Abstract
Rationale & Objective
A number of serologic tests for immunoglobulin G (IgG) against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are now commercially available, including multiple lateral flow immunoassays (LFIAs), which have the advantage of being inexpensive and easy to use, without the reliance on laboratory facilities. However, data on the development of humoral immunity to SARS-CoV-2 in patients with kidney disease is limited, and the utility of an LFIA to test for antibodies in these patients has not been assessed.
Study Design
Observational study.
Setting & Participants
60 patients (40 hemodialysis and 20 kidney transplant recipients) with SARS-CoV-2 infection confirmed by viral reverse transcriptase–polymerase chain reaction (RT-PCR) testing and 88 historic negative-control samples (collected before September 2019).
Test
A commercially available LFIA to test for SARS-CoV-2 IgG in patients with infection confirmed by viral RT-PCR testing.
Outcomes
Sensitivity and specificity of the LFIA to detect SARS-CoV-2 IgG in dialysis patients and transplant recipients.
Results
56/58 (96.6%) patients (38/39 hemodialysis and 18/19 transplant recipients) tested positive for SARS-CoV-2 IgG. 5/7 (71.4%) patients who were negative on preliminary testing had detectable IgG when retested more than 21 days postdiagnosis. Median times to first and second tests after diagnosis were 17 (interquartile range, 15-20) and 35 (interquartile range, 30-39) days, respectively. Calculation of test characteristics gave sensitivity of 96.6% (95% CI, 88.3%-99.4%) and specificity of 97.7% (95% CI, 92.0-99.6%).
Limitations
Possible exposure to other beta-coronaviruses that may cross-react with the antigen used in the LFIA cannot be excluded.
Conclusions
Symptomatic dialysis patients and transplant recipients commonly develop an immune response against SARS-CoV-2 infection that can be detected using an LFIA. Used diligently, an LFIA could be used to help screen the dialysis populations or confirm exposure on a patient level, especially in facilities in which laboratory resources are limited.
Index Words: SARS-CoV-2 antibodies, COVID-19, lateral flow immunoassays (LFIAs), hemodialysis, transplant
Graphical abstract
Plain-Language Summary.
This study investigates the use of a point-of-care test to detect severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antibodies in 40 maintenance hemodialysis patients and 20 transplant recipients with confirmed coronavirus disease 2019 (COVID-19) infection. All patients were symptomatic at the time of diagnosis. Most patients, 38/39 (97.4%) hemodialysis patients and 18/19 (94.7%) transplant recipients, tested positive for SARS-CoV-2 immunoglobulin G antibody using the point-of-care test. 2/88 (2.3%) control samples, taken from individuals pre-pandemic, were positive. The results show that a point-of-care test can detect serologic responses in patients with end-stage kidney disease with clinically meaningful accuracy. Because these tests do not require laboratory resources, they may be used to enable equity of access to serologic testing in patients with kidney disease globally.
Patients with chronic kidney disease have been severely affected by the coronavirus disease 2019 (COVID-19) pandemic. Not only have they had to contend with the inability to effectively shield, resulting in a high risk for infection and its sequelae, but they have also had to face the anxiety associated with disruption of treatment regimens.1, 2, 3, 4, 5, 6, 7 As a consequence, there has been a unified call to governing bodies from professional nephrological societies around the globe to ensure that patients with kidney disease are not disadvantaged in terms of access to treatment (medication or dialysis), personal protective equipment, and COVID-19 testing.4
The current gold-standard diagnostic test for acute infection is identifying viral RNA with reverse transcription-polymerase chain reaction (RT-PCR) of isolates from upper respiratory tract swabs, using oligonucleotides directed to nucleocapsid or viral RNA-dependent RNA polymerase genes.8,9 Access to PCR testing around the world has not been uniform. However, even in countries in which there are no restrictions on testing, there are several potential limitations to the use of nucleic acid tests in diagnosing COVID-19. These limitations include both the need for specialized laboratory staff to perform molecular diagnostic techniques and the potential for false-negative test results, which may be linked to inadequate nasopharyngeal sampling. Hence the sensitivity and specificity of PCR from nasopharyngeal swabbing is thought to be 80% to 90% and 100%, respectively.10 In addition to these limitations, PCR testing does not generate information on prior disease or assess the development of immunity, which requires serologic testing.
