Abstract
There is increasing evidence that ACE2 gene polymorphism can modulate the interaction between ACE2 and the SARS-CoV-2 spike protein affecting the viral entry into the host cell, and/or contribute to lung and systemic damage in COVID-19. Here we used in silico molecular docking to predict the effects of ACE2 missense variants on the interaction with the spike protein of SARS-CoV-2. HDOCK and FireDock simulations identified 6 ACE2 missense variants (I21T, A25T, K26R, E37K, T55A, E75G) with higher affinity for SARS-CoV-2 Spike protein receptor binding domain (RBD) with respect to wild type ACE2, and 11 variants (I21V, E23K, K26E, T27A, E35K, S43R, Y50F, N51D, N58H, K68E, M82I) with lower affinity. This result supports the hypothesis that ACE2 genetic background may represent the first “genetic gateway” during the disease progression.
Keywords: COVID-19, SARS-CoV-2 Spike protein, ACE2 polymorphism, In silico modeling, SARS-CoV-2 infectiousness, SARS-CoV-2 severity of infection
Highlights
-
•
Significant aspects of SARS-CoV-2 infectivity remain obscure.
-
•
Host-pathogen interactions are determinants of infectivity and clinical course.
-
•
Host genetics might explain heterogeneity in disease susceptibility and severity.
-
•
ACE2 polymorphisms might account for differences in receptor-ligand affinity.
-
•
We show in silico evidence of ACE2 polymorphisms with higher Spike affinity.
1. Introduction
Different phases can be distinguished during the progression of COVID-19 [1,2]. During the first phase, after the incubation period lasting 6 days in average, the onset of disease may be characterized by influenza-like symptoms, from mild to moderate. The second phase, which is known as the pulmonary phase and involves ∼30% of all SARS-CoV-2 infected subjects, is characterized by progressive respiratory involvement with onset of pneumonia-like symptoms. The third phase, which develops in ∼15% of all patients, is known as the pro-inflammatory phase, and is characterized by severe interstitial pneumonia with focal and systemic iper-inflammation, which may lead to acute respiratory distress syndrome, and systemic inflammatory response syndrome. The fourth phase of COVID-19, which is known as the pro-thrombotic phase, develops in ∼5% of patients, and is characterized by the onset of microvascular and macrovascular thrombosis possibly promoted by strong focal and/or systemic inflammation. During this phase patients require medical treatment in intensive care units, and most of them do not survive.
SARS-CoV-2 infection susceptibility and severity seem to be influenced by environmental factors (climate, pollution, cultural, social and economic inequalities, climate, health care system organizations), co-morbidities (high blood pressure, cardiovascular disease, other heart and lung conditions, diabetes, cancer, or compromised immune system), and inter-individual genetic differences [[3], [4], [5], [6]]. Inter-individual genetic differences may affect the spatial transmission dynamics of COVID-19, the susceptibility and severity of disease, and the inflammatory and immune response, and three “genetic gateways” have been proposed accounting for disease progression [7].
Specifically, there is evidence that angiotensin-converting enzyme 2 (ACE2) is the human cell receptor of SARS-CoV-2 [[8], [9], [10]], and it was speculated [5,7,[11], [12], [13], [14], [15], [16], [17]] that ACE2 gene polymorphism may modulate the interaction between ACE2 and the Spike protein of SARS-CoV-2 during the virus entry into the host cell. In particular, differential affinity of a number of ACE2 missense variants for Spike protein was predicted using different computational approaches [12,[18], [19], [20], [21]]. Moreover, since ACE2 regulates the renin-angiotensin-aldosterone system [22], ACE2 missense variants or expression quantitative trait loci (eQTL) variants may contribute to pulmonary and systemic injury by fostering vasoconstriction, inflammation, oxidation and fibrosis, thereby affecting the clinical outcome [4,11,15,[23], [24], [25]]. The possible association between specific ACE2 gene variants and COVID-19 susceptibility, severity, and clinical outcomes is supported by massive genomic data from general population [26], while large-scale genome-wide association studies are urgently needed to firmly establish the causal link [27].
In this study we have used in silico molecular docking to analyze the possible effects of ACE2 single nucleotide polymorphisms (SNPs) leading to missense variants on the interaction between ACE2 and SARS-CoV-2 Spike protein. Molecular docking was performed with HDOCK, a powerful pipeline for integrated protein-protein docking, which is based on hybrid docking algorithm of template-based modeling and ab initio free docking to optimize the adjustment of ligand [[28], [29], [30], [31]]. The HDOCK pipeline differs from other molecular docking platforms in its ability to support amino acid sequences as inputs, and in its hybrid docking strategy in which experimental information on the protein-protein binding site and small-angle X-ray scattering are incorporated during the docking and post-docking processes [28].
With respect to the other pipelines that were previously used to model the interaction between SARS-CoV-2 Spike protein and ACE2 missense variants [5,7,11,[13], [14], [15], [16], [17]], HDOCK has the advantage of integrating two approaches with the same software, together with a remarkable simplicity of use, and it is completely automated with consequent high reproducibility.
