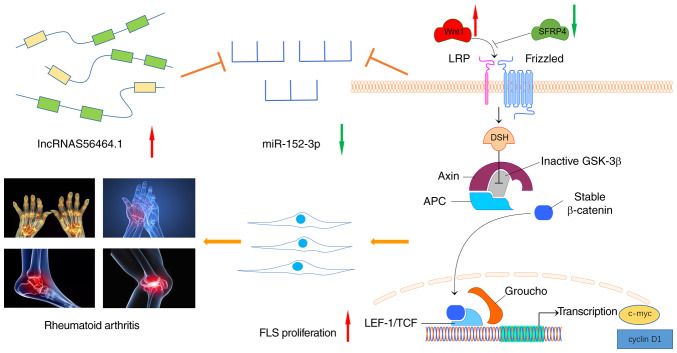
Abstract
Rheumatoid arthritis (RA) is an autoimmune disease that occurs in approximately 1.0% of the general population. In RA patients, physical disability and joint damage are the major prognostic factors, which are associated with a reduction in the quality of life and early mortality. At present, the exact molecular mechanism of RA remains elusive. Long noncoding RNAs (lncRNAs) have been revealed to play a regulatory role in the pathogenesis of RA. To reveal the function of lncRNAs in rheumatoid arthritis, lncRNAS56464.1 was screened to verify its targeting of the microRNA (miR)-152-3p/Wnt pathway and its effect on the proliferation of fibroblast-like synoviocytes (FLS). In the present study, based on the competing endogenous RNA (ceRNA) theory, siRNA was designed for transfection into FLS to calculate the lncRNAS56464.1 interference efficiency and then the effect of lncRNAS56464.1 interference on FLS proliferation was detected by MTT assay. Then, lncRNAS56464.1 targeting of the miR-152-3p/Wnt pathway was detected by a dual-luciferase reporter assay. In addition, RT-qPCR, immunofluorescence and western blotting techniques were employed to detect the expression of lncRNAS56464.1, miR-152-3p and some key genes of the Wnt signaling pathway in FLS after lncRNAS56464.1 interference. The results revealed that lncRNAS56464.1 could combine with miR-152-3p and promoted the proliferation of FLS. In addition, lncRNAS56464.1 interference could not only decrease the proliferation of FLS and the expression of Wnt1, β-catenin, c-Myc, cyclin D1, and p-GSK-3β/GSK-3β, but it also increased the expression of SFRP4. The present data indicated that lncRNAS56464.1 could target the miR-152-3p/Wnt pathway to induce synovial cell proliferation and then participate in the pathogenesis of RA.
Keywords: rheumatoid arthritis, fibroblast-like synoviocytes, lncRNAS56464.1, microRNA-152-3p, Wnt signaling pathway
Introduction
Rheumatoid arthritis (RA), as a systemic and autoimmune disease, is characterized by chronic inflammation, abnormal synovial cell proliferation, joint swelling and tenderness and can lead to limitations in joint function and the loss of the ability to work (1,2). Fibroblast-like synoviocytes (FLS) are an important type of synovial cell and are the main type of effector cell mediating joint destruction and synovitis by functioning in tissue shaping in the physiological state and in matrix remodeling in pathological injury (3-6). At present, the specific pathogenesis of RA remains elusive, and may be related to immune factors, environmental factors, heredity, as well as other factors (7-9). Therefore, it is of great theoretical and practical significance to actively explore the pathogenesis of RA and search for potential diagnostic markers.
Long noncoding RNAs (lncRNAs), which are a recently discovered class of noncoding RNAs, are transcribed from noncoding sequences in the genome, are longer than 200 nt and do not have the ability to be translated into proteins (10,11). LncRNAs have become a novel research focus since numerous lncRNAs have been discovered that are closely related to the occurrence of a great number of diseases due to their extensive regulatory effects on epigenetics (12), transcription (13), and protein translation (14) and modification (15). Therefore, a systematic study of the regulatory role of lncRNAs in the pathogenesis of RA will help in more comprehensively understanding the mechanism of RA and providing new insight and targets for its clinical diagnosis and treatment. According to previous research by our group, we revealed that lncRNAS56464.1 is a crucial gene related to the pathogenesis of experimental arthritis, which can be used as a potential biomarker for diagnosis and a clinical treatment target (16). However, the specific mechanism of lncRNAS56464.1 in RA development and progression remains in its infancy.
In 2011, Salmena et al (17) proposed a new mechanism for describing RNA interactions: The competitive endogenous RNA (ceRNA) hypothesis. The hypothesis holds that RNA transcripts such as lncRNAs and circRNAs regulate the expression level of mRNAs by competing with miRNAs to bind miRNA response elements (MREs). miRNAs are important genetic molecules in organisms that participate in the regulation of posttranscriptional gene expression. Their aberrant expression is related to the occurrence and development of numerous diseases. miR-152-3p was revealed through miRNA screening in our previous study, to be the key gene associated with RA (16). miR-152-3p belongs to the miR-152 family and participates in biological processes such as cell proliferation, differentiation and apoptosis (18-20). A study has revealed that miR-152 can target Wnt1 to regulate the Wnt signaling pathway and affect the development of RA (21). However, the specific relationship between lncRNAS56464.1, miR-152-3p, the Wnt signaling pathway and the occurrence and development of RA remains to be studied. Therefore, in the present study, based on the ceRNA theory, it was investigated whether lncRNAS56464.1 can target the miR-152-3p/Wnt pathway and affect the proliferation of FLS in RA.
Materials and methods
Induction of arthritis and the culture and identification of synovial cells
In the present study, 40 male SPF Sprague-Dawley (SD) rats (6-8 weeks old; 200±20 g) were purchased from the Anhui Experimental Animal Center. The rats were housed individually under a set temperature (18-22°C) and humidity (40-60%) with a 12-h light/dark cycle, free access to standard laboratory food and water, and environmental noise kept to a minimum range. All animal experiments were approved by the Animal Ethics Committee of Anhui University of Chinese Medicine (Anhui, China).
After one week of adaptive feeding, all rats were randomly divided into a control group (n=10) and a model group (n=30). Except for the control group, all rats were intracutaneously injected with 0.1 ml Freund's complete adjuvant (FCA) into the right hind metatarsal footpad to induce adjuvant-induced arthritis (AA) and 0.9% saline was administered to the control group rats as a placebo. The rats were anesthetized with 1.0% sodium pentobarbital by a 60 mg/kg intraperitoneal injection and were sacrificed after blood was collected from the abdominal aorta on the 20th day after immunization (22,23).
Then, the synovial tissue was obtained and cultured in complete DMEM medium with 20% fetal bovine serum (lot no. 1715752; cat. no. 10099141C; Gibco; Thermo Fisher Scientific, Inc.) at 37°C and 5% CO2 by the tissue explant method (24). After a large number of cells were dissociated near the synovial tissue, the tissue mass was discarded and the cells continued to be cultured. When the cell density reached 80-90%, the cells were digested with 0.25% trypsin and passaged at 1:2 and FLS used in the experiment were 3-5 generations. The primary and sub-cultured FLS were observed under an inverted phase contrast microscope (Olympus Corporation), the sub-cultured synovial cells were identified by immunofluorescence staining for vimentin, and the specific operation was carried out according to the instructions of the kit as previously described (25,26). The cells were fixed 20 min with 4% paraformaldehyde (PFA), permeabilized 20 min using 0.1% Triton X-100 and then blocked in 5% BSA for 30 min; all operations aforementioned were performed at room temperature. The cells were then incubated with anti-vimentin (1:100; product code ab92547; Abcam) overnight at 4°C, followed by a further incubation at room temperature for 1 h with FITC-labeled Goat Anti-Rabbit IgG (1:500; cat. no. A0562; Beyotime Institute of Biotechnology). Nuclear DNA was labelled in blue with DAPI. The cell viability was detected by trypan blue staining as follows: Cell survival rate (percentage)=nonblue-stained cells/(blue-stained cells + nonblue-stained cells) ×100%.