The development of serologic tests for immunoglobulin G (IgG) against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus that causes COVID-19, has been an area of intense investigation and a number of tests are now commercially available. Due to the clinical urgency, independent validation of these tests has been occurring postmarketing, and there is a lack of data for special patient populations such as those with kidney disease. Although evidence is emerging that enzyme-linked immunosorbent assays (ELISAs) are more sensitive than lateral flow immunoassays (LFIAs) or point-of-care tests, the latter are inexpensive, are fast, and do not rely on laboratory facilities.11,12 They may therefore be an option to enable access to rapid SARS-CoV-2 testing in patients with kidney disease, including those where laboratory resources are limited.
In this study, we assess the sensitivity and specificity of a commercially available LFIA to detect IgG against SARS-CoV-2 in patients with kidney disease with confirmed SARS-CoV-2 infection.
Methods
Participants
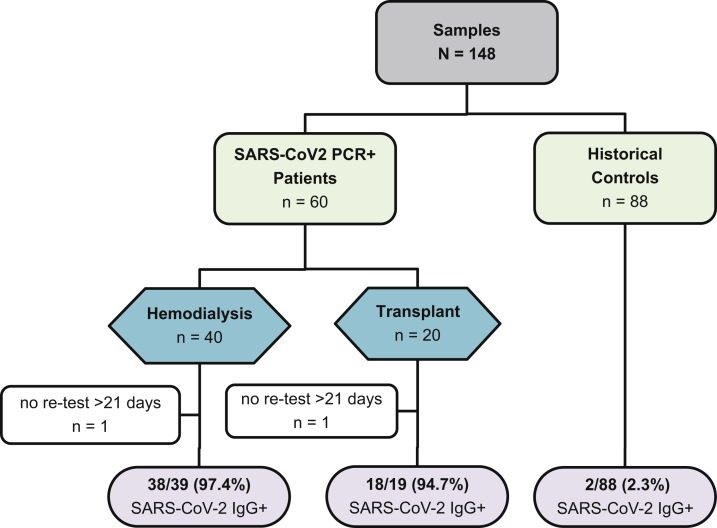
All participants were prospectively recruited from Imperial College Renal and Transplant Centre, London, and provided written informed consent before participation. The study was approved by the Health Research Authority Research Ethics Committee (reference: 20/WA/0123-The Impact of COVID19 on Patients With Renal disease and Immunosuppressed Patients). A study flow diagram may be seen in Fig 1.
Figure 1.
Study flow diagram. Abbreviations: IgG, immunoglobulin G; PCR, polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Samples
Sixty samples were collected from maintenance hemodialysis patients and kidney transplant recipients. Patients were identified by screening inpatient renal wards and outpatient renal clinics for individuals who were a minimum of 7 days post–positive PCR test results until 40 hemodialysis patients and 20 transplant recipients had been recruited. Twenty-five patients were recruited as outpatients, whereas 35 patients were inpatients at the time of enrollment. All patients had undergone RT-PCR testing due to symptoms. Patients who tested negative for IgG antibodies underwent repeat testing if the first test was performed on a sample taken 21 or fewer days after confirmation of infection.
Disease severity was classified according to the World Health Organisation (WHO) as severe (respiratory rate ≥ 30 breaths/min, blood oxygen saturation ≤ 93%, Pao2 to fraction of inspired oxygen ratio< 300 or infiltrates affecting 50% of the lung field within 24 to 48 hours) or critical (respiratory failure, septic shock, and/or multiple organ dysfunction/failure).13 All other cases were classified as mild-moderate and we subdivided this group into mild disease (those who received outpatient care only) and moderate disease (those who required admission to the hospital).
Eighty-eight plasma samples were collected from individuals in the United Kingdom before September 2019 and were used as controls (Table S1). Cases were first identified from paired tissue bank samples for all patients with SARS-CoV-2 infection included in the study cohort. When these samples were exhausted, we identified sequentially collected samples working back from the inclusion date.
