Abstract
The facilitates chromatin transcription (FACT) complex is a histone H2A/H2B chaperone, which represses endogenous retroviruses (ERVs) and transcription of ERV-chimeric transcripts. It binds to both transcription start site and gene body region. Here, we investigated the downstream targets of FACT complex to identify the potential regulators of MERVL, which is a key 2-cell marker gene. H3K36me2 profile was positively correlated with that of FACT component Ssrp1. Among H3K36me2 deposition enzymes, Nsd2 was downregulated after the loss of Ssrp1. Furthermore, we demonstrated that Nsd2 repressed the expression of ERVs without affecting the expression of pluripotency genes. The expression of MERVL and 2-cell genes was partially rescued by Nsd2 overexpression. The enrichment of H3K36me2 decreased on MERVL-chimeric gene in ESCs without Ssrp1. Our study discovers that Nsd2 is a repressor of MERVL, and FACT partially represses MERVL expression by regulating the expression of Nsd2 and its downstream H3K36me2.
1. Introduction
Endogenous retroviruses are important components of the mammalian genome [1]. They are usually silenced by host cells to maintain genome stability. However, studies also show that ERVs are functional during development and in mouse embryonic stem cells (ESCs) [2–7]. For example, MERVL marks the 2-cell (2C) embryos and a minority of 2C-like cells within the ESC population [8, 9]. MERVL can be silenced by various epigenetic regulators, such as histone H3 variants, H3K9 methyltransferases, and histone chaperones [10–15]. Recently, we found that H2A/H2B histone chaperone FACT (facilitates chromatin transcription) complex participated in the repression of MERVL and MERVL-derived cryptic transcripts in ESCs [16]. FACT functions partially through Usp7 to remove H2Bub on MERVL and MERVL-fused genes [16]. However, the impact of Usp7 depletion on MERVL induction is weaker than the loss of FACT complex itself [16]. This implies that there are other ways present for FACT complex to repress the expression of MERVL and its chimeric transcripts. Therefore, in this study, we aim to identify indirect pathways downstream of FACT complex in repressing the expression of MERVL.
We and others previously found that Ssrp1 binding was enriched around transcription start sites and on gene body region [16, 17]. Gene body region can be marked by H3K36me3 and H3K36me2 [18, 19]. An important H3K36 methyltransferase family is Nsd family. Here, we examine the role of Nsd family members at the downstream of FACT complex in repressing the ERV expression.
2. Methods
2.1. Cell Culture
E14 mouse embryonic stem cells (ESCs) were cultured on plates coated with sterile 0.1-0.2% gelatin (G1890, Sigma) in medium containing Dulbecco's modified Eagle's medium (Hyclone), 15% fetal bovine serum (Hyclone), 10 ng/ml leukaemia inhibitory factor (LIF) (Z03077, GenScript), 1% penicillin/streptomycin (P1400, Solarbio), 2 mM L-glutamine (Gibco), 0.1 mM nonessential amino acids (Gibco), and 0.1 mM β-mercaptoethanol (Sigma). ESCs were passaged every two days for maintenance.
2.2. Analysis of ChIP-seq Data
For ChIP-seq data analysis, all reads were first processed with Cutadapt to trim adaptor sequences and low-quality reads and subsequently mapped to the mouse mm10 genome assembly using Bowtie2. The correlation coefficient between Ssrp1, H3K4me3, H3K36me2, and H3K36me3 was determined by plotCorrelation from Deeptools. The ChIP-seq signal enrichment file was obtained by bamCompare from Deeptools, and the ChIP signal line plot was also generated by Deeptools. Gene structure information was inferred from Gencode.vM21 annotation file.
2.3. Reverse Transcription and qPCR
Total RNA was isolated from cells by RNAiso Reagent (B9109, Takara) in DEPC water (B501005, Sangon Biotech) following by DNase I treatment in RNase-free tubes (401001, NEST Biotechnology). Reverse transcription was performed with 1 μg purified RNA using Transcriptor First Strand cDNA Synthesis Kit (4897030001, Roche) as described previously [20]. qPCR analysis was carried out using SYBR Green qPCR Master Mix (H97410, Yeasen) and a qPCR detection system (CFX384 Real-Time System, Bio-Rad) according to standard protocols. Primers are synthesized by Sangon Biotech and included in Table 1.