Cell transfection
To suppress lncRNAS56464.1, specific small interfering RNAs (siRNAs) targeting lncRNAS56464.1 were synthesized and purchased from Shanghai GenePharma Co., Ltd., and were as follows: siRNA1 forward sequence, 5′-GCU ACU UUG UGG UAU CAA UTT-3′ and reverse sequence, 5′-AUU GAU ACC ACA AAG UAG CUU-3′; siRNA2 forward sequence, 5′-GAG GAG AUA GGU AGA ACC UTT-3′ and reverse sequence, 5′-AGG UUC UAC CUA UCU CCU CUU-3′; siRNA negative control (NC) forward sequence, 5′-UUC UUC GAA CGU GUC ACG UTT-3′ and reverse sequence, 5′-ACG UGA CAC GUU CGG AGA ATT-3′. The cells were inoculated on a 6-well plate, and the cell density reached 60-80% confluence before the transfection of siRNA (20, 50, 100 and 200 nM). The mixture of transfection reagents of Lipofectamine 2000 (cat. no. 11668027; Invitrogen; Thermo Fisher Scientific, Inc.), Opti-MEM (cat. no. 31985070; Gibco; Thermo Fisher Scientific, Inc.) and siRNA was slowly dripped into the 6-well plate. After being cultured at 37°C and 5% CO2 for 4-6 h, the mixed transfection reagents were replaced with complete DMEM (without antibiotics), and the follow-up experiment was carried out after 24 h of transfection as previously described (27).
Cell proliferation assay
The effect of lncRNAS56464.1 on FLS proliferation was detected by an MTT assay as previously described (28). The transfected FLS (1×105/ml) were cultured in complete DMEM medium with 20% fetal bovine serum for 12, 24 and 48 h in 96-well plates. After culturing, 20 µl MTT (5 mg/ml) (cat. no. ST316; Beyotime Institute of Biotechnology,) was added into each well and cultured at 37°C and 5% CO2 for 4 h. Then, 150 µl dimethyl sulfoxide (DMSO) was utilized to dissolve the formazan after discarding the liquid in each well as previously described (29,30). Determination of the absorbance at 490 nm was performed using a microplate reader (BioTek Instruments, Inc.). The cell proliferation inhibition rate was calculated as follows: Inhibition rate = [1−(OD siRNA group-OD blank control group)/(OD control group-OD blank control group)] ×100%. The blank control group consisted of MTT reagents without FLS and the control group refers to FLS treated with DEPC water, which is the solvent of siRNA.
Prediction of miRNA targets of lncRNAS56464.1 and validation by a luciferase reporter assay
The full-length sequences of lncRNAS56464.1 and mature miRNAs such as miR-152-3p, miR-20a-3p, miR-17-2-3p as well as others which were differentially expressed key miRNAs in the experimental arthritis in our previous study (16), were obtained from NCBI Refseq and miRbase (31), respectively. We predicted the miRNA target of lncRNAS56464.1 using OG-local tools based on the RNAhybrid algorithm (32) and revealed that miR-152-3p to lncRNAS56464.1 had the highest binding degree and lowest free energy. Then, the full fragment of lncRNAS56464.1 with the binding site of miR-152-3p was amplified through PCR, including the wild-type or mutant lncRNAS56464.1 sequence binding site. Then, the constructed plasmid and miR-152-3p mimic with the Lipofectamine 2000 reagent were co-transfected into 293T functional cells (cat. no. FH0244; Shanghai FuHeng Biology). The cells were collected at 48 h post-transfection, and the luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega Corporation) and normalized to the Renilla luciferase activity as previously described (33).
Reverse transcription-quantitative (RT-q)PCR
Total RNA was extracted from FLS using TRIzol (lot no. 90802; cat. no. 15596018; Invitrogen; Thermo Fisher Scientific, Inc.) and reverse transcribed into cDNAs using the Reverse Transcription kit (K1621; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. RT-qPCR was performed to amplify the cDNA templates by 2X Universal SYBR-Green Fast qPCR Mix (cat. no. RK21203; ABclonal) and using the PIKOREAL 96 fluorescence quantitative PCR instrument (Thermo Fisher Scientific, Inc.). The thermocycling conditions were as follows: Pre-denaturation at 95°C for 3 min, 40 cycles at 95°C for 5 sec and 60°C for 30 sec. The relative expression of each target gene was quantified and normalized by calculating (cycle of quantification) the values and the level of each index as previously described (34,35). The primers were synthesized by Shanghai Sangon Biotech Co., Ltd., and are presented in Table I.
Table I.
Sequences of primers for RT-qPCR.
| Targeted gene | Forward sequence and reverse sequence | Product length (bp) |
|---|---|---|
| Wnt1 | F: 5′-AGAAACCGCCGCTGGAACT-3′ | |
| R: 5′-GGAGGTGATTGCGAAGATAAACG-3′ | 105 | |
| β-catenin | F: 5′-CCACGACTAGTTCAGCTGCTTGTAC-3′ | |
| R: 5′-ACTGCACAAACAGTGGAATGGTATT-3′ | 225 | |
| c-Myc | F: 5′-CTGGAGTGAGAAGGGCTTTG-3′ | |
| R: 5′-CAGCAGCTCGAATTTCTTCC-3′ | 480 | |
| Cyclin D1 | F: 5′-TGGAGCCCCTGA AGAAGAG-3′ | |
| R: 5′-AAGTGCGTTGTGCGGTAGC-3′ | 424 | |
| GSK-3β | F: 5′-TACCCATACGATGTTCCAGAT-3′ | |
| R: 5′-ACCCTGCCCAGGAGTTGCCAC-3′ | 120 | |
| SFRP4 | F: 5′-AAGTCTTTGTCACCTATCCCTCG-3′ | |
| R: 5′-CGGCTGGCTATCTGCTTCTT-3′ | 162 | |
| β-actin | F: 5′-CAGCGGAACCGCTCATTGATGG-3′ | |
| R: 5′-TCACCCACACTGTGCCCAACGA-3′ | 300 |
RT-qPCR, reverse transcription-quantitative PCR; F, forward; R, reverse.
