Abstract
Antiviral drug development for coronavirus disease 2019 (COVID-19) is occurring at an unprecedented pace, yet there are still limited therapeutic options for treating this disease. We hypothesized that combining drugs with independent mechanisms of action could result in synergy against SARS-CoV-2, thus generating better antiviral efficacy. Using in silico approaches, we prioritized 73 combinations of 32 drugs with potential activity against SARS-CoV-2 and then tested them in vitro. Sixteen synergistic and eight antagonistic combinations were identified; among 16 synergistic cases, combinations of the US Food and Drug Administration (FDA)-approved drug nitazoxanide with remdesivir, amodiaquine, or umifenovir were most notable, all exhibiting significant synergy against SARS-CoV-2 in a cell model. However, the combination of remdesivir and lysosomotropic drugs, such as hydroxychloroquine, demonstrated strong antagonism. Overall, these results highlight the utility of drug repurposing and preclinical testing of drug combinations for discovering potential therapies to treat COVID-19.
Keywords: SARS-CoV-2, drug combinations, drug synergy, in silico design, knowledge mining, nitazoxanide remdesivir combo, CPE assay, drug repurposing, combination therapy, COVID-19
Graphical Abstract
Sixteen synergistic drug combinations against SARS-CoV-2 were identified in silico and confirmed experimentally. Combinations of nitazoxanide with remdesivir, amodiaquine, or umifenovir exhibited significant synergy against SARS-CoV-2. Remdesivir combined with hydroxychloroquine or other lysosomotropic drugs resulted in strong antagonism.
Introduction
At the start of the coronavirus disease 2019 (COVID-19) pandemic, few antiviral drug combinations were explored as options for treating the disease despite past evidence that show they are especially effective in treating viral diseases. Namely, drug combinations are useful in treating viral infections due to the fact that they can substantially lower the risk of the development of resistance to any one drug.1 Additionally, the antiviral action of the combination may be stronger than either drug alone, a phenomenon known as synergy. Antiviral synergy has been previously illustrated in combinations tested against Ebola and other viruses.2 Often the reason why synergism occurs remains unclear—it is difficult to provide explanations for existing synergistic/antagonistic drug combinations without prior, extensive, experimental investigations.3
Some evidence suggests that combinations of antiviral drugs are more likely to be synergistic if they (1) are of different classes, (2) have independent mechanisms of action, or (3) act upon different stages of the virus life cycle.1,4 Not only could the drugs under study work through a mechanism of action (MoA) that directly inhibits portions of the viral life cycle, they could also interact with components of host-regulated pathways involved in viral replication to hinder viral replication. For example, arbidol and its analogs have been noted to disrupt the interaction between the spike glycoprotein of the SARS-CoV-2 and its host receptor ACE2, as well as the host proteases furin and TMPRSS2, which are important in proteolytic activation of the virus.5,6 According to ClinicalTrials.gov, though there have been many compounds tested against SARS-CoV-2 in vitro, many of these drugs are only being evaluated as single agents in enzymatic and cellular assays to determine antiviral activity.7 Comparatively, there has been limited systematic screening of drug combinations.8
Meanwhile, the situation in clinical trials is somewhat different. Of the 2,844 clinical trials relevant to COVID-19 as of July 31, 2020, ca. 150 describe drug combinations. However, many of these trials evaluate the same combinations, e.g., lopinavir-ritonavir and azithromycin-hydroxychloroquine. Perhaps the earliest, most noteworthy antiviral drug combination was lopinavir-ritonavir—a phase II trial for a triple antiviral therapy combining interferon-β1b, lopinavir-ritonavir, and ribavirin was shown to shorten the duration of viral shedding and hospital stay in patients with mild to moderate COVID-19.9 However, another study assessing the effectiveness of lopinavir-ritonavir alone in treating COVID-19 found that its administration in COVID-19 patients had no benefit.10 As of July 31, 2020, no combination therapy has yielded positive results in phase III randomized clinical trials.11
There are many ongoing or upcoming clinical trials testing combinations to treat COVID-19, but few have undergone extensive preclinical studies prior to their application on patients. Due to a lack of such studies, more information is needed on the combinatorial use of existing drugs against SARS-CoV-2 in order to (1) more efficiently prioritize synergistic combinations for translation into clinical use and (2) flag antagonistic combinations for further preclinical evaluation prior to clinical trials. To this point, we used data- and text-mining approaches to propose drug combinations for repurposing against SARS-CoV-2,12 operating on the assumption that combinations of drugs with differing mechanisms might exhibit synergistic activity. The goal of this study is to report the antiviral activity, synergy, and antagonism of 73 binary combinations of 31 drugs identified earlier12 as demonstrated in an in vitro SARS-CoV-2 cytopathic assay.
Results
Performance of Matrix Screening
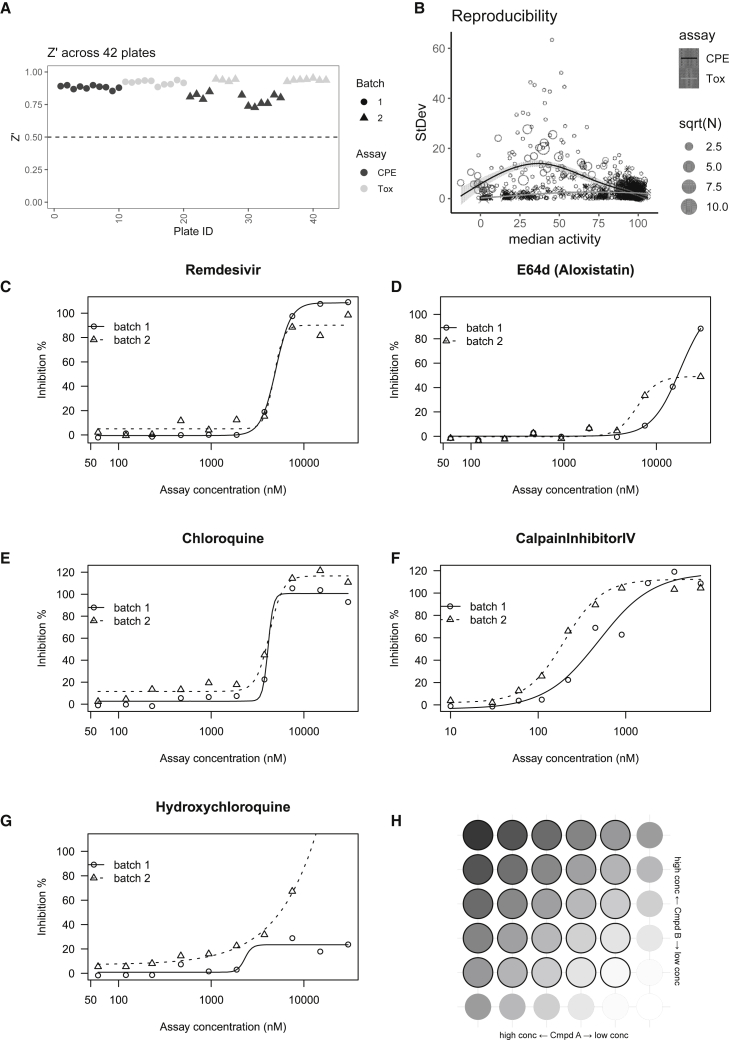
In total, we screened 73 pairwise combinations in a 6 × 6 dose matrix format, which involved two biological batches (cell and SARS-CoV-2 virus) and two assays (cytopathic effect and cytotoxicity against Vero E6 cells) across 42 384-well plates including replicates (Tables S1 and S4). The Z′ factor was robust across batches and assays (all Z′ > 0.7; Figure 1A). Each batch was assessed by a benchmark compound collection including five known antivirals, performed in biosafety level-3 (BSL-3). We did not observe significant drift of potencies or efficacies between batches, except for hydroxychloroquine, which consistently resulted in inconclusive dose-response curves (Figures 1C–1G; Table S2). In addition, we performed a third QC to check the reproducibility across all available replicates in CPE (cytopathic effect) or Tox (toxicity) assays (Figure 1B). CPE activity showed a biphasic trend between median activity and standard deviation: most reproducible (SD < 20) when activity is extreme (activity < 20 or > 75) but less reproducible in between (Figure 1B, ◯ points). This is probably due to the high sensitivity to technical/biological variations when the concentration is close to the EC50 (half maximal effective concentration). This biphasic reproducibility of CPE activity also highlights the importance of using a dose matrix, instead of a single dose combination, to enhance the confidence of synergism/antagonism findings. In contrast, Tox activity is highly reproducible regardless of the median activity (Figure 1B, ✕ points).