LFIA Antibody Testing
We tested a commercially available LFIA (Biomedomics Inc) according to the manufacturer’s instruction. The test uses SARS-CoV-2 antigen MK201027, which is located in the receptor binding domain of the spike protein.14 The test is validated for use with whole blood, serum, and plasma; in this study, both whole blood and plasma samples were included in the analysis (because all historic samples were plasma or serum). To ensure reproducibility of results between whole blood, plasma, and serum, a cohort of 6 people was tested using whole blood, plasma, serum, and plasma/serum that had been freeze thawed to −80 °C (the method of storage of historic control samples). There was agreement between tests in all cases with the same intensity of positive bands. The assay was carried out strictly to the manufacturer’s instructions by applying 20 μL of sample to the test, followed by 2 to 3 drops of buffer. Assays were observed to a maximum of 10 minutes, then results were assessed blindly by 2 independent observers. Tests were scored as IgG positive or negative, and any band was considered positive regardless of intensity.
Statistical Analysis
Statistical analysis was carried out using Prism, version 8 (GraphPad). For test characteristics, the Wilson-Brown method was used to compute CIs. Mann-Whitney test was used to compare nonparametric data, and t test, for normally distributed data. Data are reported as median and interquartile range. Fisher exact test or χ2 test was used for proportional assessments. The 2-sided level of significance was set at P < 0.05.
Results
With a median time to testing of 17 (interquartile range, 15-20) days after a positive RT-PCR result, 51/60 (85.0%) patients were IgG positive. All 9 patients who were IgG negative were tested 21 or fewer days after PCR diagnosis. Seven of these patients had further samples available for retesting at more than 21 days, and the remaining 2 patients were excluded from subsequent analysis. Five of 7 (71.4%) patients who were retested at more than 21 days had IgG antibodies on their second test. Median time from diagnosis to the second test was 35 (interquartile range, 30-39) days. SARS-CoV-2 IgG antibodies were therefore detected in 56/58 (96.6%) patients after 21 days after confirmation of SARS-CoV-2 infection.
Of the 2 patients who failed to develop IgG antibodies, 1 was a transplant recipient who had received a kidney transplant with alemtuzumab induction within 6 weeks of acquiring SARS-CoV-2 infection. This patient had a mild COVID-19 course by WHO criteria. The second patient was a maintenance hemodialysis patient who was not currently receiving immunosuppression treatment, although had previously been treated with chemotherapy for breast cancer more than 5 years previously. This second patient had critical disease severity by WHO criteria.
Clinical characteristics of study participants together with the clinical manifestations of their corresponding SARS-CoV-2 infection are shown in Table 1. Although there was no difference in the proportion of hemodialysis patients and transplant recipients who developed antibodies, 38/39 (97.4%) and 18/19 (94.7%), respectively, P = 0.54, there were several baseline differences between the 2 patient cohorts. Hemodialysis patients were older, P = 0.004, and more likely to have end-stage kidney disease secondary to diabetic nephropathy than transplant recipients, P = 0.02, as shown in Table 1. Intuitively, transplant recipients were more likely to be receiving immunosuppression. However, 5/39 (12.8%) hemodialysis patients were also receiving immunosuppression therapy at the time of SARS-CoV-2 infection; the indications for which were the presence of an in situ kidney transplant (3 patients) or to treat an underlying multisystemic autoimmune condition (2 patients). Seventeen of 18 (89.5%) kidney transplant recipients had received alemtuzumab induction, 5 of whom had undergone transplantation less than 1 year before having SARS-CoV-2 infection diagnosed. Compared with transplant recipients, the hemodialysis population was less likely to have mild disease, with 12 (63.2%) and 6/39 (15.4%) patients, respectively, having mild disease as classified by WHO, P < 0.001.
Table 1.