Western blotting
FLS was extracted using RIPA lysis buffer (product no. P0013B; Beyotime Institute of Biotechnology) containing PMSF (100 mM) (product no. ST506; Beyotime Institute of Biotechnology). FLS were washed three times with cold PBS and digested with 100 µl protein lysate. The protein concentration was determined using the BCA method. The lysates (20 µg of protein loaded per lane) were separated by 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to a polyvinylidene fluoride membrane (PVDF membrane). The PVDF membrane was washed with 1X TBST for 2 min, sealed with 5% skim milk at room temperature for 2 h, and incubated with anti-Wnt1 (1:5,000; product no. ab63934; Abcam), anti-β-catenin (1:500; ab68183; Abcam), anti-c-Myc (1:500; BM4042; Boster Biological Technology), anti-cyclin D1 (1:500; ab40754), anti-GSK3β (1:2,000; product code ab93926), anti-GSK3β (phospho S9) (1:10,000; product code ab75814), anti-SFRP4 (1:200; ab154167), and anti-β-actin (1:1,000; ab8226; all from Abcam) antibodies overnight at 4°C. Then, the membrane was washed three times with TBST and incubated with a second anti-mouse or anti-rabbit antibody HRP (1:10,000; cat. nos. AS003 and AS014, respectively; ABclonal) for 2 h at room temperature. The protein was detected with an enhanced chemiluminescence kit (EMD Millipore). Each protein band was a representative picture of three replicates. Western blotting data were quantified with ImageJ software (version 1.52, National Institutes of Health) (30).
Immunofluorescence
The cells were fixed in 4% paraformaldehyde for 30 min, then 50-100 µl 0.25% Triton X-100 was added and incubated for 10 min; all steps aforementioned were performed at room temperature. The sections were washed with PBS for three times, and immunostained with primary antibodies, and incubated overnight in a wet chamber at 4°C. The dilutions of the antibodies (as aforementioned) were as follows: Wnt1, 1:100; β-catenin, 1:200; c-Myc, 1:200; cyclin D1, 1:500; p-GSK-3β (Ser9), 1:200; SFRP4, 1:200; and β-actin, 1:200. Then, the sections were stained with the corresponding secondary antibody (as aforementioned) (1:10,000), and incubated at room temperature for 50 min. The nucleus was stained for 5 min with DAPI at room temperature. Six fluorescence images for semi-quantitative analysis and the average optical density were used to reflect the fluorescence intensity of immune response of different indexes as previously described (36).
Statistical analysis
The data were inputted into SPSS17.0 software (SPSS, Inc.) for statistical processing, and the results were expressed as the mean ± SD (standard deviation) . The data were analyzed using one-way ANOVA (one-way analysis of variance) followed by Tukey's multiple comparison test to detect the differences between groups. The result was considered statistically significant when P<0.05.
Results
Identification and activity detection of FLS
After more than two passages, the characteristics of the FLS were stable, the species were mainly fibroblasts, the shape was mainly fusiform, and the polar processes were slender, as revealed in Fig. 1A. Under a fluorescence microscope, the passage of FLS was positive for vimentin antibody labeling, the cells were fusiform, and the nucleus was oval and located in the center of the cells, as revealed in Fig. 1B. In addition, the results of trypan blue staining revealed that the survival rate of the FLS was more than 90% (data not shown).
Figure 1.
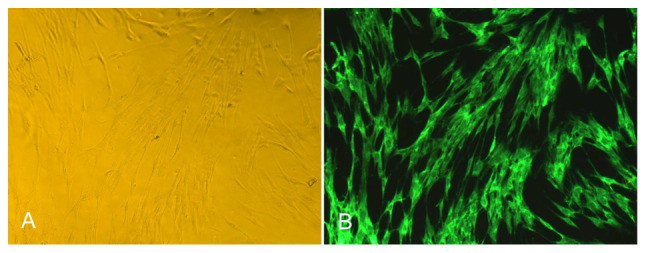
Morphological observation of synovial cells by optical microscopy and fluorescence microscopy. (A) The morphological observation of transmembrane FLS under optical microscope (×200). (B) The morphological observation of transmembrane FLS under fluorescence microscope (×200). FLS, fibroblast-like synoviocytes.
Negative regulation of FLS proliferation under lncRNAS56464.1 interference
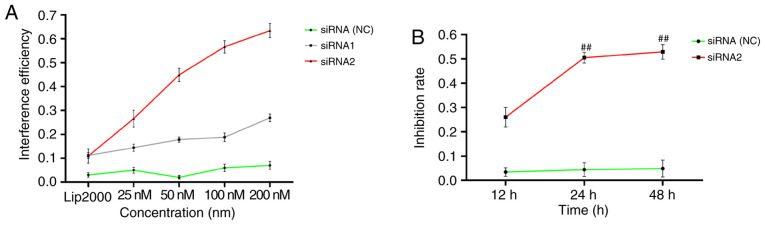
As revealed in Fig. 2A, the highest efficiency of lncRNAS56464.1 siRNA1 interference on FLS was ~30%; the highest efficiency of lncRNAS56464.1 siRNA2 interference on FLS was >60%, and the interference efficiency of 100 nM was more than 50%. According to the interference efficiency, lncRNAS56464.1 siRNA2 (100 nM) was selected for the following experimental study. As revealed in Fig. 2B, with the increasing interference time of the lncRNAS56464.1, the inhibition rate of FLS increased continuously. The inhibition rate significantly increased between 12 and 24 h as well as 12 and 48 h. However, there was no significant increase between 24 and 48 h. Thus, 24 h was selected as the stimulation time for the follow-up experiment.
Figure 2.
Interference efficiency of lncRNAS56464.1 siRNA and the effect on FLS proliferation. (A) Interference efficiency of lncRNAS56464.1 siRNA. (B) Effect of lncRNAS56464.1 interference on FLS proliferation. siRNA, small interfering RNA; FLS, fibroblast-like synoviocytes; NC, negative control. ##P<0.01 compared with the 12 h group.
lncRNAS56464.1 can combine with miR-152-3p
lncRNAs are considered to bind competitively with miRNAs, resulting in a series of biological effects (37,38). In Fig. 3A, the present results revealed using RNAhybrid, that miR-152-3p was identified as having a potential binding site of lncRNAS56464.1. Then, in the subsequent luciferase experiment, the luciferase constructor containing the wild-type or mutant lncRNAS56464.1 was co-transfected with miR-152-3p or control miRNAs to verify whether lncRNAS56464.1 could target miR-152-3p. As revealed in Fig. 3B, upon overexpressing miR-152-3p in FLS, the luciferase activity of wild-type lncRNAS56464.1 was lower than that of mutant lncRNAS56464.1, which also indicated that lncRNAS56464.1 could directly target miR-152-3p and combine with it.
Figure 3.
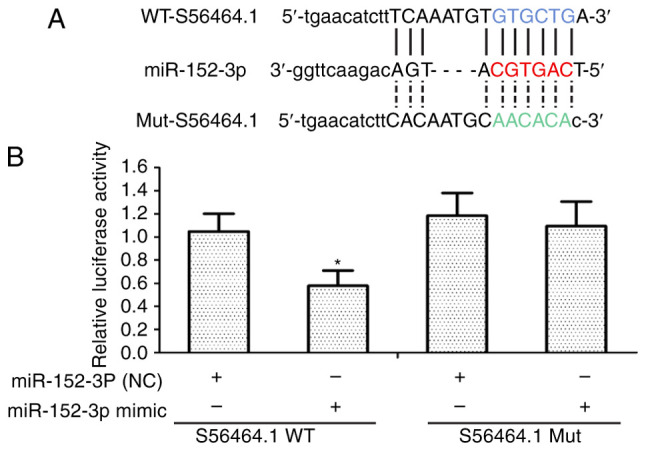
miR-152-3p is a potential target gene of lncRNAS56464.1. (A) Schematic comparison between lncRNAS56464.1 and the 'seed sequence' in miR-152-3p. (B) miR-152-3p inhibited the luciferase activity of WT lncRNAS56464.1 rather than that of Mut lncRNAS56464.1 in FLS. *P<0.05 compared with the NC group. WT, wild-type; Mut, mutant; FLS, fibroblast-like synoviocytes; NC, negative control.