Figure 1.
Performance of Matrix Screening
(A) Z′ factor on different assays (CPE or Tox) and biological batches. (B) Reproducibility across all replicates (defined as a compound at certain concentration). Number of replicates (n) may vary, e.g., more single-agent replicates were performed due to matrix setting. (C–G) Dose response curves from an independent benchmark set performed at a different site. (H) Layout of a 6 × 6 dose matrix. Wells with (or without) bold border represent dose combination (or single agent alone).
Overview of Hits
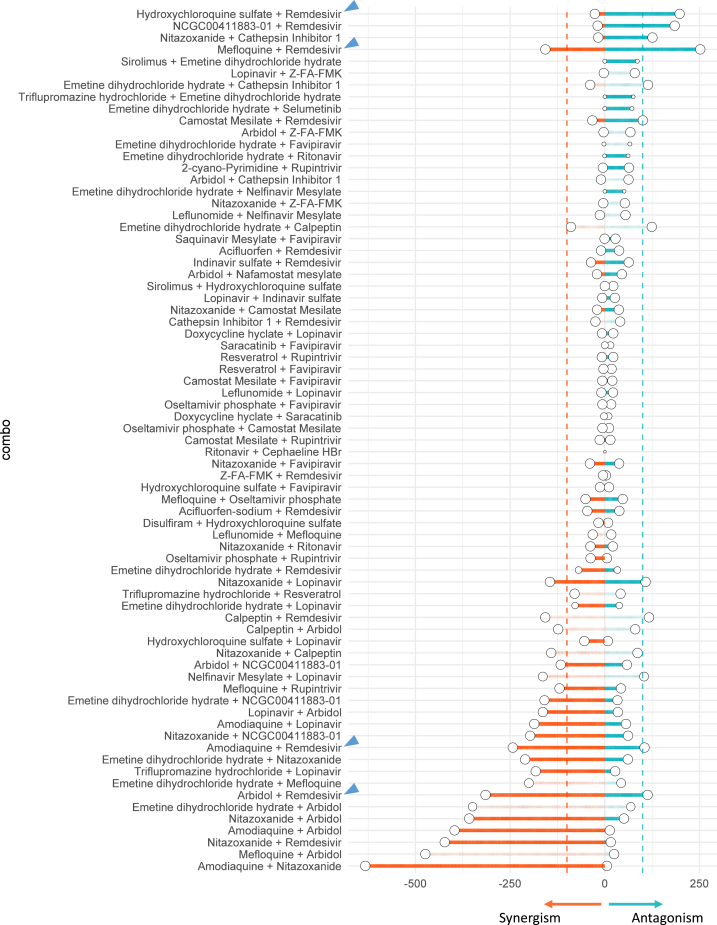
Since synergism and antagonism might occur simultaneously in a concentration-dependent manner, we employed the highest single agent (HSA) synergy model and analyzed synergism and antagonism separately (Figure 2). Within 73 binary combinations of 32 compounds, we identified 16 synergistic and 8 antagonistic combinations, four of which displayed both synergistic and antagonistic interactions at different compound concentrations (Figures 3 and 4). There are 29 combinations with inconclusive determinations that require additional validation due to cytotoxicity or the roughness of the activity landscape (see Materials and Methods). A summary of the screening result is available in Table S4. A map of drug combinations depicting their synergism/antagonism outcomes is shown in Figure 4.
Figure 2.
Summary of Synergism or Antagonism across 73 Combinations
Due to biphasic dose response, synergism was separated from antagonism. Synergism is calculated as the sum of HSA.neg values from non-toxic dose combinations (Tox > 50%) and vice versa. The size of circle reflected the confidence of the observed synergism/antagonism (bigger circle = less doses were excluded due to toxicity). The inconclusive blocks (nnon-toxic < 25 or rough activity landscape) were shown in transparent points and lines. Two dashed lines indicated the cutoff of HSA synergism (−100) or antagonism (100). Blue arrows highlighted the combinations between remdesivir and tertiary amine compounds from conclusive blocks.
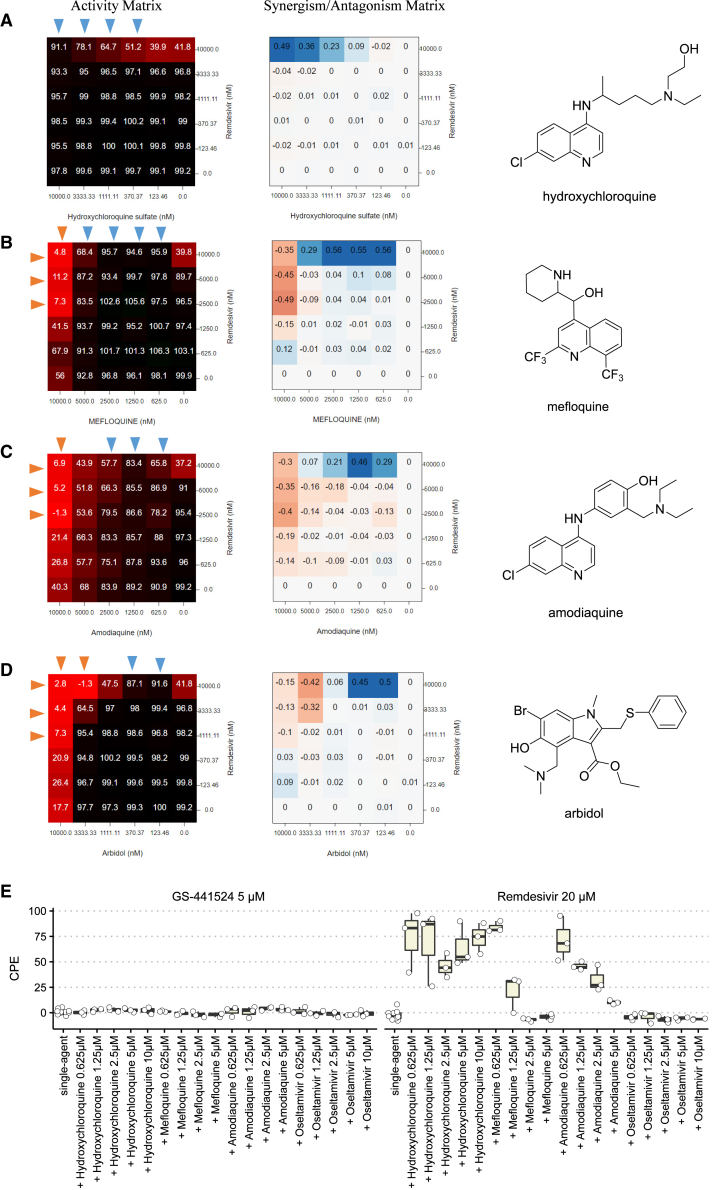
Figure 3.