Table 1.
Primers for qPCR analysis.
| Gene | Forward | Reverse |
|---|---|---|
| Nsd1 | TCCGGTGAATTTAGATGCCTCC | CGGTAACTGCATAGTACACCCAT |
| Nsd2 | GGTGATCCTGGCACAGACAA | GAGCAGAGCCTGTGGACTTT |
| Nsd3 | CCGAGGTTGTGCCAAAGAAG | ACGGAGCTGTCACTGAATCTG |
| MERVL | AAGAGCCAAGACCTGCTGAG | TCCTCGTTTCTGCAACTGGT |
| LINE1 | GGACCAGAAAAGAAATTCCTCCCG | CTCTTCTGGCTTTCATAGTCTCTGG |
| SINEB1 | GTGGCGCACGCCTTTAATC | GACAGGGTTTCTCTGTGTAG |
| Oct4 | GTGGAAAGCAACTCAGAGG | GGTTCCACCTTCTCCAACT |
| Sox2 | GCGGAGTGGAAACTTTTGTCC | CGGGAAGCGTGTACTTATCCTT |
| Nanog | TTGCTTACAAGGGTCTGCTACT | ACTGGTAGAAGAATCAGGGCT |
| Zscan4 | GAGATTCATGGAGAGTCTGACTGATGAGTG | GCTGTTGTTTCAAAAGCTTGATGACTTC |
| Dux | CCCAGCGACTCAAACTCCTTC | GGACTTCGTCCAGCAGTTGAT |
| ChIP Control |
GATTAGCAGCTCCACAGGA | TGGACAATGTGGCCTGTTTA |
|
Zfp809
ChIP |
AAGCTGGCTGACTGTAGTGG | GTGAGCCTTCCAATTCCGGA |
| Foxa2 | CCCTACGCCAACATGAACTCG | GTTCTGCCGGTAGAAAGGGA |
| Sox17 | GATGCGGGATACGCCAGTG | CCACCACCTCGCCTTTCAC |
| Gata4 | CCCTACCCAGCCTACATGG | ACATATCGAGATTGGGGTGTCT |
| Nkx2.5 | GACAAAGCCGAGACGGATGG | CTGTCGCTTGCACTTGTAGC |
| Msx1 | TGCTGCTATGACTTCTTTGCC | GCTTCCTGTGATCGGCCAT |
| Pax6 | TACCAGTGTCTACCAGCCAAT | TGCACGAGTATGAGGAGGTCT |
| Foxd3 | GAGTTCATCAGCAACCGTTTTC | CGAAGCTCTGCATCATCAGC |
| Gata3 | CTCGGCCATTCGTACATGGAA | GGATACCTCTGCACCGTAGC |
2.4. shRNA-Mediated Gene Depletion
The shRNAs targeting Nsd2 were designed by an online tool (http://sirna.wi.mit.edu/) [21]. The targeting sequences of shRNAs are CCTGGTGCTCATGATACTAAA for shRNA1 and GAGCTGACTTTCAACTATAA for shRNA2. The shRNAs were synthesized by GENEWIZ corporation and cloned into pSuper-puro. 1 μg plasmid was transfected into mouse ESCs with Polyjet (SignaGen). The cells were further cultured for three days under puromycin selection (1 μg/ml) and harvested for RNA extraction.
2.5. Chromatin Immunoprecipitation (ChIP) Coupled qPCR
ChIP-qPCR was performed as described before [16]. Briefly, ESCs were harvested and crosslinked with 1% formaldehyde, and cell fixation was ceased with the addition of glycine. The cells were primarily lysed, and chromatin extracts were collected and sonicated for obtaining soluble chromatin fragments. The chromatin samples were incubated with specific antibody and immunoprecipitated on protein G magnetic beads (GenScript, the USA). The immunoprecipitated DNA was next eluted, decrosslinked, and analyzed by qPCR. For immunoprecipitation, the antibody used was anti-H3K36me2 (ab9049, Abcam).