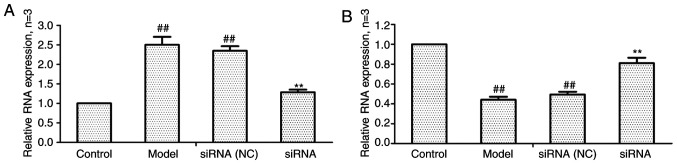
Effect of lncRNAS56464.1 interference on the expression of lncRNAS56464.1 and miR-152-3p mRNA in FLS
As revealed in Fig. 4, compared with the control group, the expression of lncRNAS56464.1 was significantly increased (Fig. 4A) and that of miR-152-3p was significantly decreased (Fig. 4B) in the model group. Compared with the model group, the expression of lncRNAS56464.1 was significantly decreased (Fig. 4A) and that of miR-152-3p significantly increased (Fig. 4B) in the siRNA group.
Figure 4.
Changes of lncRNAS56464.1 and miR-152-3p expression in FLS after siRNA treatment. (A) The change of lncRNAS56464.1 expression in FLS after siRNA treatment. (B) The change of miR-152-3p expression in FLS after siRNA treatment. ##P<0.01 compared with the control group; **P<0.01 compared with the model group. Control, FLS of the normal group; model, FLS of the AA model group. FLS, fibroblast-like synoviocytes; siRNA, small interfering RNA; siRNA (NC), small interfering RNA negative control; AA, adjuvant-induced arthritis.
Effect of lncRNAS56464.1 interference on the Wnt signaling pathway
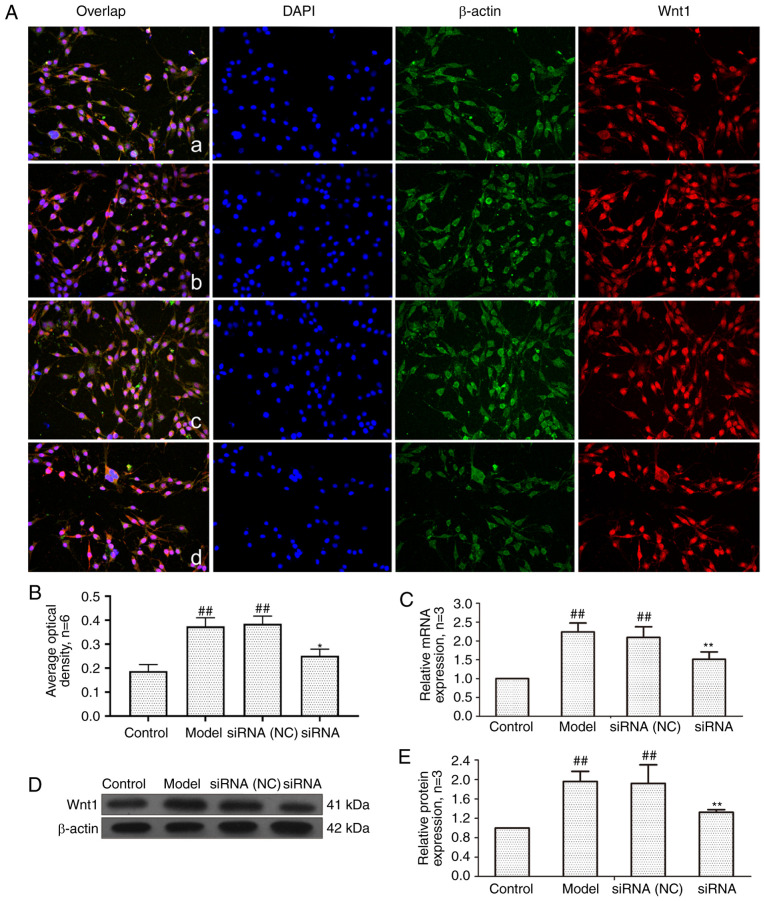
Effect of lncRNAS56464.1 interference on Wnt1
It has been demonstrated that Wnt1 can directly target miR-152-3p (39) and thus Wnt1 was selected as the detection indicator. As revealed in Fig. 5, compared with the control group, the mRNA and protein expression of Wnt1 in the model group and the siRNA (NC) group was significantly increased. Compared with the model group, Wnt1 expression in the siRNA group was significantly decreased.
Figure 5.
Changes of Wnt1 protein and mRNA expression in FLS of AA rats after siRNA treatment. (A) The changes of Wnt1 protein expression were observed by an immunofluorescence technique (×400); a, control group; b, model group, c, siRNA (NC) group, d, siRNA group. (B) Semiquantitative analysis of fluorescence intensity. (C) The changes of Wnt1 mRNA expression were observed by RT-qPCR. (D) The changes of Wnt1 protein expression were observed by western blotting. (E) Semiquantitative analysis of Wnt1 protein. ##P<0.01 compared with the control group; *P<0.05 and **P<0.01 compared with the model group. Control, FLS of the normal group; Model, FLS of the AA model group. siRNA (NC), small interfering RNA negative control; siRNA, small interfering RNA; FLS, fibroblast-like synoviocytes; AA, adjuvant-induced arthritis; RT-qPCR, reverse transcription-quantitative PCR.
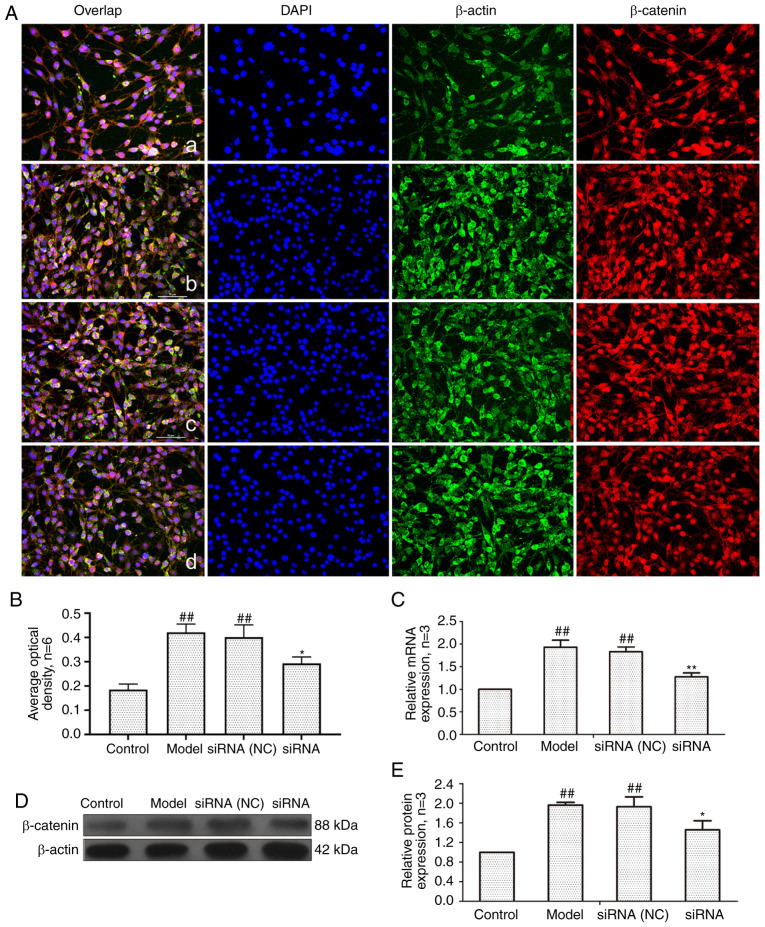
Effect of lncRNAS56464.1 interference on the expression of β-catenin in FLS of AA rats
β-catenin is a key gene in the classical Wnt signaling pathway, which plays a central role in signal transduction (40). As revealed in Fig. 6, the mRNA and protein expression of β-catenin in the model group and siRNA (NC) group was significantly higher than that in the control group. The expression levels of β-catenin at both the protein and mRNA level were significantly lower in the siRNA group than that in the model group.