Matrix Blocks from Remdesivir + Amine Drugs in CPE Assay
The activity was normalized so that 100 corresponded to full cytopathic effect and 0 corresponded to no cytopathic effect. Red arrow, the concentrations that synergize with the partner compound. Blue arrow, the concentrations that antagonize against the partner compound. Chemical structures were shown on the right. (A) Hydroxychloroquine, (B) mefloquine, (C) amodiaquine, (D) arbidol. (E) CPE data from single-dose combination (5 μM GS-441524 or 20 μM remdesivir ± amine compounds; nsingle-agent RdRp inhibitor = 12 and ncombination = 3). The lower and upper box hinges correspond to the first and third quartiles, and the whiskers extend from upper or lower hinges to 1.5 * IQR (inter-quartile range).
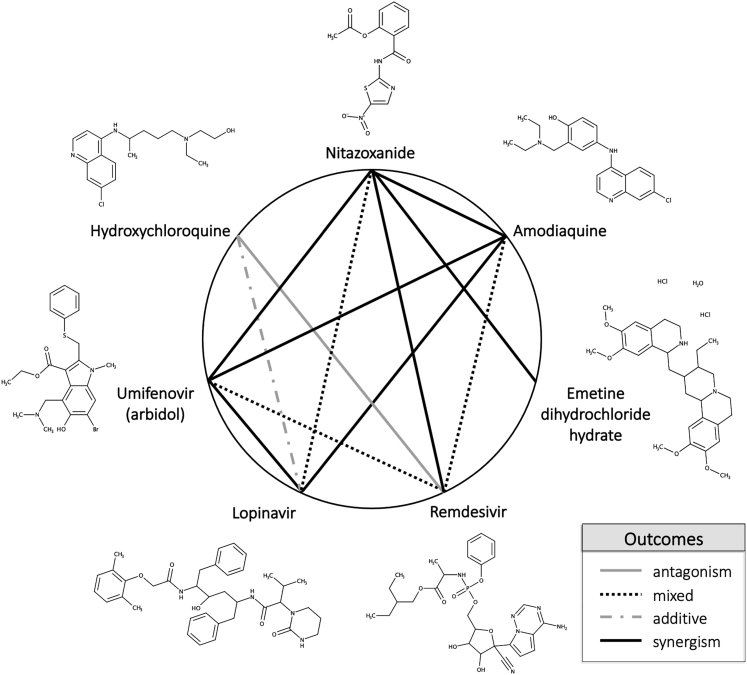
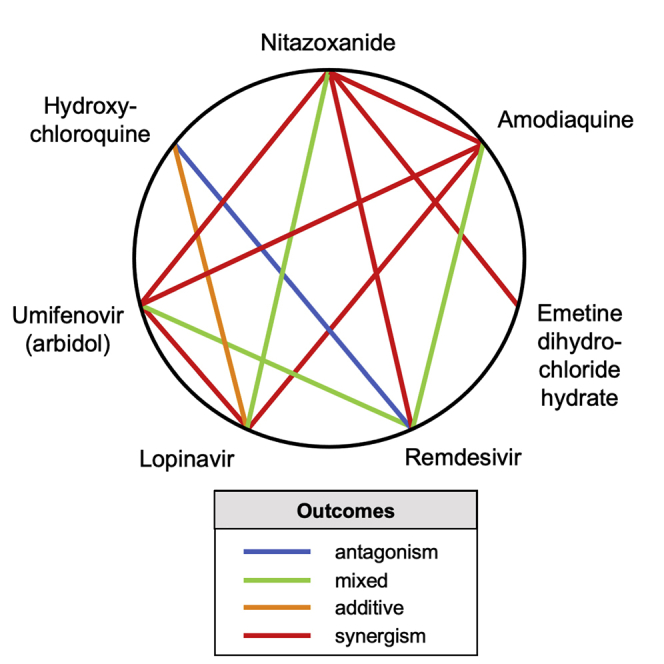
Figure 4.
Heptagonal Polygonogram Depicting Some of the Binary Combinations Tested in the Study
Degrees of synergism/antagonism were ascertained from Figure 3. The definitions were defined based on the degree of HSA synergism/antagonism determined in the CPE assay.
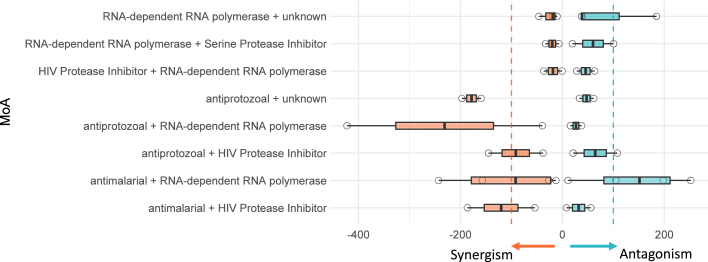
Next, we investigated whether antiviral synergism or antagonism is attributable to combination of different MoAs. A previous study demonstrated that synergism/antagonism is predictable based on MoA in an oncology screening.13 Unfortunately, we found limited evidence of MoA-associated synergism/antagonism (up to 10 μM) for SARS-CoV-2 (Figure 5). The most antagonistic MoA combination came from the combination of remdesivir (an RNA-dependent RNA polymerase [RdRp] inhibitor) and an antimalarial drug, in which three (hydroxychloroquine, amodiaquine, and mefloquine) out of four appeared to be antagonistic (Figure 2). However, the current data is insufficient to conclusively infer any MoA-associated synergism/antagonism.
Figure 5.
Summary of Synergism or Antagonism over Different Mechanism of Action (MoA) Combination
Inconclusive blocks or singleton MoA were excluded. Two dashed lines indicated the cutoff of HSA synergism (−100) or antagonism (100). The lower and upper box hinges correspond to the first and third quartiles, and the whiskers extend from upper or lower hinges to 1.5 * IQR (inter-quartile range).
Antagonism between Remdesivir and Lysosomotropic Agents
Most notably, our results demonstrate a strong antagonistic effect between remdesivir and the antimalarial drugs hydroxychloroquine, mefloquine, and amodiaquine (Figure 3). The most striking antagonism was observed in the combination of the only two drugs ever approved with US Food and Drug Administration (FDA) emergency use authorization (EUA) to treat COVID-19: hydroxychloroquine and remdesivir (the EUA for hydroxychloroquine has since been withdrawn by the FDA). Our results showed that 10 μM hydroxychloroquine could completely extinguish the antiviral activity of remdesivir in vitro. The antagonistic effect exerted by hydroxychloroquine could be observed at a concentration as low as 0.37 μM (Figure 3A). Other aminoquinoline drugs, mefloquine (Figure 3B) and amodiaquine (Figure 3C), showed similar antagonism at low concentrations, but they could synergize with remdesivir at high concentrations.
Likewise, we observed a similar antagonism against remdesivir from another drug with a tertiary amine moiety, umifenovir (Arbidol), at low concentrations (123–370 nM; Figure 3D). Umifenovir synergized with remdesivir at high concentrations (3–10 μM). Hydroxychloroquine, mefloquine, and amodiaquine are known lysosomotropic agents,14 and umifenovir is a lipophilic weak base (cLogP = 5.5), suggesting an association between reduced antiviral efficacy of remdesivir by lysosomotropic amines.