2.6. Establishment of ESC Cell Lines Overexpressing Nsd2
Mouse Nsd2 coding region was cloned into pCAG-3HA vector with hygromycin resistance and purified with a kit (1211-01, Biomiga). Ssrp1−/− ESCs were transfected with 1 μg plasmid expressing Nsd2 via Polyjet reagent (SL100688, SignaGen) following the manufacturer's recommended protocol. The ESCs were continuously selected with 800 μg/ml hygromycin B for 14 days. After selective cell culture, ESCs were collected for downstream experiments.
3. Results
3.1. FACT Complex Binding Is Correlated with H3K36 Methylation
Previously, we found that FACT complex interacted with both promoter and gene body regions, which are marked by H3K36me2/3. Interestingly, the genomic distribution profile of H3K36me2 and H3K36me3 was positively correlated with that of Ssrp1 (Figure 1(a)), in contrast with the lower correlation strength of Ssrp1 with H3K4me3 (Figure 1(a)). Moreover, it was noteworthy that H3K36me3 enrichment on the gene body continuously increased from transcription start site (TSS) to transcription end site (TES) whereas the H3K36me2 was preferentially associated with TSS region and gradually decayed from TSS to TES (Figure 1(b)). The distribution profile of H3K36me2 was more similar to that of FACT complex than H3K36me3 (Figures 1(a) and 1(b)). Therefore, we further examined the expression of Nsd family genes (Nsd1, Nsd2, and Nsd3), which are known to mediate H3K36 methylation. Nsd1 was expressed highest in ESCs while the expression of Nsd2 and Nsd3 was lower (Figure 1(c)). The expression of Nsd1 and Nsd3 remained unchanged or slightly upregulated in ESCs without FACT complex (Figure 1(d)). However, the Nsd2 expression was downregulated in Ssrp1−/− ESCs (Figure 1(d)), implying Nsd2 as a potential downstream target gene of FACT complex.
Figure 1.
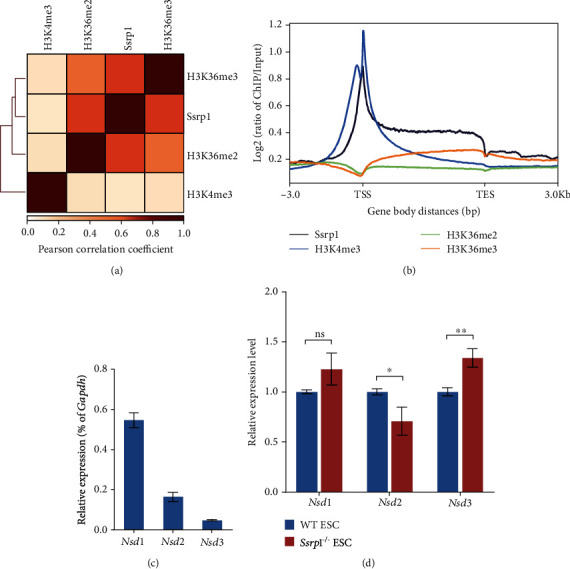
Nsd2-mediated H3K36me2 is correlated with Ssrp1 binding. (a) Genomic distribution profile of H3K36me2, H3K36me3, H3K4me3, and Ssrp1. Color scale represents the strength of Pearson's correlation. (b) ChIP-seq signal density enrichment of Ssrp1, H3K4me3, H3K36me2, and H3K36me3 (Y-axis) on gene body from TSS to TES (X-axis). H3K36me3 enrichment increased from TSS to TES. (c) Relative mRNA expression of Nsd1, Nsd2, and Nsd3 in ESCs according to qPCR. (d) qPCR analysis of Nsd1, Nsd2, and Nsd3 expression levels in WT ESC and Ssrp1−/− ESC; data are shown as mean ± s.e.m.; n = 3 biologically independent repeats. ns: nonsignificant; ∗p < 0.05; ∗∗p < 0.01.