Figure 6.
Changes of β-catenin protein and mRNA expression in FLS of AA rats after siRNA treatment. (A) The changes of β-catenin protein expression were observed by an immunofluorescence technique (×400); a, control group; b, model group, c, siRNA (NC) group, d, siRNA group. (B) Semiquantitative analysis of fluorescence intensity. (C) The changes of Wnt1 mRNA expression were observed by RT-qPCR. (D) The changes of β-catenin protein expression were observed by western blotting. (E) Semiquantitative analysis of β-catenin protein. ##P<0.01 compared with the control group; *P<0.05 and **P<0.01 compared with the model group. Control, FLS of the normal group; Model, FLS of the AA model group. siRNA (NC), small interfering RNA negative control; siRNA, small interfering RNA; FLS, fibroblast-like synoviocytes; AA, adjuvant-induced arthritis; RT-qPCR, reverse transcription-quantitative PCR.
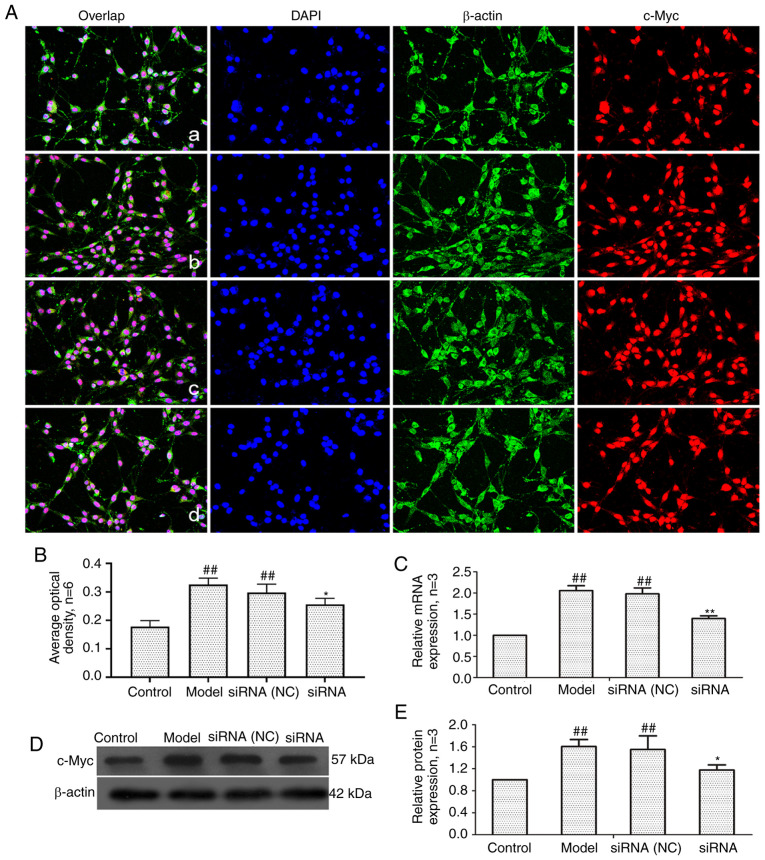
Effect of lncRNAS56464.1 interference on the expression of c-Myc in FLS of AA rats
c-Myc is a proto-oncogene that is a downstream target gene of the Wnt signaling pathway (41). As revealed in Fig. 7, compared with the control group, the mRNA and protein expression of c-Myc in the model group and siRNA (NC) group was significantly increased. After lncRNAS56464.1 interference, compared with the model group, the mRNA and protein expression of c-Myc was significantly decreased.
Figure 7.
Changes of c-Myc protein and mRNA expression in FLS of AA rats after siRNA treatment. (A) The changes of c-Myc protein expression were observed by an immunofluorescence technique (×400); a, control group; b, model group, c, siRNA (NC) group, d, siRNA group. (B) Semiquantitative analysis of fluorescence intensity. (C) The changes of c-Myc mRNA expression were observed by RT-qPCR. (D) The changes of c-Myc protein expression were observed by western blotting. (E) Semiquantitative analysis of c-Myc protein. ##P<0.01 compared with the control group; *P<0.05 and **P<0.01 compared with the model group. Control, FLS of the normal group; Model, FLS of the AA model group. siRNA (NC), small interfering RNA negative control; siRNA, small interfering RNA; FLS, fibroblast-like synoviocytes; AA, adjuvant-induced arthritis; RT-qPCR, reverse transcription-quantitative PCR.
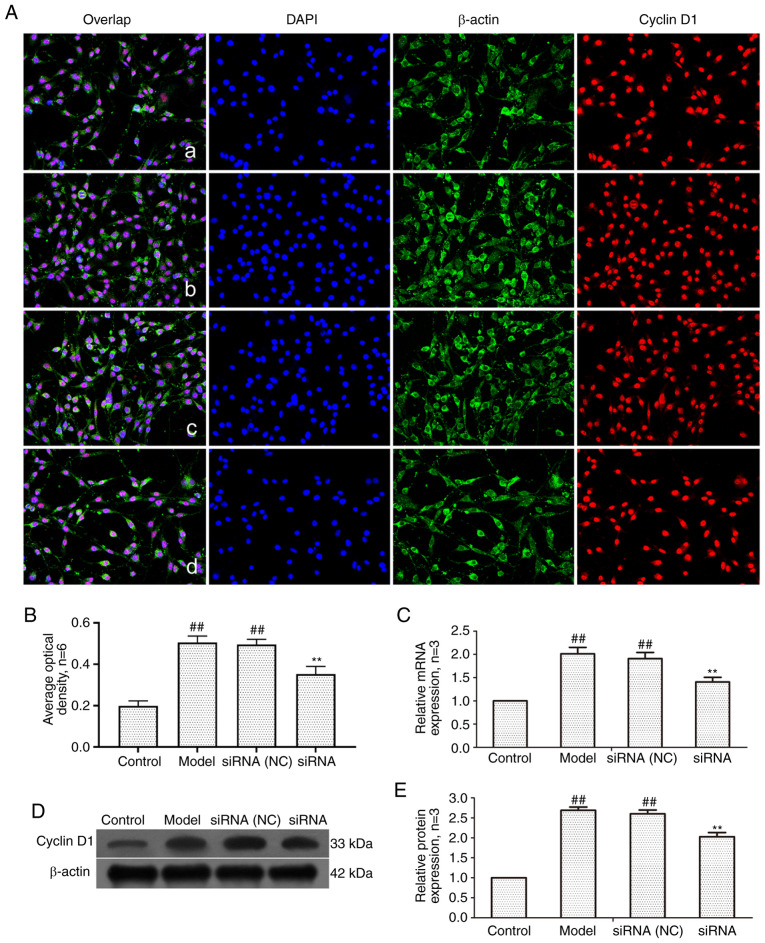
Effect of lncRNAS56464.1 interference on the expression of cyclinD1 in FLS of AA rats
The main function of cyclin D1 is to promote cell proliferation, and its overexpression can lead to cell loss of control (42). To detect the level of cyclin D1, RT-qPCR and western blotting were employed. As revealed in Fig. 8, the mRNA and protein expression of cyclin D1 in the model group and siRNA (NC) group was significantly higher than that in control group. The expression of cyclin D1 protein and mRNA in the siRNA group was significantly lower than that in the model group.