To further investigate the mechanism, we tested GS-441524 (a remdesivir analog absent of McGuigan prodrug moiety) to see if antagonism could be maintained. Both GS-441524 (5 μM) and remdesivir (20 μM) alone completely rescued the cytopathic effect (CPE ≈ 0) by SARS-CoV-2. However, hydroxychloroquine (0.625–10 μM), mefloquine (0.625–1.25 μM), and amodiaquine (0.625–5 μM) significantly antagonized against remdesivir, but not GS-441524 (Figure 3E). The non-lysosomotropic amine drug oseltamivir (cLogP = 2.1) demonstrated no antagonism with remdesivir (Figure 3E). Our results suggest that lysosomotropic agents antagonize remdesivir by impairing its upstream activation (e.g., esterase-mediated hydrolysis) rather than the formation of triphosphate active agent.
Synergism between Nitazoxanide and Remdesivir, Amodiaquine, or Umifenovir
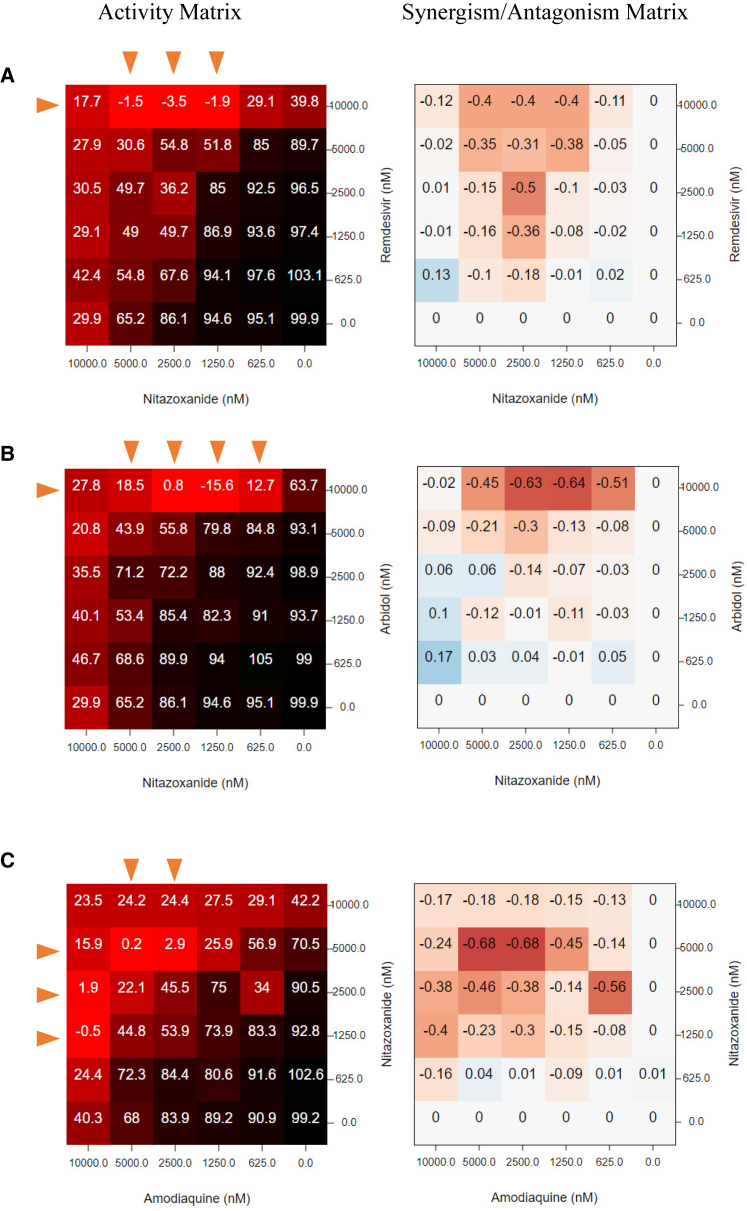
Among 16 synergistic combinations, we observed an enrichment of nitazoxanide, an FDA-approved broad-spectrum antiviral and antiparasitic drug. The three most synergistic combinations with nitazoxanide, i.e., nitazoxanide + remdesivir/umifenovir/amodiaquine are shown in Figure 6. A complete rescue of CPE could be observed from 0.625–5 μM of nitazoxanide when combined with remdesivir/umifenovir/amodiaquine, where any of these drugs alone could only achieve a maximum 40%–60% rescue (Figures 6A–6C). Nitazoxanide is not cytotoxic when concentration is below 5 μM. However, we observed a mild toxicity (∼20%) of nitazoxanide to Vero E6 cells at 10 μM (Table S3), which may explain the vanishment of synergy at this concentration.
Figure 6.
Matrix Blocks from Three Synergistic Combinations Involving Nitazoxanide
(A) Nitazoxanide + remdesivir; (B) nitazoxanide + arbidol; (C) nitazoxanide + amodiaquine. Red arrow, the concentrations that synergize with the partner compound.
Discussion
Combining modern computational techniques and experimental approaches, we have identified 16 synergistic antiviral combinations in vitro (Figure 2). Somewhat unexpectedly, our results also revealed an antagonism between remdesivir and hydroxychloroquine, the two drugs approved with FDA EUA for COVID-19 (the EUA for hydroxychloroquine has since been revoked, as of June 15, 2020). Other lysosomotropic agents, such as mefloquine, amodiaquine, and umifenovir, also antagonize remdesivir, suggesting a general mechanism. These findings demonstrate the importance of preclinical research investigating antiviral drug combinations prior to their application in patients, as well as the utility of data- and text-mining approaches to explore MoAs underlying synergism/antagonism within the context of COVID-19. Lack of preclinical studies on combinations prior to their administration in patients may significantly increase the risk of antagonism and undesirable side effects. The matrix screening platform presented in this study is an efficient, data-driven method for prioritizing synergistic combinations and flagging undesirable pharmacological interactions.
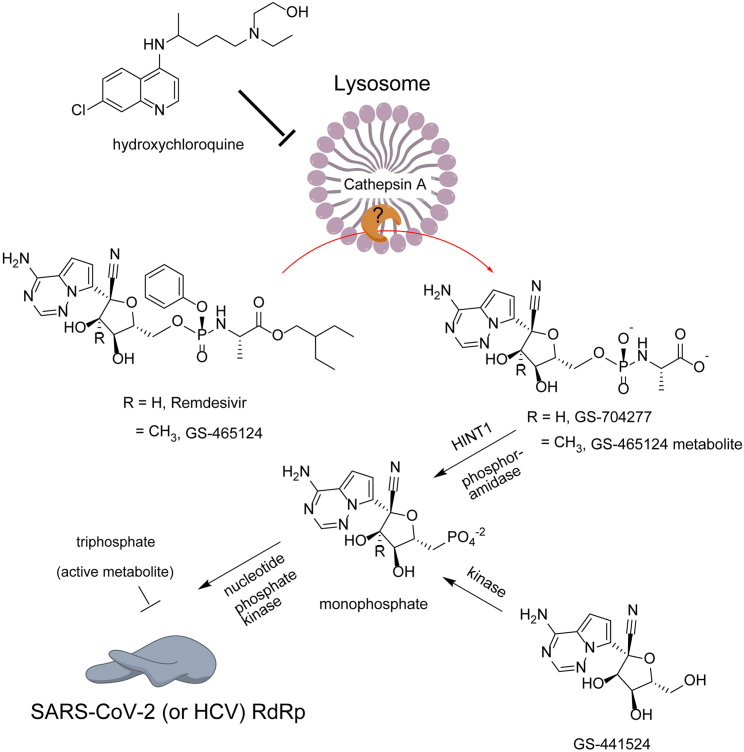
Remdesivir is a nucleotide analog McGuigan prodrug, which inhibits SARS-CoV-2 RdRp through inducing delayed chain termination.15 Although the key enzymes required to activate remdesivir into the active triphosphate form have not been fully revealed, we hypothesize that remdesivir shares, at least partially, a similar activation pathway with GS-465124, a metabolite from nucleotide prodrug GS-6620 for hepatitis C virus, based on the chemical similarity (Figure 7).16 Both remdesivir and GS-465124 are adenosine-like phosphoramidates, with the only difference being a methyl group on the 2′ pentose ring. Therefore, remdesivir is also likely to be hydrolyzed into alanine metabolite (GS-704277) intracellularly by cathepsin A, akin to GS-465124.16 Since cathepsin A is an acidic pH-dependent serine protease strictly located in lysosome, the lysosomotropic agents such as hydroxychloroquine may reduce the amount of monophosphate intermediate by increasing lysosomal pH.17 This mechanism is in line with our finding that lysosomotropic agents do not antagonize GS-441254, which does not need cathepsin A for activation (Figures 3E and 7). More mechanistic studies of remdesivir are in progress at NCATS, utilizing label-free mass spectra to elucidate the exact mechanisms underlying this striking antagonism.