3.2. Nsd2 Represses MERVL in ESCs
In agreement with the close resemblance of Ssrp1 binding profile and H3K36me2, the main chromatin-regulatory activity of Nsd2 is mediating the dimethylation of histone H3 at lysine 36 (H3K36me2) [19]. Hence, we depleted Nsd2 in ESCs with two independent shRNAs to examine whether Nsd2 can regulate the expression of ERVs (Figure 2(a)). The depletion of Nsd2 did not affect the cell morphology of ESCs (Figure 2(b)). Also, the expression of pluripotency genes (Oct4, Sox2, Nanog) was not disturbed by two Nsd2 shRNAs at the same time (Figure 2(c)). The suppression of Nsd2 by two independent shRNAs did not disrupt the expression of differentiation markers for endoderm (Foxa2 and Sox17), mesoderm (Gata4 and Nkx2.5), ectoderm (Msx1 and Pax6), and trophectoderm (Foxd3 and Gata3) at the same time (Figures 2(d)–2(g)), suggesting that ESCs remain undifferentiated without Nsd2. Intriguingly, the expression of MERVL was activated to ~2 folds by Nsd2 depletion (Figure 2(h)), but other retrotransposons (LINE1 or SINE B1) were less activated or downregulated, confirming that Nsd2 acts downstream of FACT complex to repress the ERV expression. These results suggest that Nsd2 represses the expression of MERVL without affecting ESC pluripotency.
Figure 2.
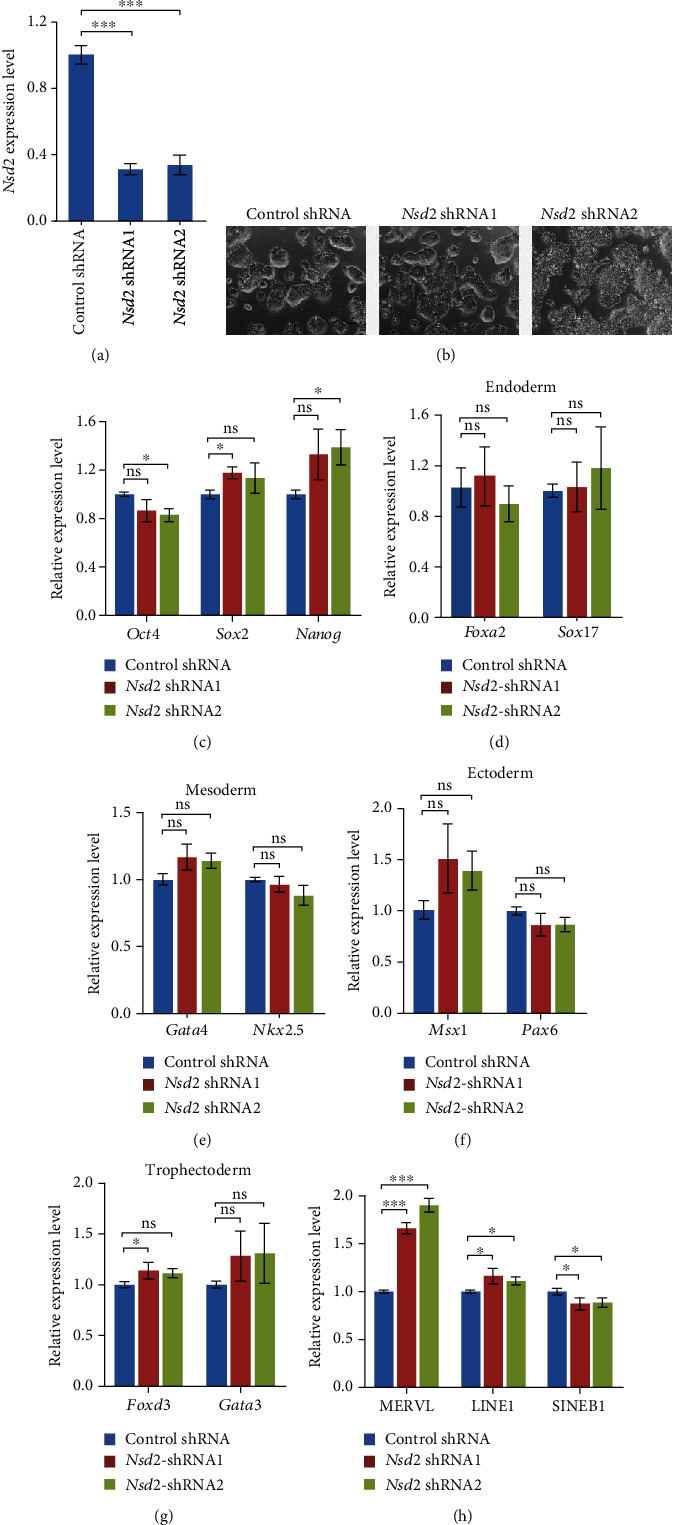
Depletion of Nsd2 in ESCs. (a) qPCR analysis of the relative expression levels of Nsd2 after Nsd2 depletion in ESCs with normalization to Gapdh. Data were plotted as mean ± s.e.m., n = 3 biological repeats. (b) Cell morphology of Nsd2 depletion in ESCs. Scale bar, 100 μm. (c) qPCR analysis of pluripotent genes (Oct4, Sox2, and Nanog) in ESCs with Nsd2 depleted. (d–g) qPCR analysis of endoderm markers Foxa2 and Sox17 (d), mesoderm markers Gata4 and Nkx2.5 (e), ectoderm markers Msx1 and Pax6 (f), and trophectoderm markers Foxd3 and Gata3 (g). (h) qPCR analysis of the expression levels of MERVL and other retrotransposons (LINE1, SINE B1) after Nsd2 depletion in ESCs. The results were normalized to Gapdh. Data were shown as mean ± s.e.m.; n = 3 biologically independent replicates; ns: nonsignificant; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
3.3. Nsd2 Overexpression Rescues MERVL Expression in Ssrp1−/− ESCs
Since Nsd2 represses the MERVL expression and Nsd2 is downregulated in Ssrp1−/− ESCs, we next asked whether restoration of the Nsd2 expression in Ssrp1−/− ESCs could rescue the expression of MERVL. Hence, we established an Ssrp1−/− ESC line with Nsd2 overexpressed. Our qPCR results showed that Nsd2 was successfully overexpressed in Ssrp1−/− ESCs (Figure 3(a)). Moreover, overexpression of Nsd2 could partially reduce the expression of MERVL in Ssrp1−/− ESCs (Figure 3(b)). Furthermore, the expression of 2-cell marker genes (Zscan4 and Dux) was partially restored (Figure 3(c)). These results suggest that Nsd2 is an important downstream target gene of Ssrp1 in repressing ERVs and 2-cell genes.
Figure 3.
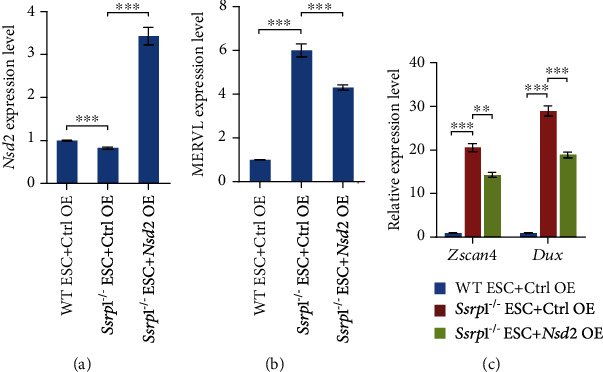
Nsd2 overexpression rescues the expression of MERVL and 2-cell genes. (a) qPCR analysis of Nsd2 expression after overexpression of Nsd2 in Ssrp1−/− ESC. The results were shown as mean ± s.e.m., n = 3 biologically independent replicates and normalized to Gapdh. (b) qPCR analysis of the expression of MERVL after overexpression of Nsd2 in Ssrp1−/− ESC. Mean ± s.e.m., n = 3. (c) qPCR analysis of the expression of 2-cell marker genes Zscan4 and Dux after overexpression of Nsd2 in Ssrp1−/− ESC. Ctrl: control; OE: overexpression; mean ± s.e.m., n = 3; ∗∗p < 0.01; ∗∗∗p < 0.001.