2. Materials and methods
2.1. Databases
3D structures of proteins were downloaded from Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB) (http://www.rcsb.org/) [32]. We focused our analysis on structures of SARS-COV-2 Spike Receptor Binding Domain (RBD)/ACE2 complexes 6M17 (10.2210/pdb6M17/pdb) [33] 6LZG (10.2210/pdb6LZG/pdb) [34], and 6M0J (10.2210/pdb6M0J/pdb) [35] models. The Single Nucleotide Polymorphism Database (dbSNP) [36,37] was used to identify the ACE2 SNPs leading to missense variants. Functional information of ACE2 was acquired by UniProt database (Q9BYF1, ACE2_HUMAN) [38]. ACE2 SNP frequencies were obtained by the database GnomAD-Exomes (https://gnomad.broadinstitute.org/). ACE2 bat sequences were downloaded from NCBI database. Multiple alignments of human and bat sequences were carried out by Clustal Omega [39].
2.2. Molecular docking and statistical analysis
HDOCK server (http://hdock.phys.hust.edu.cn/) was used to carry out molecular docking between receptor binding domain (RBD) of SARS-CoV-2 Spike protein and ACE2 wild type or missense variants from the dbSNP. We focused our analysis on two ACE2 N-terminal alpha helices that form the major binding interface with SARS-CoV-2 Spike protein RBD based on X-ray crystallography [[33], [34], [35]]. In our analysis we used, as a receptor, the amino acid sequence of ACE2 wild type or missense variants, and, as a ligand, the SARS-CoV-2 Spike protein RBD models (6LZG chain B, 6M0J chain E or 6M17 chain E) downloaded from RCSB-PDB database. 6M17 was the most complete structure because it contains the ACE2 collectrin-like domain [33]. Although this domain is far from the binding interface, it could still affect the geometry of the protein. Since HDOCK only provides score for ab initio free docking, to compare the complex scores obtained by ab initio free docking and template-based modeling we used FireDock (http://bioinfo3d.cs.tau.ac.il/FireDock/) [40,41].
Results of HDOCK/FireDock simulations were confirmed by submitting HDOCK-generated ACE2/Spike protein RBD complexes to PRODIGY [42]. Furthermore, wild type ACE2 and K26R ACE2 models were also built by using MODELLER 9.25 [43] via Chimera [44], and these models were used as receptors in SwarmDock simulations [45]. HDOCK/FireDock pipeline was also used to evaluate the impact of SARS-CoV-2 Spike protein RBD variants on binding to wild type or K26R ACE2. QMEANDisCo (SwissDock) [46] and MolProbity [47] were used for bad bonds and angles metrics. A detailed protocol of the computational workflow is depicted in Supplementary Fig. S1.
3. Results
3.1. Molecular docking of SARS-CoV-2 Spike protein RBD and ACE2 wild type or missense variants
Two ACE2 N-terminal alpha helices form the major binding interface with SARS-CoV-2 Spike protein RBD based on X-ray crystallography [[33], [34], [35]]. In this region 25 SNPs causing leading to ACE2 missense variants are listed in the dbSNP.
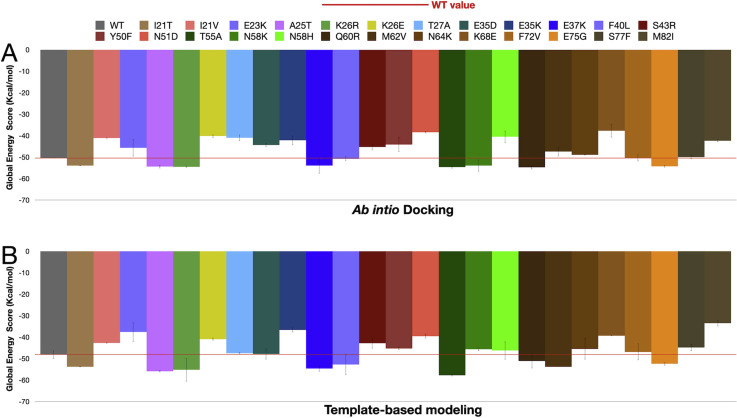
HDOCK and FireDock pipelines were used for molecular docking. For each ACE2 missense variant, three docking simulations were carried, each with a different PDB model (6M17, 6LZG, 6M0J), and the results obtained with the two methods (template-based modeling and ab initio free docking) were analyzed separately.
Before proceeding with the simulations, the quality of the models generated by HDOCK was analyzed and compared with the quality of the corresponding models generated by MODELLER. The analysis with QMEANDisCo demonstrated good quality of all models with global scores similar to those of the control PDB models (6LZG, 6M0J and 6M17). Moreover, percentage of bad angles according to MolProbity was slightly lower with HDOCK (wild type ACE2 = 0.8%; K26R ACE2 = 0.77) compared to MODELLER (1.24% for both wild type and K26R ACE2) (Supplementary Fig. S2).
Overall, HDOCK/FireDock results with the different PDB models and methods were concordant in 92% of cases (Table 1 ; Supplementary Table S1; Fig. S3). For only two polymorphisms (N58K and M62V), the template-based method produced results that differed from those produced by ab initio docking.