Clinical Characteristics of Study Patients Assessed by LFIA
| Variable | HD Patients (N = 39) | Transplant Recipients (N = 19) | P |
|---|---|---|---|
| Time of IgG+ test post PCR diagnosis, d | 17 [13-26] | 18 [14-23] | 0.43 |
| Age, y | 64 [58-76] | 55 [47-62] | 0.004a |
| Men | 24 (61.5%) | 12 (63.2%) | 0.91 |
| Ethnicity | |||
| White | 9 (23.7%) | 3 (16.7%) | 0.52 |
| BAME | 30 (76.9%) | 16 (84.2%) | |
| Cause of ESKD | |||
| APKD | 1 (2.6%) | 2 (10.5%) | 0.02a |
| Diabetes mellitusb | 19 (48.7%) | 3 (15.8%) | |
| Glomerulonephritis | 5 (12.8%) | 3 (15.8%) | |
| Unknown | 8 (20.5%) | 8 (42.1%) | |
| Other | 6 (15.4%) | 3 (15.8%) | |
| Baseline immunosuppression | |||
| Nob | 35 (89.7%) | 0 (0%) | <0.001a |
| Prednisolone | 1 (2.6%) | — | |
| FK only | 1 (2.6%) | 3 (16.7%) | |
| Predisolone, FK | 1 (2.6%) | — | |
| Prednisolone, FK, MMF | 1 (2.6%) | 6 (33.3%) | |
| FK, MMF | — | 7 (38.9%) | |
| Sirolimus, prednisolone, MMF | — | 2 (11.1%) | |
| Historic cytotoxic, B- or T-cell monoclonal antibody use | |||
| Nob | 34 (89.7%) | 0 (0%) | <0.001a |
| CyP | 1 (2.6%) | — | |
| CyP/Ritux | 1 (2.6%) | — | |
| Alemtuzumab | 2 (5.1%) | 16 (84.2%) | |
| IL-2R blocker | — | 1 (5.3%) | |
| CyP/bortezomib | 1 (2.6%) | — | |
| Alemtuzumab/Ritux | — | 1 (5.3%) | |
| Unknown | — | 1 (5.3%) | |
| Disease severity | |||
| Mildb | 6 (15.4%) | 12 (63.2%) | <0.001a |
| Moderate | 19 (48.7%) | 6 (31.6%) | |
| Severe | 11 (28.2%) | 1 (5.3%) | |
| Critical | 3 (7.7%) | — | |
| Care level | |||
| Outpatientb | 5 (12.8%) | 15 (78.9%) | <0.001a |
| Inpatient ward | 31 (79.5%) | 3 (15.8%) | |
| Intensive care unit | 3 (7.7%) | 1 (5.3%) | |
| Current patient status | |||
| Alive | 36 (92.3%) | 19 (100%) | 0.22 |
| Died | 3 (7.7%) | — |
Note: Values expressed as median [interquartile range] or number (percent).
Abbreviations: APKD, adult polycystic kidney disease; BAME, Black, Asian, and minority ethnic; CyP, cyclophosphamide; FK, tacrolimus; HD, hemodialysis; IgG, immunoglobulin G; IL-2R, interleukin 2 receptor; LFIA, lateral flow immunoassay; MMF, mycophenolate mofetil; PCR, polymerase chain reaction; Ritux, rituximab.
Statistically significant.
Comparator.
As a negative-control cohort, we used 88 saved plasma samples collected before September 2019. Two of 88 (2.3%) of these samples gave a false-positive IgG reading. Twenty-five of the 88 (28.4%) samples were historic samples stored from the current study patients with confirmed SARS-CoV-2 infection, all 25 of these samples tested negative for IgG. The false-positive samples were taken from patients who were receiving immunosuppression at the time of sampling, 1 for treatment of antineutrophil cytoplasmic antibody–associated vasculitis and the other from a hemodialysis patient with a failed transplant in situ.
Results from the study cohort and historic controls were used to calculate test characteristics for IgG detection at more than 21 days after confirmation of SARS-CoV-2 infection by PCR (Tables 2 and 3), giving sensitivity of 96.6% (95% CI, 88.3%-99.4%) and specificity of 97.7% (95% CI, 92.0-99.6%).
Table 2.
Antibody Results for SARS-CoV-2 IgG More Than 21 Days After PCR Test
| RT-PCR+ Cases | Historic Controls | Total | |
|---|---|---|---|
| Antibody result | |||
| Positive | 56 | 2 | 58 |
| Negative | 2 | 86 | 88 |
| Total | 58 | 88 | 146 |
Abbreviations: IgG, immunoglobulin G; RT-PCR, reverse transcriptase–polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Table 3.
Test Characteristics for SARS-CoV-2 IgG More Than 21 Days After PCR Test
| Statistic | Value (95% CI) |
|---|---|
| Sensitivity | 96.6% (88.3%-99.4%) |
| Specificity | 97.7% (92.0%-99.6%) |
| Positive predictive value | 96.6% (88.3%-99.4%) |
| Negative predictive value | 97.7% (92.1%-99.6%) |
| Accuracy | 97.3% (93.1%-99.2%) |
Abbreviations: IgG, immunoglobulin G; RT-PCR, reverse transcriptase–polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Discussion
In this study we describe performance characteristics of an LFIA to detect SARS-CoV-2 IgG in a cohort of hemodialysis patients and kidney transplant recipients with SARS-CoV-2 infection confirmed by RT-PCR.