Figure 8.
Changes of cyclin D1 protein and mRNA expression in FLS of AA rats after siRNA treatment. (A) The changes of cyclin D1 protein expression were observed by an immunofluorescence technique (×400); a, control group; b, model group, c, siRNA (NC) group, d, siRNA group. (B) Semiquantitative analysis of fluorescence intensity. (C) The changes of cyclin D1 mRNA expression were observed by RT-qPCR. (D) The changes of cyclin D1 protein expression were observed by western blotting. (E) Semiquantitative analysis of cyclin D1 protein. ##P<0.01 compared with the control group; **P<0.01 compared with the model group. Control, FLS of the normal group; Model, FLS of the AA model group. siRNA (NC), small interfering RNA negative control; siRNA, small interfering RNA; FLS, fibroblast-like synoviocytes; AA, adjuvant-induced arthritis; RT-qPCR, reverse transcription-quantitative PCR.
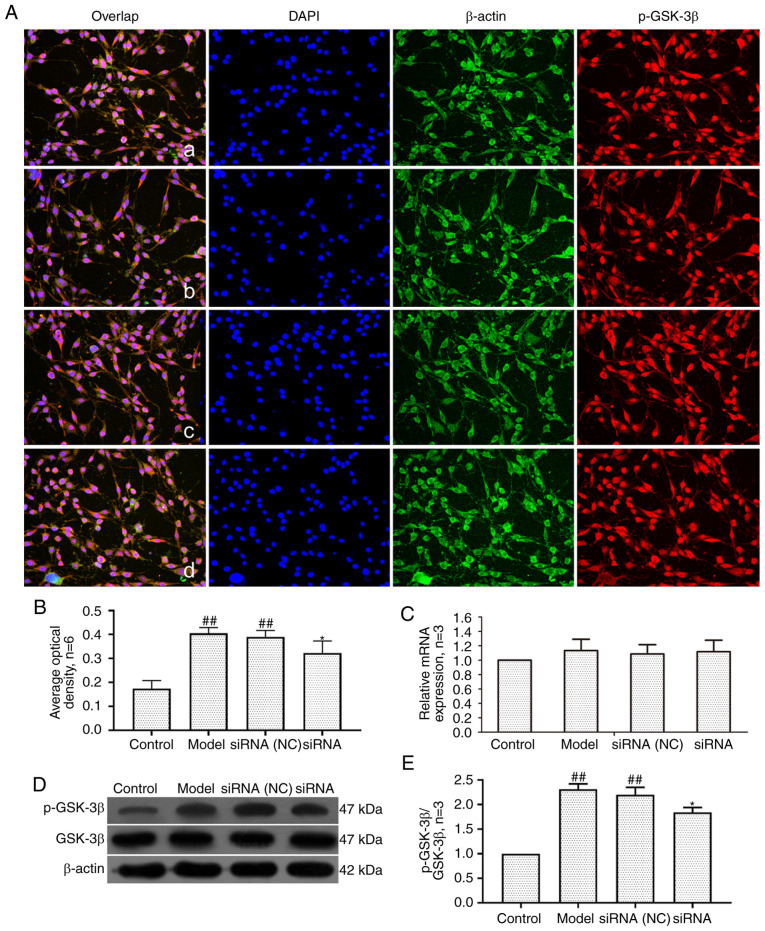
Effect of lncRNAS56464.1 interference on the expression of GSK-3β in synovial cells of AA rats
GSK-3β is a serine/threonine kinase that can regulate cell proliferation and differentiation (43). As revealed in Fig. 9, compared with the control group, the mRNA and protein expression of GSK-3β in the model group and siRNA (NC) group had no significant change, but the protein level of p-GSK-3β (Ser9)/GSK-3β was significantly increased. Compared with the model group, the mRNA and protein expression of GSK-3β in the siRNA group had no significant change, but the protein level of p-GSK-3β (Ser9)/GSK-3β was significantly decreased.
Figure 9.
Changes of GSK-3β and p-GSK-3β (Ser9) protein and mRNA expression in FLS of AA rats after siRNA treatment. (A) The changes of p-GSK-3β(Ser9) protein expression were observed by an immunofluorescence technique (×400); a, control group; b, model group, c, siRNA (NC) group, d, siRNA group. (B) Semiquantitative analysis of fluorescence intensity. (C) The changes of p-GSK-3β(Ser9) mRNA expression were observed by RT-qPCR. (D) The changes of GSK-3β and p-GSK-3β(Ser9) protein expression were observed by western blotting. (E) Semiquantitative analysis of p-GSK-3β(Ser9)/GSK-3β protein. ##P<0.01 compared with the control group; *P<0.05 compared with the model group. Control, FLS of the normal group; Model, FLS of the AA model group. siRNA (NC), small interfering RNA negative control; siRNA, small interfering RNA; FLS, fibroblast-like synoviocytes; AA, adjuvant-induced arthritis; RT-qPCR, reverse transcription-quantitative PCR.
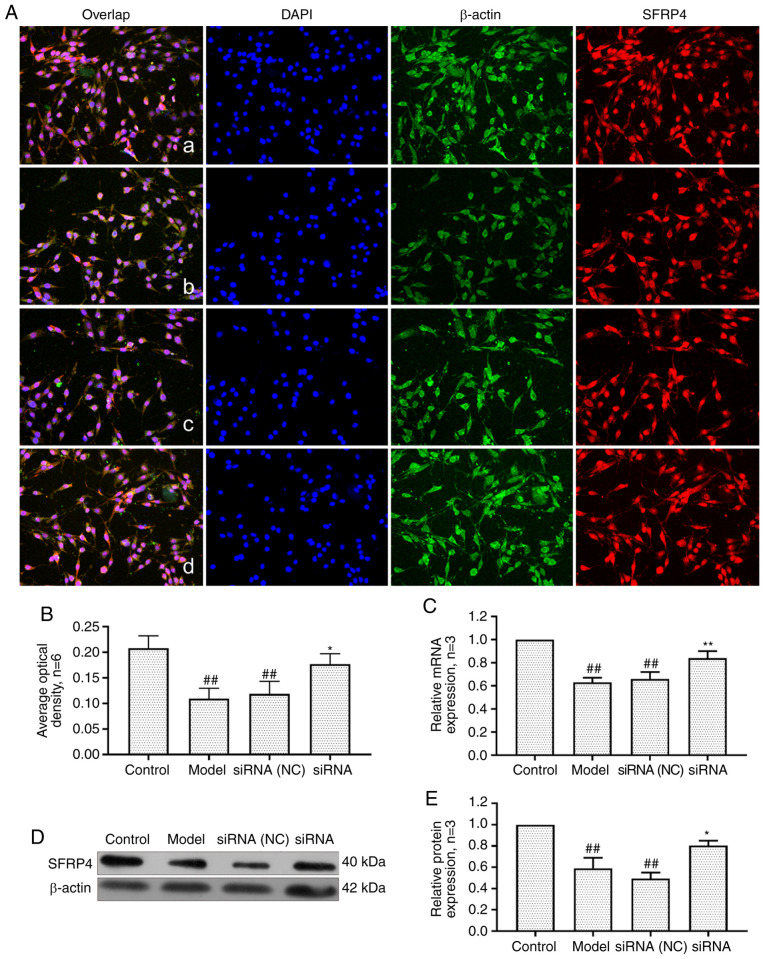
Effect of lncRNAS56464.1 interference on the expression of SFRP4 in FLS of AA rats
SFRP4 is an antagonist of the Wnt signaling pathway, which is closely related to cell proliferation and differentiation (44). As revealed in Fig. 10, the mRNA and protein expression of SFRP4 in the model group and the siRNA (NC) group was significantly lower than that in the control group. After lncRNAS56464.1 interference, the expression of SFRP4 at both protein and mRNA levels was significantly higher than that in the model group.