Table 1.
ACE2 SNPs analyzed in this study, their frequencies, and Global Energy Score (GES, Kcal/mol) of the interaction between wild type ACE2 or ACE2 missense variants and SARS-Cov-2 Spike protein.
| ACE2 wild type or missense variant | dbSNP ID | Frequency |
Template-based modeling GES (Kcal/mol)a |
Ab initio docking GES (Kcal/mol)a |
Total GES (Kcal/mol)b |
|||
|---|---|---|---|---|---|---|---|---|
| GnomAD Exomes | Average | SD | Average | SD | Average | SD | ||
| Wild type | - | – | −48.15 | 1.83 | −50.45 | 0 | −49.3 | 1.71 |
| I21T | rs1244687367 | 0.000005 | −53.83 | 4.35 | −53.96 | 3.93 | −53.9 | 3.71 |
| I21V | rs778030746 | 0.000011 | −42.76 | 0.43 | −41.17 | 0.65 | −41.97 | 1 |
| E23K | rs756231991 | 0.000005 | −37.66 | 5.38 | −45.68 | 0.43 | −41.67 | 5.56 |
| A25T | rs1434130600 | – | −55.89 | 0.54 | −54.43 | 0.62 | −55.16 | 0.95 |
| K26R | rs4646116 | 0.003971 | −55.2 | 0.36 | −54.51 | 1.43 | −54.86 | 1.01 |
| K26E | rs1299103394 | 0.000005 | −40.94 | 2.48 | −40.24 | 0.69 | −40.59 | 1.67 |
| T27A | rs781255386 | 0.000011 | −47.51 | 1.01 | −40.99 | 2.07 | −44.25 | 3.86 |
| E35D | rs778500138 | – | −47.87 | 1.24 | −44.42 | 3.63 | −46.14 | 3.08 |
| E35K | rs1348114695 | 0.000016 | −36.68 | 4.85 | −42.19 | 1.03 | −39.43 | 4.35 |
| E37K | rs146676783 | 0.000033 | −54.65 | 2.38 | −53.92 | 1.33 | −54.29 | 1.77 |
| F40L | rs924799658 | 0.000016 | −52.76 | 0.44 | −50.75 | 3.34 | −51.76 | 2.4 |
| S43R | rs1447927937 | 0.000005 | −42.82 | 0.99 | −45.28 | 0.39 | −44.05 | 1.5 |
| Y50F | rs1192192618 | 0.000005 | −45.33 | 0.32 | −44.12 | 0.61 | −44.72 | 0.79 |
| N51D | rs760159085 | 0.000005 | −39.55 | 0.72 | −38.45 | 2.67 | −39 | 1.85 |
| T55A | rs775273812 | 0.000006 | −57.8 | 4.06 | −54.67 | 2.7 | −56.24 | 3.53 |
| N58K | rs771621249 | 0.000011 | −45.68 | 3.24 | −54 | 0.84 | −49.84 | 5.03 |
| N58H | rs1222417695 | 0.000011 | −46.26 | 0.06 | −40.53 | 2.16 | −43.39 | 3.42 |
| Q60R | rs759162332 | 0.000011 | −51.12 | 4.87 | −54.81 | 0.12 | −52.96 | 3.68 |
| M62V | rs1325542104 | 0.000006 | −53.77 | 0.03 | −47.37 | 3.07 | −50.57 | 4.01 |
| N64K | rs1199100713 | 0.000005 | −45.51 | 3.81 | −48.97 | 1.46 | −47.24 | 3.2 |
| K68E | rs755691167 | 0.000011 | −39.34 | 0.63 | −37.75 | 0.45 | −38.54 | 1 |
| F72V | rs1256007252 | 0.000005 | −46.91 | 1.51 | −50.35 | 0.88 | −48.63 | 2.18 |
| E75G | rs867318181 | – | −52.47 | 1.29 | −54.27 | 0.44 | −53.37 | 1.31 |
| S77F | rs1234981462 | N.D. | −44.85 | 4.78 | −49.97 | 0.24 | −47.41 | 4.13 |
| M82I | rs766996587 | 0.000011 | −33.49 | 2.7 | −42.39 | 1.05 | −37.94 | 5.21 |
aValues are averages of GES obtained with the three different 3D PDB SRARS-CoV-2 Spike protein RDB structures (6LZG chain B, 6M0J chain E or 6M17 chain E), which were used as ligands. SD, standard deviation. bValues are averages of GES obtained with the two methods (template-based modeling or ab initio docking) and the three different 3D PDB structures.