There has been a massive surge in the development of serologic tests for IgM and IgG against SARS-CoV-2. Numerous different assays are now available in addition to the LFIA point-of-care test; including ELISAs, chemiluminescence enzyme immunoassays, fluorescence immunoassays, and pseudovirus neutralization assays.12,14,15 As assays are refined and developed, it is likely that the accuracy will improve, and the optimal serologic test has not yet been established.
The 96.6% sensitivity and 97.7% specificity of the LFIA we tested falls within the published ranges of other LFIA validation studies in the general population.11,12 Although these test characteristics are insufficient to meet Public Health England approval status in the United Kingdom, which stipulate a requirement of 98.0% sensitivity and specificity, they provide findings that should be of interest to the nephrology community.12 Given that these tests are simple to use, are inexpensive to manufacture, and give rapid results, they could play a role in population-level screening in high-risk cohorts, such as in-center hemodialysis patients.16 One potential use would be to estimate prevalence across different dialysis facilities to allow planning for further outbreaks. Although it is important to highlight that while this study has investigated the use of an LFIA to detect antibodies in patients with symptomatic SARS-CoV-2 infection, its validity in patients with asymptomatic disease has not been established. LFIA antibody testing could also potentially be used to aid diagnosis.17 Given the false-negative rates seen with RT-PCR secondary to sampling error, if clinically the pretest probability of SARS-CoV-2 infection is high, antibody testing may be used to help confirm or refute a diagnosis.17,18 In dialysis facilities with no access to RT-PCR testing, interpretation of LFIA results in conjunction with clinical symptoms may enable a confirmatory diagnosis.
The use of IgM serologic testing would be preferable to IgG to help with the diagnosis of acute infection. We excluded the interpretation of IgM in this study because SARS-CoV-2 IgG assays are recognized to be more specific than IgM.12,19 Five of 88 (5.7%) historic controls had false-positive IgM antibodies. Conversely, of the 2 IgG-negative patients who were unavailable to retest at more than 21 days, both had IgM antibodies that we were able to determine as new because both patients had historic samples in the control group that were IgM negative. It is therefore likely that the IgM detected in these 2 particular patients was in response to the acute SARS-CoV-2 infection, which would be useful information clinically if the test was accurate. However, incorporating IgM antibody data to calculate the LFIA test characteristics would have resulted in a significant reduction of the test performance, with sensitivity of 96.7% (95% CI, 88.5%-99.6%) and specificity of 84.1% (95% CI, 74.8%-91.0%), confirming an unacceptable IgM false-positive rate.
At the time of writing, this is the first report of the use of a SARS-CoV-2 LFIA in an immunosuppressed population. Patients with kidney disease are commonly considered to be immunosuppressed, either iatrogenically through the prescription of immunosuppressive medications or functionally from the effects of uremia.20 Humoral responses to viruses may be impaired in these patients and it is well described that antibody responses to viral vaccines such as hepatitis B and influenza are impaired in both kidney transplant recipients and those with chronic kidney disease.21, 22, 23 Although the serologic response to SARS-CoV-2 infection tested by ELISAs in patients receiving dialysis has been reported in 2 small studies, to our knowledge, no previous study has reported the immune response against SARS-CoV-2 in a cohort of kidney transplant recipients.24,25 Although it is not yet known whether patients who have developed antibody responses to SARS-CoV-2 are protected from reinfection, proof of an immune response is reassuring and demonstration that this immune response may be detected by an LFIA is a novel finding.
This study has several limitations, including sample size and lack of serial testing to estimate the optimal time for antibody detection. In addition, we do not know if the study population (cases and controls) had prior exposure to other beta-coronaviruses that may have the potential to cross-react with the antigen used in this LFIA. Studies have shown, using micro-array, that there is only low-level cross-reactivity with the spike protein antigen. However, it is possible that the false-positive IgG results in our historic samples were due to prior exposure to another beta-coronavirus.26 On a final note, LFIAs are nonquantitative tests so it was not possible to determine the effect of kidney disease or immunosuppression on antibody levels, which would be of interest and warrants further investigation.