3.4. Nsd2-Mediated H3K36me2 Is Reduced on MERVL-Fused Genes in Ssrp1−/− ESCs
We further investigated whether the target of Nsd2, H3K36me2, was affected at MERVL-fusion genes in ESCs without FACT by ChIP-qPCR. Our ChIP-qPCR results revealed that H3K36me2 was enriched on MERVL-fused gene such as Zfp809 (Figure 4(a)) but not on the control region (Figure 4(b)). However, this enrichment was decreased on MERVL-fusion genes in ESC without the Ssrp1 expression (Figures 4(a) and 4(b)). Together, these suggest that the decreased enrichment of H3K36me2 on MERVL-fused genes may explain the activation of MERVL-fused genes after Nsd2 downregulation in Ssrp1−/− ESCs.
Figure 4.
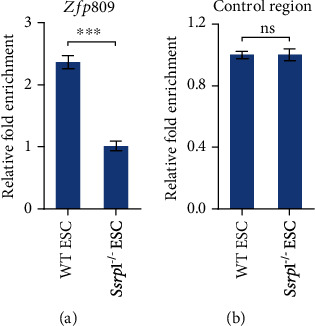
H3K36me2 enrichment on MERVL-fused genes. (a, b) ChIP-qPCR analysis of H3K36me2 enrichment on MERVL-fused gene Zfp809 (a) and control region (b) in WT ESC and Ssrp1−/− ESC with normalization to Gapdh and input; data shown as mean ± s.e.m. (n = 3 extracts). ns: nonsignificant; ∗∗∗p < 0.001.
4. Discussion
In summary, we discovered that Nsd2 was a repressor of MERVL and MERVL-fused 2C genes, and the downregulation of Nsd2 worked as a secondary regulatory route to activate MERVL after the loss of Ssrp1. It is interesting to see that only Nsd2 (Figure 1(d)), but neither Nsd1 nor Nsd3, is downregulated by the disruption of FACT function, given that all three Nsd genes participate in H3K36 methylation. Nsd2 is an important H3K36me2 methyltransferase [19, 22]. Loss of Nsd2 mimics H3.3K36M mutation, but not Nsd1 or Setd2 mimics the effects of H3.3K36M on adipogenesis [23], implicating a unique role of Nsd2 among Nsd members in gene expression regulation. H3K6me2 was associated with both activation and repression of the gene expression [24]. It was recently reported that Nsd1/Nsd2-mediated intergenic H3K36me2 recruited Dnmt3a for DNA methylation [25, 26]. In yeast cells, H3K36me1/2/3 was also shown to repress cryptic transcription [27]. Moreover, H3K36me2 can recruit the Rpd3s histone deacetylase to repress spurious transcription [28]. These are consistent with our finding that H3K36me2 decreased on MERVL-fused genes after the loss of Ssrp1 (Figures 4(a) and 4(b)), implying a potential repression role of H3K36me2.
Nsd2 is not only involved in gene transcription regulation. It participates in regulating genome stability and methylates non-histone proteins as well. Nsd2-mediated H3K36me2 promotes nonhomologous end-joining at unprotected telomeres and thereby enhances genomic instability caused by telomere dysfunction [29]. Human NSD2-mediated PTEN methylation regulates cell responses to DNA damage [30]. It is recently discovered that DNA damage is induced by the depletion of MERVL activator Zscan4 [3, 31]. Responses of ATR and CHK1 to replication stresses activate Zscan4 and MERVL [20], implying that DNA damage-induced replication stress and Zscan4 reciprocally regulate each other. It will be interesting to investigate whether Nsd2 is involved DNA damage repair and its relationship with Zscan4 in the future.
5. Conclusion
In conclusion, we found that Nsd2, as a downstream gene of FACT, repressed MERVL, without influencing ESC pluripotency. The decreased Nsd2 in Ssrp1−/− ESCs was accompanied by reduced H3K36me2 on MERVL-fused genes while overexpression of Nsd2 partially rescued the expression of MERVL. These findings establish Nsd2 as an important repressor of MERVL in ESCs and during the loss of FACT function.