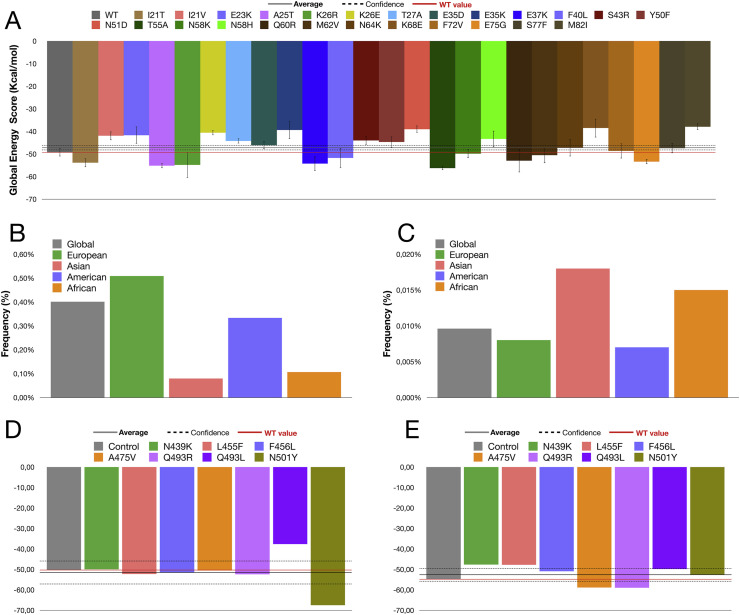
We performed 156 docking simulations (i.e., 26 ACE2 SNPs, S19AQP ACE2, wild type ACE2, all multiplied by three ligands and two methods). Global energy score (GES) average with all simulations was −47.20 kcal/mol (Fig. 1 A, gray line), total standard deviation was 6.39 kcal/mol and confidence interval was ±1.0035 kcal/mol (Fig. 1A, dotted line). The highest GES was −37.94 kcal/mol (M82I), while the lowest one was −56.24 (T55A). We used total GES average, GES value with wild type ACE2 (Fig. 1A, red line), and confidence interval as a threshold to screen the SNPs, considering as relevant only the SNPs that affected significantly the binding with SARS-CoV-2 Spike protein RBD. By using this approach, we found 6 out of the 25 ACE2 missense variants (24%) (I21T, A25T, K26R, E37K, T55A, E75G) that showed higher affinity for SARS-CoV-2 Spike protein RBD with respect to wild type ACE2, and 11 variants (44%) (I21V, E23K, K26E, T27A, E35K, S43R, Y50F, N51D, N58H, K68E, M82I) that exhibited lower affinity in silico (Fig. 1A).
Fig. 1.
Molecular docking simulations and frequencies of ACE2 SNPs that either enhance or the affinity with SARS-Cov-2 Spike protein. A) Global Energy Score (GES) (Kcal/mol) of the interaction between wild type ACE2 or ACE2 missense variants and SARS-Cov-2 Spike protein. Molecular docking simulation were carried out by using HDOCK, which is based on two methods: template-based modeling and ab initio free docking. GES here shown are an average of the GES with the two methods. As reference values are shown: the average GES obtained with all simulations (solid line), the confidence interval (dotted line), and the GES obtained with wild type ACE (red line). B–C) Frequencies of ACE2 SNPs that either enhance (I21T, K26R, E37K, T55A) (B), or reduce (I21V, E23K, K26E, T27A, E35K, S43R, Y50F, N51D, N58H, K68E, M82I) (C) the affinity with SARS-Cov-2 Spike protein, based on GES. D–E) Molecular docking simulations of affinity of known RBD mutations of the Spike protein showing that 5 of the 7 RBD mutations increased binding affinity for wild type ACE2 (D), while 5 of the 7 RBD mutations decreased the binding affinity for K26R ACE2 compared to wild type RBD (E).
3.2. Geographical distribution of ACE2 SNPs affecting binding to SARS-CoV-2 spike protein RBD
GnomAD-Exomes database was used to gain information about frequencies of the examined ACE2 SNPs worldwide (Table 1; Supplementary Table S2). K26R is the most diffused one with a global frequency of 0.3971%. The large diffusion of this SNP is also confirmed by others database: 0.4579% in TOPMED; 0.595% in 4ALFA Project; 0.368% in ExAC; 0.315% in GnomAD; 0.511% in GO-ESP; 0.21% in 1000G; 0.62% in TWINSUK; 0.93% in ALSPAC. In particular, the K26R occurs with highest frequency in European (0.503%) and American (0.329%) populations with maximum value in Ashkenazi Jewish (1.2%), while it is less common in both African (0.099%) and Asian (0.079%) populations.
Cumulative frequency analysis of ACE SNPs demonstrated that ACE2 missense variants exhibiting increased affinity for SARS-CoV-2 Spike protein were more common in European and American populations (Fig. 1B), while those exhibiting reduced affinity were more common in African and Asian populations (Fig. 1C). The frequencies of each ACE2 missense variants were plotted individually in Supplementary Fig. S4.
3.3. Impact of SARS-CoV-2 Spike protein RBD variants on binding to ACE2
A number of missense variants affecting the SARS-CoV-2 Spike protein have recently been identified worldwide, and listed in a comprehensive database [48]. In particular, some of these variants including N439K, L455F, F456L, A475V, Q493R, Q493L and N501Y, fall into the interfaces of binding of RBD. The impact of these RBD variants on binding to wild type ACE2 or K26R ACE2 was then evaluated. We focused on K26R missense variant because of its high frequency in the general population.