In conclusion, we have shown that an LFIA can detect SARS-CoV-2 IgG antibody in venous blood of symptomatic hemodialysis patients and transplant recipients with clinically meaningful sensitivity and specificity. Used diligently, an LFIA could be used to help screen dialysis populations or aid diagnosis on a patient level, especially in facilities in which laboratory resources are limited. LFIAs could therefore enable equity of access to SARS-CoV-2 serology testing for patients with kidney disease across the globe.
Article Information
Authors’ Full Names and Academic Degrees
Maria Prendecki, PhD, Candice Clarke, MRCP, Tom McKinnon, PhD, Liz Lightstone, FRCP, Matthew C. Pickering, FRCP, David C. Thomas, PhD, Stephen P. McAdoo, PhD, and Michelle Willicombe, MD.
Authors’ Contributions
Research idea, study design and data analysis: MP, CC, SM, MW. Data acquisition: MP, CC, SM, MW, DT, TM, LL, MCP. MP and CC contributed equally to this work. Each author contributed important intellectual content during manuscript drafting or revision and accepts accountability for the overall work by ensuring that questions pertaining to the accuracy or integrity of any portion of the work are appropriately investigated and resolved.
Support
This research was supported by the National Institute for Health Research Biomedical Research Centre based at Imperial College Healthcare NHS Trust and Imperial College London.
Financial Disclosure
Dr Clarke is funded by an Auchi Clinical Research Fellowship. Dr Pickering has received personal fees from Achillion Pharma and Alexion Pharma. All remaining authors have nothing to disclose.
Acknowledgements
The authors thank the patients and staff within Imperial College Healthcare NHS Trust (ICHNT) and the ICHNT Renal COVID Group for their contribution. The ICHNT renal COVID group comprises senior clinicians at the ICRTC Centre engaged in the care and management of patients during this period: Ashby DR, Brown EA, Cairns T, Charif R, Condon M, Corbett RW, Dor F, Duncan N, Frankel A, Griffith M, Herbert P, Hill P, Goodall D, Kousios A, Levy JB, Loucaidou M, Lucisano G, Mclean A, Medjeral-Thomas N, Muthusamy A, Palmer A, Papalois V, Parsons D, Salisbury E, Sandhu E, Storey R, Tanna A, Tansey K, Tomlinson J, and Webster P. The authors also thank the West London Kidney Patient Association for their support.
Peer Review
Received July 13, 2020. Evaluated by 2 external peer reviewers, with direct editorial input from the Statistical Editor, an Associate Editor, and the Editor-in-Chief. Accepted in revised form September 13, 2020.
Footnotes
Complete author and article information provided before references.
Table S1: Clinical characteristics of historic controls.
Supplementary Material
Table S1.
References
- 1.Wu J., Li J., Zhu G. Clinical features of maintenance hemodialysis patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. Clin J Am Soc Nephrol. 2020;15(8):1139–1145. doi: 10.2215/CJN.04160320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valeri A.M., Robbins-Juarez S.Y., Stevens J.S. Presentation and outcomes of patients with ESKD and COVID-19. J Am Soc Nephrol. 2020;31(7):1409–1415. doi: 10.1681/ASN.2020040470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basile C., Combe C., Pizzarelli F. Recommendations for the prevention, mitigation and containment of the emerging SARS-CoV-2 (COVID-19) pandemic in haemodialysis centres. Nephrol Dial Transplant. 2020;35(5):737–741. doi: 10.1093/ndt/gfaa069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jha V. Nephrology Societies call for ensuring optimal care to patients with kidney diseases during the COVID-19 pandemic. www.theisn.org/covid-19#modal-close Accessed June 1, 2020.
- 5.National Cener for Immunization and Respiratory Diseases Interim additional guidance for infection prevention and control recommendations for patients with suspected or confirmed COVID-19 in outpatient hemodialysis facilities. https://www.cdc.gov/coronavirus/2019-ncov/hcp/dialysis Accessed June 8, 2020.
- 6.National Institute for Health and Care Excellence COVID-19 rapid guideline: dialysis service delivery. https://www.nice.org.uk/guidance/ng160 Accessed June 8, 2020. [PubMed]
- 7.The Renal Association Initial analysis of the impact of covid19 infection on patients with advanced chronic kidney disease in the UK. https://renal.org/covid-19/data/ Accessed May 14, 2020.