Acknowledgments
This work is supported by grants from the National Key Research and Development Program of China (2018YFA0107000), the National Natural Science Foundation of China (32070858, 31671352, 31871488), and the Fundamental Research Funds for the Central Universities (63151113, 63161150).
Data Availability
Published ChIP-seq data we analyzed are GSE141791 for Ssrp1 [16], GSE117155 for H3K36me2 [32], GSE110321 for H3K36me3 [33], and GSE90893 for H3K4me3 [34].
Conflicts of Interest
We declare that there is no conflict of interest present for this study.
Authors' Contributions
X.L. conceived and designed the study. T.G., F.C., and W.Z. performed most experiments. X.Z. did bioinformatics analysis. T.G., X.H., and X.L. wrote the manuscript.
References
- 1.Enriquez-Gasca R., Gould P. A., Rowe H. M. Host gene regulation by transposable elements: the new, the old and the ugly. Viruses. 2020;12(10):p. 1089. doi: 10.3390/v12101089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu X., Sachs F., Ramsay L. A., et al. The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nature Structural & Molecular Biology. 2014;21(4):423–425. doi: 10.1038/nsmb.2799. [DOI] [PubMed] [Google Scholar]
- 3.Zhang W., Chen F., Chen R., et al. Zscan4c activates endogenous retrovirus MERVL and cleavage embryo genes. Nucleic Acids Research. 2019;47(16):8485–8501. doi: 10.1093/nar/gkz594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Göke J., Lu X., Chan Y. S., et al. Dynamic transcription of distinct classes of endogenous retroviral elements marks specific populations of early human embryonic cells. Cell Stem Cell. 2015;16(2):135–141. doi: 10.1016/j.stem.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 5.Grow E. J., Flynn R. A., Chavez S. L., et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015;522(7555):221–225. doi: 10.1038/nature14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mi S., Lee X., Li X. P., et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403(6771):785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 7.Peaston A. E., Evsikov A. V., Graber J. H., et al. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Developmental Cell. 2004;7(4):597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 8.Macfarlan T. S., Gifford W. D., Driscoll S., et al. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487(7405):57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kigami D., Minami N., Takayama H., Imai H. MuERV-L is one of the earliest transcribed genes in mouse one-cell embryos. Biology of Reproduction. 2003;68(2):651–654. doi: 10.1095/biolreprod.102.007906. [DOI] [PubMed] [Google Scholar]
- 10.Elsässer S. J., Noh K. M., Diaz N., Allis C. D., Banaszynski L. A. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature. 2015;522(7555):240–244. doi: 10.1038/nature14345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatanaka Y., Inoue K., Oikawa M., et al. Histone chaperone CAF-1 mediates repressive histone modifications to protect preimplantation mouse embryos from endogenous retrotransposons. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(47):14641–14646. doi: 10.1073/pnas.1512775112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishiuchi T., Enriquez-Gasca R., Mizutani E., et al. Early embryonic-like cells are induced by downregulating replication- dependent chromatin assembly. Nature Structural & Molecular Biology. 2015;22(9):662–671. doi: 10.1038/nsmb.3066. [DOI] [PubMed] [Google Scholar]
- 13.Yang B. X., el Farran C. A., Guo H. C., et al. Systematic identification of factors for provirus silencing in embryonic stem cells. Cell. 2015;163(1):230–245. doi: 10.1016/j.cell.2015.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rowe H. M., Jakobsson J., Mesnard D., et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463(7278):237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- 15.Maksakova I. A., Thompson P. J., Goyal P., et al. Distinct roles of KAP1, HP1 and G9a/GLP in silencing of the two-cell-specific retrotransposon MERVL in mouse ES cells. Epigenetics & Chromatin. 2013;6(1):p. 15. doi: 10.1186/1756-8935-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen F., Zhang W., Xie D., Gao T., Dong Z., Lu X. Histone chaperone FACT represses retrotransposon MERVL and MERVL-derived cryptic promoters. Nucleic Acids Research. 