Figure 10.
Changes of SFRP4 protein and mRNA expression in FLS of AA rats after siRNA treatment. (A) The changes of SFRP4 protein expression were observed by an immunofluorescence technique (×400); a, control group; b, model group, c, siRNA (NC) group, d, siRNA group. (B) Semiquantitative analysis of fluorescence intensity. (C) The changes of SFRP4 mRNA expression were observed by RT-qPCR. (D) The changes of SFRP4 protein expression were observed by western blotting. (E) Semiquantitative analysis of SFRP4 protein. ##P<0.01 compared with the control group; *P<0.05 and **P<0.01 compared with the model group. Control, FLS of the normal group; Model, FLS of the AA model group. siRNA (NC), small interfering RNA negative control; siRNA, small interfering RNA; FLS, fibroblast-like synoviocytes; AA, adjuvant-induced arthritis; RT-qPCR, reverse transcription-quantitative PCR.
Discussion
RA is a type of autoimmune disease characterized by synovial hyperplasia and bone erosion. Patients exhibit some joint swelling at the beginning of the course of the disease, which can develop into systemic joint deformities at the later stage of the course, causing serious damage to the body and psychology of a patient. The pathological process of RA involves numerous types of cells, among which FLS are the most important (45-47). FLS contain a large amount of endoplasmic reticulum and have the main characteristics of a protein secretory phase. Moreover, FLS can be activated continuously in both an inflammatory and noninflammatory environment, exhibiting tumor-like proliferation and high invasive ability, which are closely related to the destruction of articular cartilage and joint injury in RA (48-50).
lncRNAs, play a regulatory role in epigenetic, pre-transcriptional and post-transcriptional aspects and widely participate in the biological functions of the body and are closely related to the progression of numerous diseases (51,52). In our previous study, we confirmed that lncRNAS56464.1, as an antisense noncoding RNA, was found to be located on chromosome 4 and exhibited increased expression in RA. Moreover, bioinformatics analysis indicated that lncRNAS56464.1 is a key gene in RA (16). In this study, it was revealed that lncRNAS56464.1 interference inhibited the proliferation of FLS, indicating that lncRNAS56464.1 can promote the proliferation of FLS.
Since lncRNAS56464.1 is newly discovered as revealed in our pervious study (16), determination of the specific mechanism of lncRNAS56464.1 in RA development and progression remains in its infancy. At present, it is widely accepted that lncRNAs can regulate the abundance of miRNAs by binding and sequestering them, acting as the so-called miRNA sponges, and then regulating the expression of target mRNAs (53,54). Thus, it has been revealed to be efficient to infer the potential functions of lncRNAs by studying the related miRNAs and mRNAs, whose functions have been annotated (55-57). Through bioinformatics prediction, it was revealed that there was a high degree of binding between miR-152-3p and lncRNAS56464.1. Thereafter, through luciferase reporter assays, it was also revealed that the luciferase activity of lncRNAS56464.1-WT was lower than that of lncRNAS56464.1-Mut, indicating that miR-152-3p was the downstream target of lncRNAS56464.1 and can be combined with it.
miR-152 originates from chromosome 17 (17q21.32) of chr10:84719319-84719403 (58). Miao et al (59) revealed that the expression of miR-152 was significantly decreased in rats with adjuvant arthritis, and the upregulation of miR-152 expression could inhibit the activation of the Wnt signaling pathway, thus reducing the excessive proliferation of FLS. A recent study reported that miR-152-3p could directly target Wnt1 and activate the Wnt signaling pathway (60).
Large amounts of evidence indicate that the Wnt signaling pathway can participate in the occurrence and development of RA by regulating inflammation, proliferation, apoptosis, articular cartilage maturation and bone destruction (61,62). Wnt1 plays a regulatory role in the Wnt pathway, participates in biological processes such as cell differentiation, proliferation and migration and is closely related to the pathogenesis of RA (63,64). Sen et al and Nakamura et al (65,66) revealed that the expression levels of Wnt and fibronectin in the synovium of patients with RA were significantly increased. The present study revealed that under the condition of lncRNAS56464.1 interference, the expression of Wnt1 was significantly decreased, suggesting that lncRNAS56464.1 could affect the proliferation of FLS by affecting the Wnt pathway.
β-catenin is the key gene of the classical Wnt signaling pathway, which plays a central role in signal transduction and is directly related to the abnormal proliferation of RA synovial cells (67). Yoshioka et al (68) demonstrated that β-catenin siRNAs could significantly inhibit the proliferation of FLS stimulated by IL-1β. c-Myc and cyclin D1 are proto-oncogenes, which participate in cell proliferation, differentiation and apoptosis and are closely related to the occurrence and development of numerous diseases (69,70). Recent studies have revealed that c-Myc and cyclin D1, as important target genes downstream of the Wnt signaling pathway, are highly expressed in FLS and involved in the pathogenesis of RA (71,72). GSK-3β is a widely expressed serine/threonine family kinase, in which the abnormal activity of the GSK-3β subtype has been demonstrated to be involved in the occurrence and development of numerous diseases (73,74). Studies have revealed that the stability and nuclear translocation of intracellular β-catenin are landmark molecular events in the activation of the Wnt signaling pathway. β-catenin, is the classical phosphorylation substrate of GSK-3β, and its degradation is regulated by GSK-3β. When GSK-3β is phosphorylated to p-GSK-3β (Ser9), degradation of β-catenin by GSK-3β is inhibited, and then affects the signal transduction of the Wnt signaling pathway (75). The present study revealed that after lncRNAS56464.1 interference, the expression levels of β-catenin, c-Myc, cyclin D1 and p-GSK-3β (Ser9)/GSK-3β in FLS were significantly decreased.