Figure 7.
The Putative Mechanism of the Antagonism between Remdesivir and the Lysosomotropic Agent
We hypothesize that lysosomotropic agents antagonize remdesivir by impairing its upstream activation (e.g., esterase-mediated hydrolysis) rather than the formation of triphosphate active agent.
We have observed a notable synergism for combinations of nitazoxanide with remdesivir, umifenovir, or amodiaquine (Figure 6). Nitazoxanide is an FDA approved, bioavailable, broad-spectrum anti-infective drug, which recently has been investigated for use against SARS-CoV-2 owing to its previously established anticoronaviral activities.18 It was originally identified as a potential antiviral drug-repurposing candidate against SARS-CoV-2 with an IC50 of about 2 μM in a focused compound screening including remdesivir and chloroquine.19 Previous studies have suggested a complex antiviral mechanism for nitazoxanide, including activating retinoic-acid-inducible protein I (RIG-I) signaling20 and blocking entry distinct from that of endosomal pH-neutralizing agents.21 The concentration at which we observed synergy with remdesivir (>1.25 μM, equivalent to 0.383 mg/L) is achievable in plasma and lung trough even at low dose.22 Nitazoxanide is well tolerated, and there is no report of any significant adverse effects from healthy adults.23 As of July 31, 2020, ClinicalTrials.gov shows 17 clinical trials using nitazoxanide to treat COVID-19, either in combination with other drugs (ivermectin, hydroxychloroquine, atazanavir) or as a standalone therapy. Recently, a randomized placebo-controlled trial of 392 participants has demonstrated modest efficacy of nitazoxanide with respect to 7-day viral load and symptom resolution in an outpatient setting.24 Therefore, the combination of nitazoxanide with remdesivir (or its metabolite GS-441524) looks the most promising from a clinical perspective because both drugs would potentially be available for use for the treatment of COVID-19 (nitazoxanide is FDA approved and remdesivir has an EUA).
Despite the synergism and antagonism demonstrated by drug combinations reported in this study, we emphasize that these results require further validation. Vero E6 cells do not express the serine proteases TMPRSS2/4, the two proteases crucial for viral entry through the early membrane fusion pathway.25 Vero E6 also expresses P-glycoprotein (P-gp),26 indicating the observed synergy could be simply due to P-gp inhibition, which enhances the exposure of remdesivir in vitro. Likewise, it has been noted that while working with SARS-CoV-2, passaging repeatedly through Vero E6 cells can result in the deletion of a putative furin-cleavage site in the spike glycoprotein of the virus.27 This is problematic, as this furin-cleavage site has been noted to be important in determining the host range and pathogenesis of SARS-CoV-2;28,29 thus, this mutation could produce in inconclusive results in CPE studies performed in this cell line. Thus, all synergistic and antagonistic combinations need to be verified in other cell lines to determine how penetrant the reported synergies are, for example, in a TMPRSS2/4-expressing cell line or primary airway cell model. The synergy between amodiaquine and nitazoxanide in Vero E6 may not translate to Calu-3 (TMPRSS2+) where single-agent amodiaquine is >10-fold less potent.25 Moreover, the observed synergism/antagonism, especially those occurring only at high concentration (e.g., umifenovir, maximum serum concentration [Cmax] = 467 ng/mL, equivalent to 0.98 μM30), may not be achievable in vivo due to complex pharmacokinetics. Meanwhile, antagonism sometimes can be circumvented by altering dosing schedule or formulation. Therefore, more in-depth in vivo validation and pharmacokinetic modeling are still necessary.
Principally, these findings suggest that lysosomotropic agents such as hydroxychloroquine should be prescribed and used with increased caution in COVID-19 patients due to their relatively long half-life (usually weeks). This is consistent with a statement issued by the FDA on June 15, 2020, warning that combinations of chloroquine/hydroxychloroquine with remdesivir may reduce the antiviral effectiveness of remdesivir against SARS-CoV-2. In regard to the possible clinical relevancy of any of the synergistic drug combinations discovered herein, it is important to note that many of these (e.g., remdesivir and mefloquine, amodiaquine, or umifenovir) are dose dependent, demonstrating antagonism at lower concentrations and synergism at higher concentrations. This is relevant to any clinical dosing regimen that could be derived for these drugs, as determining an appropriate dosing combination is likely more difficult in vivo than the in vitro studies presented here. Additionally, higher concentrations of these drugs in combination could result in more exaggerated or markedly different biological/cytotoxic effects than those observed in vitro. Thus, we suggest that testing of these drug combinations in animal models of SARS-CoV-2 should be tried after further validation in different human cell lines.
Of the binary combinations tested, we identified 16 synergistic and 8 antagonistic combinations, with four of them exhibiting both synergy and antagonism. All together, our findings demonstrate the utility of in silico tools for rational selection of drug combinations, the importance of preclinical testing of drug combinations prior to their administration in patients, and the overall promise of using drug repurposing and combination therapies against SARS-CoV-2.