Preliminary, PRODIGY was used to confirm the effect of ACE2 K26R missense on wild type Spike protein RBD binding as predicted by FireDock with HDOCK complexes. Moreover, PRODIGY calculated dissociation constants (K d) that were 8.8 E−10 for the ACE2 K26R and 4.610 E−9 for wild type ACE2. The effect of K26R missense was further confirmed using models that were generated by MODELLER, and then submitted to SwarmDock obtaining an energy of −39.88 kcal/mol for wild type ACE2 and of −46.13 kcal/mol for K26R ACE2.
HDOCK/FireDock analysis was performed using the wild type or K26R ACE2 receptor and missense variants of the Spike protein RBD listed above. Results demonstrated that 5 of the 7 RBD mutations increased binding affinity for wild type ACE2 (Fig. 1D), while 5 of the 7 RBD mutations decreased the binding affinity for K26R ACE2 compared to wild type RBD (Fig. 1E).
4. Discussion and conclusions
The present study supports the hypothesis that ACE2 gene polymorphism may contribute to the genetic susceptibility to COVID-19 affecting the SARS-CoV-2 entry into the host cells, thus representing the first “genetic gateways” during disease progression [7]. Our results broaden the list of ACE2 missense variants that can affect the interaction with the SARS-CoV-2 spike protein [7, 18, 19, 21]. Specifically, we focused our attention on ACE2 SNPs affecting two N-terminal alpha helices that form the major binding interface with SARS-CoV-2 Spike protein [[33], [34], [35]].
We did not include in our analysis the S19P variant because it falls into the cleavage site of ACE2 precursor, and it may affect the N-terminal sequence of the mature protein. Besides, there is evidence that the S19P may reduce the affinity for SARS-CoV-2 Spike protein [19]. Results about K26R that is expected to increase the affinity for Spike protein is noteworthy because this variant is relatively frequent in European people with a frequency about 0.5%, which would correspond to a potential target population of 2,230,000 people at the European Union level [12]. In this study we confirmed the results of K26R by using HDOCK that also allowed us to identify additional missense variants (I21T, A25T, E37K, T55A, E75G) with higher affinity for SARS-CoV-2 Spike protein, and 11 variants (I21V, E23K, K26E, T27A, E35K, S43R, Y50F, N51D, N58H, K68E, M82I) with lower affinity.
It is worth of noticing the K26R variant of ACE2 was identified in a COVID-19 patient but not in control subjects in Italy, in a recent genome-wide association study enrolling a cohort of 131 patients and 258 controls [27], further reinforcing the hypothesis that this missense variant may be associated with clinical susceptibility to disease.
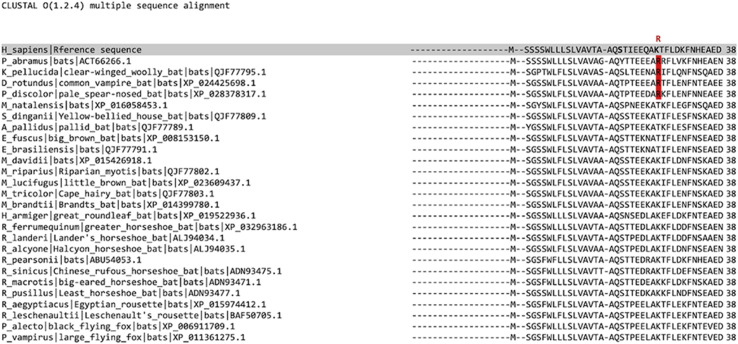
Beside, it may be also relevant to note is rather common in different families of bats including Vespertilionidae and Phyllostomidae (Supplementary Fig. S5). Specifically, Phyllostomidae are diffused in South America (Desmodus rotundus XP_024425698.1, Phyllostomus discolor XP_028378317.1), while Phyllostomidae are very common in China (Pipistrellus abramus ACT66266.1) and Indochina (Kerivoula pellucida QJF77795.1), and the presence of P. abramus was confirmed in the Wuhan area [49].
It is conceivable that the polymorphisms responsible for a higher affinity may be responsible for a greater severity of the disease in humans, especially when very high affinity receptors are overexpressed due to the environmental and pharmacological factors. Of course, underlying diseases would contribute to an even more severe course of the disease, with an intense viral replication capable of infecting in turn a large number of persons, including some individuals with similar ACE2 polymorphisms, and so on. Another aspect to consider is the co-evolution of Spike protein. Indeed, missense mutations in the Spike RBD may have conflicting effects on binding affinity for wild type and K26R ACE2 (Fig. 1D and E).
Polymorphisms in genes coding for proteases from the respiratory tract belonging to the transmembrane protease/serine subfamily (TMPRSS) may also contribute to inter-individual differences in susceptibility and severity of disease [26,50]. Indeed, there is evidence that TMPRSS proteolytic activity induces SARS-CoV Spike protein fusogenic activity, and, notably, SARS-CoV-2 cell entry is dependent on TMPRSS2, and blocked by protease inhibitors [51].
