug design and development. Curr. Top. Med. Chem. 2011;11:760–770. doi: 10.2174/156802611795165098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meanwell N.A. Drug-target interactions that involve the replacement or displacement of magnesium ions. Bioorg. Med. Chem. Lett. 2017;27:5355–5372. doi: 10.1016/j.bmcl.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 27.McCoy L.S., Roberts K.D., Nation R.L., Thompson P.E., Velkov T., Li J., Tor Y. Polymyxins and analogues bind to ribosomal RNA and interfere with eukaryotic translation in vitro. CHEMBiochem Communications. 2013;14:2083–2086. doi: 10.1002/cbic.201300496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gleitsmann K.R., Sengupta R.N., Herschlag D. Slow molecular recognition by RNA. RNA. 2017;23:1745–1753. doi: 10.1261/rna.062026.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hosseini Rad SM A., McLellan A.D. Implications of SARS-CoV-2 mutations for genomic RNA structure and host microRNA targeting. Int. J. Mol. Sci. 2020;21:4807. doi: 10.3390/ijms21134807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.