Materials and Methods
Experimental Design
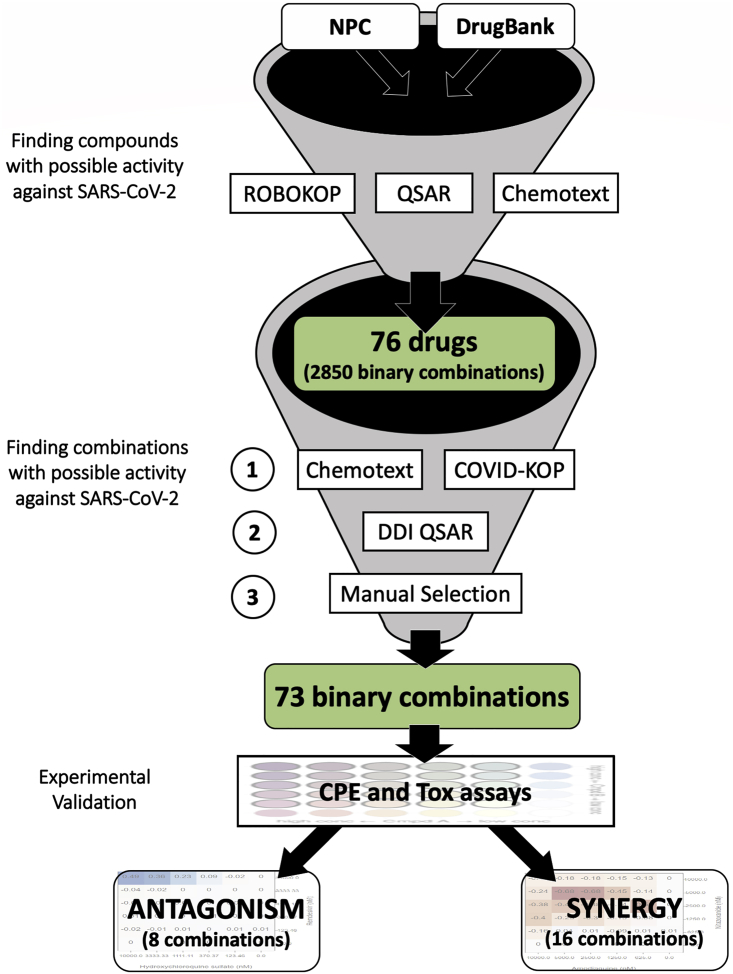
A detailed description of our study design is provided in our recent paper12 and is also outlined in Figure 8. In the beginning, we applied a combination of text mining (Chemotext),31 knowledge mining (ROBOKOP/COVID-KOP knowledge graphs),32,33 and machine learning (QSAR)34 tools to identify existing drugs with possible activities against SARS-CoV-2. Thus, we first identified a list of 76 individual drug candidates for repurposing in combination therapy against COVID-19. These drugs could result in 2,850 unique binary combinations. From this selection of possible combinations, we then prioritized combinations of drugs with different mechanisms of action and/or targeting the virus at different stages of its life cycle, which increases the probability of synergy between drugs.8,35 The rationale behind combination selection is depicted in Figure S1 through the example of umifenovir and emetine, which are suspected to act upon different stages of the viral life cycle. A combination of in silico tools such as Chemotext,31 the recently developed COVID-KOP, and QSAR models of major drug-drug interactions36 were used to determine if compounds had been previously tested together and if any negative drug-drug interactions were to be expected. Analyzing the result obtained using these tools, we finally prioritized 73 combinations of 32 drugs for testing in vitro against SARS-CoV-2. It should be noted that in addition to these, we also identified 95 ternary combinations of 15 drugs in silico, which will be described in a future study. A brief description of the aforementioned computational tools is provided below.
Figure 8.
Study Design for Selecting Possible Synergistic Drug Combinations
In this study, we report only 73 binary combinations; 95 ternary combinations identified in a similar fashion will be reported in a future study.
In Silico Tools
Chemotext
Chemotext is a publicly available web server that mines the published literature in PubMed in the form of medical subject headings (MeSH) terms.31 One capability of Chemotext is finding papers that share two search terms, in this case, the MeSH terms of compound #1 and compound #2, and returning papers that contain both these MeSH terms. These shared MeSH terms, depicting proteins, chemicals, etc., allow us to hypothesize on how these two compounds may be connected, namely via shared biochemical pathways discussed in these common papers. Chemotext was used for elucidation of the relationships between drugs, protein targets, SARS-CoV-2, and COVID-19 from the papers’ annotated MEDLINE/PubMed database.
ROBOKOP37 and COVID-KOP33
ROBOKOP is a data-mining tool developed within Biomedical Data Translator Initiative38 to efficiently query, store, rank, and explore sub-graphs of a complex knowledge graph for hypothesis generation and scoring. We have used ROBOKOP in a similar fashion as Chemotext; Chemotext can help the user to find and impute the connections among drugs, targets, and diseases, and ROBOKOP can help explore and score them. COVID-KOP, a new knowledge base integrating the existing ROBOKOP biomedical knowledge graph with information from recent biomedical literature on COVID-19 annotated in the CORD-19 collection,39 was developed midway through the project and was subsequently utilized instead of ROBOKOP.
QSAR Models
QSAR models developed by us earlier were used for selection of drugs40,41 that could be repurposed as combinations and exclusion of potential drug-drug interactions and side effects.36 All models were developed according to the best practices of QSAR modeling,34,42,43 with special attention paid for data curation44, 45, 46 and rigorous external validation.47 Mixture-specific descriptors and validation techniques48 specially developed for modeling of drug combinations were utilized for modeling of drug-drug interactions.36
Assay Protocols
All the protocols and results are freely available to the scientific community at https://opendata.ncats.nih.gov/covid19/matrix.49 In brief, 30 nL of each compound in DMSO was acoustically dispensed into assay-ready plates (ARPs). Media was then added to the plates at 5 μL/well and incubated at room temperature to dissolve the compounds. Vero E6 cells (selected for high ACE2 expression) were premixed with SARS-CoV-2 (USA_WA1/2020) at a MOI of 0.002 and were dispensed as 25 mL/well into ARPs within 5 min in a BSL-3 lab. The final cell density was 4,000 cells/well. The cells and virus were incubated with compounds for 72 hr, then viability was assayed by ATP content using CellTiter-Glo (Promega).
For the cytotoxicity assay, ARPs were prepared in the same way as for CPE assay. Then, 5 mL/well of media was dispensed into assay plates. Vero E6 cells, suspended in medium, were dispensed into assay plates at 25 µL/well for a final cell density of 4,000 cells/well. Assay plates were incubated for 72 hr at 37°C, 5% CO2, 90% humidity, before viability was assayed by ATP content using CellTiter-Glo (Promega).
ARP Production
To generate the compound source plate, a Perkin-Elmer Janus automated workstation was used to transfer compounds from 1.4 mL matrix 2D barcode tube (sample tube) to the individual wells of Echo Qualified 384-well polypropylene microplate 2.0∗∗, along with positive and neutral (DMSO) controls (Labcyte, San Jose, CA, USA). The plates were centrifuged for 2 min at 100 × g. A comma-separated file was generated that contained the unique plate and well pairings for each of the compound matrix blocks for the Labcyte acoustic dispenser (Labcyte, San Jose, CA, USA). Compounds were dispensed to generate ARPs using an access laboratory workstation with dual Echo 655 dispensers (Labcyte). Each plate was sealed with a peelable aluminum seal, remaining covered until the initiation of the biological assay, frozen at −80°C until used for screening.
Data Analysis
CPE and Tox activity were normalized using independent control wells on each plate, so activity values were not strictly bounded between [0, 100]. For CPE assay, DMSO + virus was treated as the neutral control, whereas DMSO-only (no virus) served as the positive control. A calpain inhibitor IV was used as batch control (2 μg/mL final assay concentration). Normalized CPE activity = 1 − (x - neutralCtrl)/(positiveCtrl − neutralCtrl) × 100%. For Tox assay, DMSO-only was used as the neutral control and media-only wells (no cell) as the negative control. Normalized Tox activity = (x − negativeCtrl)/(neutralCtrl − negativeCtrl) × 100%. Plate-level data was pivoted to block-level data, and replicates were median aggregated.
Synergism and antagonism from a 6 × 6 block were evaluated using the HSA model.50 Given a dose combination Aconc1 + Bconc2,
Therefore, we have
Synergism, HSA(∗) < 0
Antagonism, HSA(∗) > 0
Additivity, HSA(∗) = 0.
To account for dose-dependent synergism and antagonism, we analyzed the negative HSA (HSA.neg) and positive HSA (HSA.pos) separately. The overall synergism (or antagonism) given a 6 × 6 block was calculated as the sum of all negative (or positive) HSA(Aconc∗+Bconc∗) across the non-toxic dose combinations (defined as Tox activity > 50).
Since variation of CPE activity is heteroscedastic (Figure 1B), which made it more difficult to ascertain the reproducibility of synergy/antagonism (given limited resources), we evaluated the smoothness of the 2D activity landscape. Smoothness was calculated as the root-mean-square deviation (RMSD) between the actual and Gaussian smoothed (σ = 2) landscape. If a block had RMSD (observed, Gaussian smoothed) > 20 or < 25 non-toxic CPE values, inconclusive synergism/antagonism was recorded. Otherwise, synergism and/or antagonism were recorded if HSA.neg < −100 and/or HSA.pos > 100.