Obviously, the impact of these polymorphisms on severity of outcome should be weighted by appropriate demographic and clinical factors. If this difference were confirmed, this would pave the way for the identification, on a population scale, of healthy individuals whose molecular phenotypes would be responsible for disease that is more serious. Apart from the usual social distancing measures, targeted drug prevention strategies could be evaluated. It could be logical to assess pharmacological prophylactic interventions, as proposed in categories of healthy people at particular risk of exposure such as care-givers. The serine protease inhibitor camostat mesylate, approved in Japan to treat unrelated diseases, has been shown to block TMPRSS2 activity [52,53], and is thus an interesting candidate. Conversely, the identification of broader categories of people with lower risk of developing severe disease, could allow a safer exit from the lock-down phases, while facilitating the establishment of a faster herd immunity, and waiting reliable serological tests and effective vaccine.
Authors’ contribution
M.C. contributed to experimental set-up, pipeline development, in silico analysis; P.F., A.I., and C.B. contributed to study designing and data providing; M.A. and P.A. contributed to coordination, conception, designing and writing. M.A. and P.A. contributed equally to the work. All authors critically revised draft versions of the manuscript and approved the final version.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
We wish to thank prof. Diane Damotte (University of Paris) for advice and critical reading of the manuscript.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.biochi.2020.11.004.
Appendix A. Supplementary data
Workflow and detailed protocol of the computational simulations.
Quality control of models used in in silico simulations.
Global Energy Score (GES) of the interaction between wild type ACE2 or ACE2 missense variants and SARS-Cov-2 Spike protein. A-B) Molecular docking simulation were carried out by using HDOCK, which is based on two methods: template-based modeling and ab initio free docking. (GES) (Kcal/mol) obtained using either the ab initio docking method (A) or template-based modeling method (B) are shown. D-E) the effects of RBD missense variants on GES (Kcal/mol) in wild type ACE2 (C) or K26R ACE2 models (E).
Frequencies of ACE2 missense variants that increase or decrease the binding affinity between ACE2 receptor and SARS-CoV-2 Spike RDB (according GnomAD exome database).
Alignment between human and bat ACE2 amino acid sequences spanning the binding interface with SARS-CoV-2. The R in position 26 in several bat sequences was highlighted in red.
Multimedia component 6.
Complete results of in silico binding analysis obtained using HDOCK and FireDock.
Multimedia component 7.
Frequencies of ACE2 SNPs in the GnomAD-Exomes database.
References
- 1.Dos Santos W.G. Natural history of COVID-19 and current knowledge on treatment therapeutic options. Biomed. Pharmacother. 2020;129:110493. doi: 10.1016/j.biopha.2020.110493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lippi G., Sanchis-Gomar F., Henry B.M. COVID-19: unravelling the clinical progression of nature’s virtually perfect biological weapon. Ann. Transl. Med. 2020;8:693. doi: 10.21037/2Fatm-20-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease control and Prevention (CDC) 2020. Assessing Risk Factors for Severe COVID-19 Illness.https://www.cdc.gov/coronavirus/2019-ncov/covid-data/investigations-discovery/assessing-risk-factors.html accessed. [PubMed] [Google Scholar]
- 4.Alifano M., Alifano P., Forgez P., Iannelli A. Renin-angiotensin system at the heart of COVID-19 pandemic. Biochimie. 2020;174:30–33. doi: 10.1016/j.biochi.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alifano M., Attanasi G., Iannelli F., Cherikh F., Iannelli A. COVID-19 pandemic: a European perspective on health economic policies. JBEP COVID-19 Special Issue. 2020;4:35–43. [Google Scholar]
- 6.Iannelli A., Favre G., Frey S., Esnault V., Gugenheim J., Bouam S., Schiavo L., Tran A., Alifano M M. Obesity and COVID-19: ACE 2, the missing tile. Obes. Surg. 2020;25:1–3. doi: 10.1007/2Fs11695-020-04734-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Debnath M., Banerjee M., Berk M. Genetic gateways to COVID-19 infection: implications for risk, severity, and outcomes. FASEB J. 2020 doi: 10.1096/fj.202001115R. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5(4):562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bosso M., Thanaraj T.A., Abu-Farha M., Alanbaei M., Abubaker J., Al-Mulla F. The two faces of ACE2: the role of ACE2 receptor and its polymorphisms in hypertension and COVID-19. Mol. Ther. Methods Clin. Dev. 2020;18:321–327. doi: 10.1016/2Fj.omtm.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calcagnile M., Forgez P., Iannelli A., Bucci C., Alifano M., Alifano P. ACE2 Polymorphisms and Individual Susceptibility to SARS-CoV-2 Infection: Insights from an in Silico Study. bioRxiv. 2020;4(23) doi: 10.1101/2020.04.23.057042. [DOI] [Google Scholar]