2020;48(18):10211–10225. doi: 10.1093/nar/gkaa732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mylonas C., Tessarz P. Transcriptional repression by FACT is linked to regulation of chromatin accessibility at the promoter of ES cells. Life Science Alliance. 2018;1(3, article e201800085) doi: 10.26508/lsa.201800085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mikkelsen T. S., Ku M., Jaffe D. B., et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuo A. J., Cheung P., Chen K., et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Molecular Cell. 2011;44(4):609–620. doi: 10.1016/j.molcel.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atashpaz S., Samadi Shams S., Gonzalez J. M., et al. ATR expands embryonic stem cell fate potential in response to replication stress. eLife. 2020;9 doi: 10.7554/eLife.54756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan B., Latek R., Hossbach M., Tuschl T., Lewitter F. siRNA Selection Server: an automated siRNA oligonucleotide prediction server. Nucleic Acids Research. 2004;32(Web Server):W130–W134. doi: 10.1093/nar/gkh366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y., Trojer P., Xu C. F., et al. The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate. The Journal of Biological Chemistry. 2009;284(49):34283–34295. doi: 10.1074/jbc.M109.034462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhuang L., Jang Y., Park Y. K., et al. Depletion of Nsd2-mediated histone H3K36 methylation impairs adipose tissue development and function. Nature Communications. 2018;9(1):p. 1796. doi: 10.1038/s41467-018-04127-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wagner E. J., Carpenter P. B. Understanding the language of Lys36 methylation at histone H3. Nature reviews. Nature Reviews Molecular Cell Biology. 2012;13(2):115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weinberg D. N., Papillon-Cavanagh S., Chen H., et al. The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature. 2019;573(7773):281–286. doi: 10.1038/s41586-019-1534-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu W., Li J., Rong B., et al. DNMT3A reads and connects histone H3K36me2 to DNA methylation. Protein & Cell. 2020;11(2):150–154. doi: 10.1007/s13238-019-00672-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DiFiore J. V., Ptacek T. S., Wang Y., Li B., Simon J. M., Strahl B. D. Unique and shared roles for histone H3K36 methylation states in transcription regulation functions. Cell Reports. 2020;31(10):p. 107751. doi: 10.1016/j.celrep.2020.107751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li B., Jackson J., Simon M. D., et al. Histone H3 lysine 36 dimethylation (H3K36me2) is sufficient to recruit the Rpd3s histone deacetylase complex and to repress spurious transcription. The Journal of Biological Chemistry. 2009;284(12):7970–7976. doi: 10.1074/jbc.M808220200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Krijger I., van der Torre J., Peuscher M. H., Eder M., Jacobs J. J. L. H3K36 dimethylation by MMSET promotes classical non-homologous end-joining at unprotected telomeres. Oncogene. 2020;39(25):4814–4827. doi: 10.1038/s41388-020-1334-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J., Lee Y. R., Dang F., et al. PTEN methylation by NSD2 controls cellular sensitivity to DNA damage. Cancer Discovery. 2019;9(9):1306–1323. doi: 10.1158/2159-8290.CD-18-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srinivasan R., Nady N., Arora N., et al. Zscan4 binds nucleosomal microsatellite DNA and protects mouse two-cell embryos from DNA damage. Science Advances. 2020;6(12):p. eaaz9115. doi: 10.1126/sciadv.aaz9115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LeRoy G., Oksuz O., Descostes N., et al. LEDGF and HDGF2 relieve the nucleosome-induced barrier to transcription in differentiated cells. Science Advances. 2019;5(10):p. eaay3068. doi: 10.1126/sciadv.aay3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang H., Weng H., Zhou K., et al. Histone H3 trimethylation at lysine 36 guides m6A RNA modification co-transcriptionally. Nature. 2019;567(7748):414–419. doi: 10.1038/s41586-019-1016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chronis C., Fiziev P., Papp B., et al. Cooperative binding of transcription factors orchestrates reprogramming. Cell. 2017;168(3):442–459.e20. doi: 10.1016/j.cell.2016.12.016. e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Published ChIP-seq data we analyzed are GSE141791 for Ssrp1 [16], GSE117155 for H3K36me2 [32], GSE110321 for H3K36me3 [33], and GSE90893 for H3K4me3 [34].