Acknowledgments
Data-mining tools used in this study were developed under the Biomedical Data Translator Initiative of the National Center for Advancing Translational Sciences, National Institutes of Health (NIH) (grants OT3TR002020 and OT2R002514) and under support of the NIH (grant 1U01CA207160). This research was also supported by the Intramural Research Programs of the National Center for Advancing Translational Sciences (NCATS), NIH, United States.
Author Contributions
Conceptualization, A.V.Z. and E.N.M.; Methodology, T.B., L.C., H.G., R.T.E., C.Z.C., W.Z., S.M., A.S., M.D.H., A.V.Z., and E.N.M.; Validation, T.B., L.C., H.G., R.T.E., C.Z.C., Z.I., and P.S.; Formal Analysis, L.C. and H.G. Investigation, T.B., L.C., H.G., R.T.E., C.Z.C., Z.I., and P.S.; Data Curation, T.B., L.C., H.G., R.T.E.; Writing – Original Draft, T.B. and L.C., Writing – Review & Editing, all authors; Visualization, T.B. and L.C.
Declaration of Interests
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2020.12.016.
Contributor Information
Alexey V. Zakharov, Email: alexey.zakharov@nih.gov.
Eugene N. Muratov, Email: murik@email.unc.edu.
Supplemental Information
References
- 1.Einav S., Sobol H.D.D., Gehrig E., Glenn J.S.S. The hepatitis C virus (HCV) NS4B RNA binding inhibitor clemizole is highly synergistic with HCV protease inhibitors. J. Infect. Dis. 2010;202:65–74. doi: 10.1086/653080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun W., He S., Martínez-Romero C., Kouznetsova J., Tawa G., Xu M., Shinn P., Fisher E., Long Y., Motabar O. Synergistic drug combination effectively blocks Ebola virus infection. Antiviral Res. 2017;137:165–172. doi: 10.1016/j.antiviral.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chou T.C. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 2006;58:621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 4.Sun X., Vilar S., Tatonetti N.P. High-Throughput Methods for Combinatorial Drug Discovery. Sci. Transl. Med. 2013;5:205rv1. doi: 10.1126/scitranslmed.3006667. [DOI] [PubMed] [Google Scholar]
- 5.Choudhary S., Silakari O. Scaffold morphing of arbidol (umifenovir) in search of multi-targeting therapy halting the interaction of SARS-CoV-2 with ACE2 and other proteases involved in COVID-19. Virus Res. 2020;289:198146. doi: 10.1016/j.virusres.2020.198146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vankadari N. Arbidol: A potential antiviral drug for the treatment of SARS-CoV-2 by blocking trimerization of the spike glycoprotein. Int. J. Antimicrob. Agents. 2020;56:105998. doi: 10.1016/j.ijantimicag.2020.105998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Menden M.P., Wang D., Mason M.J., Szalai B., Bulusu K.C., Guan Y., Yu T., Kang J., Jeon M., Wolfinger R., AstraZeneca-Sanger Drug Combination DREAM Consortium Community assessment to advance computational prediction of cancer drug combinations in a pharmacogenomic screen. Nat. Commun. 2019;10:2674. doi: 10.1038/s41467-019-09799-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hung I.F.N., Lung K.C., Tso E.Y.K., Liu R., Chung T.W.H., Chu M.Y., Ng Y.Y., Lo J., Chan J., Tam A.R. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395:1695–1704. doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M. A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe Covid-19. N. Engl. J. Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., ACTT-1 Study Group Members Remdesivir for the Treatment of Covid-19—Preliminary Report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muratov E., Zakharov A. Viribus Unitis: Drug Combinations as a Treatment Against COVID-19. chemRxiv. 2020 doi: 10.26434/chemrxiv.12143355.v1. [DOI] [Google Scholar]
- 13.Richards R., Schwartz H.R., Honeywell M.E., Stewart M.S., Cruz-Gordillo P., Joyce A.J., Landry B.D., Lee M.J. Drug antagonism and single-agent dominance result from differences in death kinetics. Nat. Chem. Biol. 2020;16:791–800. doi: 10.1038/s41589-020-0510-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glaumann H., Motakefi A.-M., Jansson H. Intracellular distribution and effect of the antimalarial drug mefloquine on lysosomes of rat liver. Liver. 1992;12:183–190. doi: 10.1111/j.1600-0676.1992.tb01045.x. [DOI] [PubMed] [Google Scholar]
- 15.Eastman R.T., Roth J.S., Brimacombe K.R., Simeonov A., Shen M., Patnaik S., Hall M.D. Remdesivir: A Review of Its Discovery and Development Leading to Emergency Use Authorization for Treatment of COVID-19. ACS Cent. Sci. 2020;6:672–683. doi: 10.1021/acscentsci.0c00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murakami E., Wang T., Babusis D., Lepist E.I., Sauer D., Park Y., Vela J.E., Shih R., Birkus G., Stefanidis D. Metabolism and pharmacokinetics of the anti-hepatitis C virus nucleotide prodrug GS-6620. Antimicrob. Agents Chemother. 2014;58:1943–1951. doi: 10.1128/AAC.02350-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferner R.E., Aronson J.K. Chloroquine and hydroxychloroquine in covid-19. BMJ. 2020;369:m1432. doi: 10.1136/bmj.m1432. [DOI] [PubMed] [Google Scholar]
- 18.Rossignol J.F. Nitazoxanide, a new drug candidate for the treatment of Middle East respiratory syndrome coronavirus. J. Infect. Public Health. 2016;9:227–230. doi: 10.1016/j.jiph.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jasenosky L.D., Cadena C., Mire C.E., Borisevich V., Haridas V., Ranjbar S., Nambu A., Bavari S., Soloveva V., Sadukhan S. The FDA-Approved Oral Drug Nitazoxanide Amplifies Host Antiviral Responses and Inhibits Ebola Virus. iScience. 2019;19:1279–1290. doi: 10.1016/j.isci.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jurgeit A., McDowell R., Moese S., Meldrum E., Schwendener R., Greber U.F. Niclosamide Is a Proton Carrier and Targets Acidic Endosomes with Broad Antiviral Effects. PLoS Pathog. 2012;8:e1002976. doi: 10.1371/journal.ppat.1002976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajoli R.K., Pertinez H., Arshad U., Box H., Tatham L., Curley P., Neary M., Sharp J., Liptrott N.J., Valentijn A. Dose prediction for repurposing nitazoxanide in SARS-CoV-2 treatment or chemoprophylaxis. medRxiv. 2020 doi: 10.1101/2020.05.01.20087130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stockis A., Allemon A.M., De Bruyn S., Gengler C. Nitazoxanide pharmacokinetics and tolerability in man using single ascending oral doses. Int. J. Clin. Pharmacol. Ther. 2002;40:213–220. doi: 10.5414/cpp40213. [DOI] [PubMed] [Google Scholar]