- 13.Cao Y., Li L., Feng Z., Wan S., Huang P., Sun X., Wen F., Huang X., Ning G., Wang W. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov. 2020;6:11. doi: 10.1038/s41421-020-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darbani B. The expression and polymorphism of entry machinery for COVID-19 in human: juxtaposing population groups, gender, and different tissues. Int. J. Environ. Res. Publ. Health. 2020;17:3433. doi: 10.3390/ijerph17103433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Devaux C.A., Rolain J.M., Raoult D. ACE2 receptor polymorphism: susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J. Microbiol. Immunol. Infect. 2020;53:425–435. doi: 10.1016/2Fj.jmii.2020.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Godri Pollitt K.J., Peccia J., Ko A.I., Kaminski N., Dela Cruz C.S., Nebert D.W., Reichardt J.K.V., Thompson D.C., Vasiliou V. COVID-19 vulnerability: the potential impact of genetic susceptibility and airborne transmission. Hum. Genom. 2020;4:17. doi: 10.1186/s40246-020-00267-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lippi G., Lavie C.J., Henry B.M., Sanchis-Gomar F. Do genetic polymorphisms in angiotensin converting enzyme 2 (ACE2) gene play a role in coronavirus disease 2019 (COVID-19)? Clin. Chem. Lab. Med. 2020 doi: 10.1515/cclm-2020-0727. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 18.Hadi-Alijanvand H., Rouhani M. Studying the effects of ACE2 mutations on the stability, dynamics, and dissociation process of SARS-CoV-2 S1/hACE2 complexes. J. Proteome Res. 2020 doi: 10.1021/acs.jproteome.0c00348. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 19.Hussain M., Jabeen N., Raza F., Shabbir S., Baig A.A., Amanullah A., Aziz B. Structural variations in human ACE2 may influence its binding with SARS-CoV-2 spike protein. J. Med. Virol. 2020 doi: 10.1002/jmv.25832. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stawiski E.W., Diwanji D., Suryamohan K., Gupta R., Fellouse F.A., Sathirapongsasuti J.F., Liu J., Jiang Y.P., Ratan A., Mis M. Human ACE2 Receptor Polymorphisms Predict SARS-CoV-2 Susceptibility. bioRxiv. 2020;2020 doi: 10.1038/s42003-021-02030-3. 04.07.024752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Othman H., Bouslama Z., Brandenburg J.T., da Rocha J., Hamdi Y., Ghedira K., Srairi-Abid N., Hazelhurst S. Interaction of the spike protein RBD from SARS-CoV-2 with ACE2: similarity with SARS-CoV, hot-spot analysis and effect of the receptor polymorphism. Biochem. Biophys. Res. Commun. 2020;527:702–708. doi: 10.1016/j.bbrc.2020.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng Y.Y., Ma Y.T., Zhang J.Y., Xie X. COVID-19 and the cardiovascular system. Nat. Rev. Cardiol. 2020;17(5):259–260. doi: 10.1038/s41569-020-0360-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gemmati D., Bramanti B., Serino M.L., Secchiero P., Zauli G., Tisato V. COVID-19 and individual genetic susceptibility/receptivity: role of ACE1/ACE2 genes, immunity, inflammation and coagulation. Might the double X-chromosome in females Be protective against SARS-CoV-2 compared to the single X-chromosome in males? Int. J. Mol. Sci. 2020;21:3474. doi: 10.3390/ijms21103474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson C.P., Sama I.E., Codd V., Webb T.R., Ye S., Lang C.C., Voors A.A., Ng L.L., Samani N.J. Genetic associations with plasma angiotensin converting enzyme 2 concentration: potential relevance to COVID-19 risk. Circulation. 2020;15142(11):1117–1119. doi: 10.1161/CIRCULATIONAHA.120.049007. [DOI] [PubMed] [Google Scholar]
- 25.Strafella C., Caputo V., Termine A., Barati S., Gambardella S., Borgiani P., Caltagirone C., Novelli G., Giardina E., Cascella R. Analysis of ACE2 genetic variability among populations highlights a possible link with COVID-19-related neurological complications. Genes. 2020;11:741. doi: 10.3390/genes11070741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hou Y., Zhao J., Martin W., Kallianpur A., Chung M.K., Jehi L., Sharifi N., Erzurum S., Eng C., Cheng F. New insights into genetic susceptibility of COVID-19: an ACE2 and TMPRSS2 polymorphism analysis. BMC Med. 2020;18:216. doi: 10.1186/s12916-020-01673-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benetti E., Tita R., Spiga O., Ciolfi A., Birolo G., Bruselles A., Doddato G., Giliberti A., Marconi C., Musacchia F., Pippucci T., Torella A., Trezza A., Valentino F., Baldassarri M., Brusco A., Asselta R., Bruttini M., Furini S., Seri M., Nigro V., Matullo G., Tartaglia M., Mari F., GEN-COVID Multicenter Study. Renieri A., Pinto A.M. ACE2 gene variants may underlie interindividual variability and susceptibility to COVID-19 in the Italian population. Eur. J. Hum. Genet. 2020;17:1–13. doi: 10.1038/s41431-020-0691-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan Y., Tao H., He J., Huang S.Y. The HDOCK server for integrated protein-protein docking. Nat. Protoc. 2020;15:1829–1852. doi: 10.1038/s41596-020-0312-x. [DOI] [PubMed] [Google Scholar]
- 29.Yan Y., Wen Z., Wang X., Huang S.Y. Addressing recent docking challenges: a hybrid strategy to integrate template-based and free protein-protein docking. Proteins. 2017;85:497–512. doi: 10.1002/prot.25234. [DOI] [PubMed] [Google Scholar]