- 8.Konrad R., Eberle U., Dangel A. Rapid establishment of laboratory diagnostics for the novel coronavirus SARS-CoV-2 in Bavaria, Germany, February 2020. Eurosurveillance. 2020;25(9):2000173. doi: 10.2807/1560-7917.ES.2020.25.9.2000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zou L., Ruan F., Huang M. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382(12):1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ai T., Yang Z., Hou H. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296(2):E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitman J.D., Hiatt J., Mowery C.T. Test performance evaluation of SARS-CoV-2 serological assays. medRxiv. 2020 May 17 doi: 10.1101/2020.04.25.20074856. [DOI] [Google Scholar]
- 12.Kontou P.I., Braliou G.G., Dimou N.L., Nikolopoulos G., Bagos P.G. Antibody tests in detecting SARS-CoV-2 infection: a meta-analysis. Diagnostics. 2020;10(5):319. doi: 10.3390/diagnostics10050319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukaya S., Yasuda S., Hashimoto T. Clinical features of haemophagocytic syndrome in patients with systemic autoimmune diseases: analysis of 30 cases. Rheumatology. 2008;47(11):1686–1691. doi: 10.1093/rheumatology/ken342. [DOI] [PubMed] [Google Scholar]
- 14.Li Z., Yi Y., Luo X. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol. 2020;92(9):1518–1524. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nie J., Li Q., Wu J. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg Microbes Infect. 2020;9(1):680–686. doi: 10.1080/22221751.2020.1743767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clapham H., Hay J., Routledge I. Seroepidemiologic study designs for determining SARS-COV-2 transmission and immunity. Emerg Infect Dis. 2020;26(9):1978–1986. doi: 10.3201/eid2609.201840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao J., Yuan Q., Wang H. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis. 2020;71(16):2027–2034. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woloshin S., Patel N., Kesselheim A.S. False negative tests for SARS-CoV-2 infection — challenges and implications. N Engl J Med. 2020;383(6):e38. doi: 10.1056/NEJMp2015897. [DOI] [PubMed] [Google Scholar]
- 19.Wang Q., Du Q., Guo B. A method to prevent SARS-CoV-2 IgM false positives in gold immunochromatography and enzyme-linked immunosorbent assays. J Clin Microbiol. 2020;58(6) doi: 10.1128/JCM.00375-20. e00375-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Girndt M., Sester U., Sester M., Kaul H., Köhler H. Impaired cellular immune function in patients with end-stage renal failure. Nephrol Dial Transplant. 1999;14(12):2807–2810. doi: 10.1093/ndt/14.12.2807. [DOI] [PubMed] [Google Scholar]
- 21.DaRoza G., Loewen A., Djurdjev O. Stage of chronic kidney disease predicts seroconversion after hepatitis B immunization: earlier is better. Am J Kidney Dis. 2003;42(6):1184–1192. doi: 10.1053/j.ajkd.2003.08.019. [DOI] [PubMed] [Google Scholar]
- 22.Friedrich P., Sattler A., Müller K., Nienen M., Reinke P., Babel N. Comparing humoral and cellular immune response against HBV vaccine in kidney transplant patients. Am J Transplant. 2015;15(12):3157–3165. doi: 10.1111/ajt.13380. [DOI] [PubMed] [Google Scholar]
- 23.Rambal V., Müller K., Dang-Heine C. Differential influenza H1N1-specific humoral and cellular response kinetics in kidney transplant patients. Med Microbiol Immunol. 2014;203(1):35–45. doi: 10.1007/s00430-013-0312-3. [DOI] [PubMed] [Google Scholar]
- 24.Hains D.S., Schwaderer A.L., Carroll A.E. Asymptomatic seroconversion of immunoglobulins to SARS-CoV-2 in a pediatric dialysis unit. JAMA. 2020;323(23):2424–2425. doi: 10.1001/jama.2020.8438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Vriese A.S., Reynders M. IgG antibody response to SARS-CoV-2 infection and viral RNA persistence in patients on maintenance hemodialysis. Am J Kidney Dis. 2020;76(3):440–441. doi: 10.1053/j.ajkd.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Assis RRd, Jain A., Nakajima R. Analysis of SARS-CoV-2 antibodies in COVID-19 convalescent plasma using a coronavirus antigen microarray. bioRxiv. 2020 Apr 17 doi: 10.1101/2020.04.15.043364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.