- 24.Rocco P.R.M., Silva P.L., Cruz F.F., Junior A.C.M., Tierno P.F.G.M.M., Moura M.A., De Oliveira L.F.G., Lima C.C., Dos Santos E.A., Junior W.F. Early use of nitazoxanide in mild Covid-19 disease: randomized, placebo-controlled trial. medRxiv. 2020 doi: 10.1101/2020.10.21.20217208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ko M., Jeon S., Ryu W.-S., Kim S. Comparative analysis of antiviral efficacy of FDA-approved drugs against SARS-CoV-2 in human lung cells: Nafamostat is the most potent antiviral drug candidate. bioRxiv. 2020 doi: 10.1101/2020.05.12.090035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riva L., Yuan S., Yin X., Martin-Sancho L., Matsunaga N., Pache L., Burgstaller-Muehlbacher S., De Jesus P.D., Teriete P., Hull M.V. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020;586:113–119. doi: 10.1038/s41586-020-2577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davidson A.D., Williamson M.K., Lewis S., Shoemark D., Carroll M.W., Heesom K.J., Zambon M., Ellis J., Lewis P.A., Hiscox J.A., Matthews D.A. Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med. 2020;12:68. doi: 10.1186/s13073-020-00763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klimstra W.B., Tilston-Lunel N.L., Nambulli S., Boslett J., McMillen C.M., Gilliland T., Dunn M.D., Sun C., Wheeler S.E., Wells A. SARS-CoV-2 growth, furin-cleavage-site adaptation and neutralization using serum from acutely infected hospitalized COVID-19 patients. J. Gen. Virol. 2020;101:1156–1169. doi: 10.1099/jgv.0.001481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson B.A., Xie X., Kalveram B., Lokugamage K.G., Muruato A., Zou J., Zhang X., Juelich T., Smith J.K., Zhang L. Furin Cleavage Site Is Key to SARS-CoV-2 Pathogenesis. bioRxiv. 2020 doi: 10.1101/2020.08.26.268854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng P., Zhong D., Yu K., Zhang Y., Wang T., Chen X. Pharmacokinetics, metabolism, and excretion of the antiviral drug arbidol in humans. Antimicrob. Agents Chemother. 2013;57:1743–1755. doi: 10.1128/AAC.02282-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Capuzzi S.J., Thornton T.E., Liu K., Baker N., Lam W.I., O’Banion C.P., Muratov E.N., Pozefsky D., Tropsha A. Chemotext: A Publicly Available Web Server for Mining Drug-Target-Disease Relationships in PubMed. J. Chem. Inf. Model. 2018;58:212–218. doi: 10.1021/acs.jcim.7b00589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bizon C., Cox S., Balhoff J., Kebede Y., Wang P., Morton K., Fecho K., Tropsha A. ROBOKOP KG and KGB: Integrated Knowledge Graphs from Federated Sources. J. Chem. Inf. Model. 2019;59:4968–4973. doi: 10.1021/acs.jcim.9b00683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korn D., Bobrowski T., Li M., Kebede Y., Wang P., Owen P., Vaidya G., Muratov E., Chirkova R., Bizon C., Tropsha A. COVID-KOP: Integrating Emerging COVID-19 Data with the ROBOKOP Database. chemRxiv. 2020 doi: 10.26434/chemrxiv.12462623.V1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tropsha A. Best Practices for QSAR Model Development, Validation, and Exploitation. Mol. Inform. 2010;29:476–488. doi: 10.1002/minf.201000061. [DOI] [PubMed] [Google Scholar]
- 35.Bulusu K.C., Guha R., Mason D.J., Lewis R.P.I., Muratov E., Kalantar Motamedi Y., Cokol M., Bender A. Modelling of compound combination effects and applications to efficacy and toxicity: state-of-the-art, challenges and perspectives. Drug Discov. Today. 2016;21:225–238. doi: 10.1016/j.drudis.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 36.Zakharov A.V., Varlamova E.V., Lagunin A.A., Dmitriev A.V., Muratov E.N., Fourches D., Kuz’min V.E., Poroikov V.V., Tropsha A., Nicklaus M.C. QSAR Modeling and Prediction of Drug-Drug Interactions. Mol. Pharm. 2016;13:545–556. doi: 10.1021/acs.molpharmaceut.5b00762. [DOI] [PubMed] [Google Scholar]
- 37.Morton K., Wang P., Bizon C., Cox S., Balhoff J., Kebede Y., Fecho K., Tropsha A. ROBOKOP: an abstraction layer and user interface for knowledge graphs to support question answering. Bioinformatics. 2019;35:5382–5384. doi: 10.1093/bioinformatics/btz604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biomedical Data Translator Consortium The Biomedical Data Translator Program: Conception, Culture, and Community. Clin. Transl. Sci. 2019;12:91–94. doi: 10.1111/cts.12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.COVID-19 Open Research Dataset Challenge (CORD-19) 2020. An AI challenge with AI2, CZI, MSR, Georgetown, NIH, and the White House. Kaggle.https://www.kaggle.com/allen-institute-for-ai/CORD-19-research-challenge [Google Scholar]
- 40.Zakharov A.V., Zhao T., Nguyen D.-T., Peryea T., Sheils T., Yasgar A., Huang R., Southall N., Simeonov A. Novel Consensus Architecture To Improve Performance of Large-Scale Multitask Deep Learning QSAR Models. J. Chem. Inf. Model. 2019;59:4613–4624. doi: 10.1021/acs.jcim.9b00526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alves V.M., Bobrowski T., Melo-Filho C.C., Korn D., Auerbach S., Schmitt C., Muratov E.N., Tropsha A. QSAR modeling of SARS-CoV Mpro inhibitors identifies Sufugolix, Cenicriviroc, Proglumetacin and other drugs as candidates for repurposing against SARS-CoV-2. Mol. Inform. 2020 doi: 10.1002/minf.202000113. Published online July 28, 2020. [DOI] [PubMed] [Google Scholar]
- 42.Muratov E.N., Bajorath J., Sheridan R.P., Tetko I.V., Filimonov D., Poroikov V., Oprea T.I., Baskin I.I., Varnek A., Roitberg A. QSAR without borders. Chem. Soc. Rev. 2020;49:3525–3564. doi: 10.1039/d0cs00098a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cherkasov A., Muratov E.N., Fourches D., Varnek A., Baskin I.I., Cronin M., Dearden J., Gramatica P., Martin Y.C., Todeschini R. QSAR modeling: where have you been? Where are you going to? J. Med. Chem. 2014;57:4977–5010. doi: 10.1021/jm4004285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fourches D., Muratov E., Tropsha A. Trust, but verify: on the importance of chemical structure curation in cheminformatics and QSAR modeling research. J. Chem. Inf. Model. 2010;50:1189–1204. doi: 10.1021/ci100176x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fourches D., Muratov E., Tropsha A. Trust, but Verify II: A Practical Guide to Chemogenomics Data Curation. J. Chem. Inf. Model. 2016;56:1243–1252. doi: 10.1021/acs.jcim.6b00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fourches D., Muratov E., Tropsha A. Curation of chemogenomics data. Nat. Chem. Biol. 2015;11:535. doi: 10.1038/nchembio.1881. [DOI] [PubMed] [Google Scholar]
- 47.Golbraikh A., Tropsha A. Beware of q2! J. Mol. Graph. Model. 2002;20:269–276. doi: 10.1016/s1093-3263(01)00123-1. [DOI] [PubMed] [Google Scholar]
- 48.Muratov E.N., Varlamova E.V., Artemenko A.G., Polishchuk P.G., Kuz’min V.E. Existing and Developing Approaches for QSAR Analysis of Mixtures. Mol. Inform. 2012;31:202–221. doi: 10.1002/minf.201100129. [DOI] [PubMed] [Google Scholar]
- 49.Brimacombe K.R., Zhao T., Eastman R.T., Hu X., Wang K., Backus M., Baljinnyam B., Chen C.Z., Chen L., Eicher T. An OpenData portal to share COVID-19 drug repurposing data in real time. bioRxiv. 2020 doi: 10.1101/2020.06.04.135046. [DOI] [Google Scholar]
- 50.Foucquier J., Guedj M. Analysis of drug combinations: current methodological landscape. Pharmacol. Res. Perspect. 2015;3:e00149. doi: 10.1002/prp2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.