- 30.Yan Y., Zhang D., Zhou P., Li B., Huang S.Y. HDOCK: a web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017;45:W365–W373. doi: 10.1093/nar/gkx407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang S.Y., Zou X X. An iterative knowledge-based scoring function for protein-protein recognition. Proteins. 2008;72:557–579. doi: 10.1002/prot.21949. [DOI] [PubMed] [Google Scholar]
- 32.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1107/s0907444902003451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., Wang Q., Zhou H., Yan J., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 36.Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhagwat M. Searching NCBI’s dbSNP database. Curr. Protoc. Bioinform. 2010 Dec doi: 10.1002/2F0471250953.bi0119s32. (Chapter 1):Unit 1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.UniProt Consortium T UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2018;46:2699. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sievers F., Higgins D.G. Multiple Sequence Alignment Methods. Humana Press; Totowa, NJ: 2014. Clustal Omega, accurate alignment of very large numbers of sequences; pp. 105–116. [DOI] [PubMed] [Google Scholar]
- 40.Andrusier N., Nussinov R., Wolfson H.J. FireDock: fast interaction refinement in molecular docking. Proteins. 2007;69:139–159. doi: 10.1002/prot.21495. [DOI] [PubMed] [Google Scholar]
- 41.Mashiach E., Schneidman-Duhovny D., Andrusier N., Nussinov R., Wolfson H.J. FireDock: a web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 2008;36:W229–W232. doi: 10.1093/nar/gkn186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xue L.C., Rodrigues J.P., Kastritis P.L., Bonvin A.M., Vangone A. A. Prodigy: A web server for predicting the binding affinity of protein–protein complexes. Bioinformatics. 2016;32(23):3676–3678. doi: 10.1093/bioinformatics/btw514. [DOI] [PubMed] [Google Scholar]
- 43.Webb B., Sali A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinfor. 2016;54(1):5–6. doi: 10.1002/cpps.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 45.Torchala M., Moal I.H., Chaleil R.A., Fernandez-Recio J., Bates SwarmDock P.A. A server for flexible protein–protein docking. Bioinformatics. 2013;29(6):807–809. doi: 10.1093/bioinformatics/btt038. [DOI] [PubMed] [Google Scholar]
- 46.Studer G., Rempfer C., Waterhouse A.M., Gumienny R., Haas J., Schwede T. T. QMEANDisCo—distance constraints applied on model quality estimation. Bioinformatics. 2020;36(6):1765–1771. doi: 10.1093/bioinformatics/btz828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davis I.W., Leaver-Fay A., Chen V.B., Block J.N., Kapral G.J., Wang X., Murray L.W., Arendall W.B., Snoeyink J., Richardson J.S., Richardson D.C. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35(suppl_2):W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shaminur Rahman M., Rafiul Islam M., Nazmul Hoque M., Rubayet Ul Alam A.S.M., Akther m., Akter Puspo J., Akter S., Anwar A., Sultana M., Anwar Hossain M. Comprehensive annotations of the mutational spectra of SARS-CoV-2 spike protein: a fast and accurate pipeline. Transbound Emerg. 2020 doi: 10.1111/tbed.13834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tong C.P. HKU Theses Online (HKUTO); 2016. Distribution and preference of landscape features and foraging sites of insectivorous bats in Hong Kong urban parks. [Google Scholar]
- 50.Vargas-Alarcón G., Posadas-Sánchez R., Ramírez-Bello J. Variability in genes related to SARS-CoV-2 entry into host cells (ACE2, TMPRSS2, TMPRSS11A, ELANE, and CTSL) and its potential use in association studies. Life Sci. 2020;260:118313. doi: 10.1016/j.lfs.2020.118313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271-280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawase M., Shirato K., van der Hoek L., Taguchi F., Matsuyama S. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J. Virol. 2012;86:6537-6545. doi: 10.1128/JVI.00094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou Y., Vedantham P., Lu K., Agudelo J., Jr Carrion R., Nunneley J.W., Barnard D., Pöhlmann S., McKerrow J.H., Renslo A.R., Simmons G. Protease inhibitors targeting coronavirus and filovirus entry. Antivir. Res. 2015;116:76–84. doi: 10.1016/j.antiviral.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Workflow and detailed protocol of the computational simulations.
Quality control of models used in in silico simulations.
Global Energy Score (GES) of the interaction between wild type ACE2 or ACE2 missense variants and SARS-Cov-2 Spike protein. A-B) Molecular docking simulation were carried out by using HDOCK, which is based on two methods: template-based modeling and ab initio free docking. (GES) (Kcal/mol) obtained using either the ab initio docking method (A) or template-based modeling method (B) are shown. D-E) the effects of RBD missense variants on GES (Kcal/mol) in wild type ACE2 (C) or K26R ACE2 models (E).
Frequencies of ACE2 missense variants that increase or decrease the binding affinity between ACE2 receptor and SARS-CoV-2 Spike RDB (according GnomAD exome database).
Alignment between human and bat ACE2 amino acid sequences spanning the binding interface with SARS-CoV-2. The R in position 26 in several bat sequences was highlighted in red.