SFRP4 is a secretory protein with a size of 40 kDa, consisting of more than 300 amino acids (76). It can inhibit the activation of the Wnt signaling pathway by competitively binding with receptors related to the Wnt signaling pathway. The present data indicated that the expression levels of SFRP4 were decreased at both the mRNA and protein levels in FLS, which was in accordance with findings from other relevant research studies (77,78). Notably, after lncRNAS56464.1 interference, the expression of SFRP4 was significantly increased.
Based on the ceRNA theory (17), the effects of lncRNAS56464.1 on targeting the miR-152-3p/Wnt pathway in FLS were studied, and the relationship between RA and lncRNAs, miRNAs, and the Wnt pathway was elucidated. However, the targeting relationship between lncRNAS56464.1 and miR-152-3p in AA was only verified, thus in future studies we will explore the relationship between miR-152-3p and the Wnt signaling pathway in experimental arthritis.
In conclusion, as revealed in Fig. 11, the present study revealed that lncRNAS56464.1 could target the miR-152-3p/Wnt pathway, promote the proliferation of FLS, and then lead to the occurrence and development of RA. This study deepens our understanding of the pathogenesis of RA from the perspective of lncRNAs and provides a new direction and target for the clinical diagnosis and treatment of RA.
Figure 11.
lncRNAS56464.1 as a ceRNA promotes the proliferation of fibroblast-like synoviocytes in experimental arthritis via the Wnt signaling pathway by sponging miR-152-3p. When the body is stimulated by external damage factors, the expression of lncRNAS56464.1 increases, and it competitively binds to miRNA and decreases the expression of miR-152-3p. Therefore, it increases the expression of Wnt1, β-catenin, c-Myc, and cyclin D1 in the Wnt signaling pathway, further promoting the phosphorylation of GSK-3β, reducing the expression of SFRP4, and activating the Wnt signaling pathway and thus promoting the proliferation of FLS and affecting the occurrence and development of RA. FLS, fibroblast-like synoviocytes; RA, rheumatoid arthritis.
Acknowledgments
We are grateful to Mr Qiang Fan (Ao Ji Bio-tech Co., Ltd. Shanghai, China) for providing help with data analysis.
Funding
The present study was financially supported by the National Natural Science Foundation of China (grant no. 81873139), the Anhui Provincial Laboratory of Applied Basis and Development of Internal Medicine of Modern Traditional Chinese Medicine (grant no. 2016080503B041), and the 12th Batch of '115' Innovation Team of Anhui Province [Anhui Talent Office (2019) no. 1].
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
WL made substantial contributions to the conception and design of the study. HJ, CF and JW performed the experiments. HJ contributed to data acquisition, and data analysis and interpretation. JL revised the manuscript critically for important intellectual content. All authors agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of the work are appropriately investigated and resolved. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All animal experiments were approved by the Animal Ethics Committee of Anhui University of Chinese Medicine (Hefei, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Taylor PC. Aetiopathology of rheumatoid arthritis. Medicine. 2014;42:227–230. doi: 10.1016/j.mpmed.2014.02.010. [DOI] [Google Scholar]
- 2.Gutierrez MJ, Desiderio SV, Wang NY, Darrah E, Cappelli L, Nino G, Jones M, Bingham CO., III Soluble markers of antibody secreting cell function as predictors of infection risk in rheumatoid arthritis. J Immunol Res. 2019;2019:3658215. doi: 10.1155/2019/3658215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Onuora S. Rheumatoid arthritis: RA synovium harbours distinct fibroblast subsets. Nat Rev Rheumatol. 2018;14:250. doi: 10.1038/nrrheum.2018.47. [DOI] [PubMed] [Google Scholar]
- 4.Collison J. Rheumatoid arthritis: Features of synovium in RA remission revealed. Nat Rev Rheumatol. 2016;12:316. doi: 10.1038/nrrheum.2016.63. [DOI] [PubMed] [Google Scholar]
- 5.Liu R, Zhao P, Zhang Q, Che N, Xu L, Qian J, Tan W, Zhang M. Adiponectin promotes fibroblast-like synoviocytes producing IL-6 to enhance T follicular helper cells response in rheumatoid arthritis. Clin Exp Rheumatol. 2020;38:11–18. [PubMed] [Google Scholar]
- 6.Hanlon MM, Rakovich T, Cunningham CC, Ansboro S, Veale DJ, Fearon U, McGarry T. STAT3 mediates the differential effects of oncostatin M and TNFα on RA synovial fibroblast and endothelial cell function. Front Immunol. 2019;10:2056. doi: 10.3389/fimmu.2019.02056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adawi M, Firas S, Blum A. Rheumatoid arthritis and atherosclerosis. Isr Med Assoc J. 2019;21:460–463. [PubMed] [Google Scholar]
- 8.Ferreira CC, Campi-Azevedo AC, Peruhype-Magalhāes V, Coelho-Dos-Reis JG, Antonelli LRDV, Torres K, Freire LC, da Costa-Rocha IA, Oliveira ACV, Maia MLS, et al. Impact of synthetic and biological immunomodulatory therapy on the duration of 17DD yellow fever vaccine-induced immunity in rheumatoid arthritis. Arthritis Res Ther. 2019;21:75. doi: 10.1186/s13075-019-1854-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu M, Hou J, Zheng M, Cao Y, Alike Y, Mi Y, Zhu J. IL-21 gene rs6822844 polymorphism and rheumatoid arthritis susceptibility. Biosci Rep. 2020;40:BSR20191449. doi: 10.1042/BSR20191449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bi X, Guo XH, Mo BY, Wang ML, Luo XQ, Chen YX, Liu F, Olsen N, Pan YF, Zheng SG. lncRNA PICSAR promotes cell proliferation, migration and invasion of fibroblast-like synoviocytes by sponging miRNA-4701-5p in rheumatoid arthritis. EBioMedicine. 2019;50:408–420. doi: 10.1016/j.ebiom.2019.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang J, Chen W, Lin J. LncRNA: An all-rounder in rheumatoid arthritis. J Transl Int Med. 2019;7:3–9. doi: 10.2478/jtim-2019-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fok ET, Davignon L, Fanucchi S, Mhlanga MM. The lncRNA connection between cellular metabolism and epigenetics in trained immunity. Front Immunol. 2019;9:3184. doi: 10.3389/fimmu.2018.03184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng H, Sun M, Wang ZL, Wu Q, Yao J, Ren G, Sun XL. lncRNA RMST-mediated miR-107 transcription promotes OGD-induced neuronal apoptosis via interacting with hnRNPK. Neurochem Int. 2020;133:104644. doi: 10.1016/j.neuint.2019.104644. [DOI] [PubMed] [Google Scholar]
- 14.Guo M, Liu T, Zhang S, Yang L. RASSF1-AS1 an antisense lncRNA of RASSF1A, inhibits the translation of RASSF1A to exacerbate cardiac fibrosis in mice. Cell Biol Int. 2019;43:1163–1173. doi: 10.1002/cbin.11085. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, Du W. lncRNA FENDRR attenuates colon cancer progression by repression of SOX4 protein. Onco Targets Ther. 2019;12:4287–4295. doi: 10.2147/OTT.S195853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang H, Ma R, Zou S, Wang Y, Li Z, Li W. Reconstruction and analysis of the lncRNA-miRNA-mRNA network based on competitive endogenous RNA reveal functional lncRNAs in rheumatoid arthritis. Mol Biosyst. 2017;13:1182–1192. doi: 10.1039/C7MB00094D. [DOI] [PubMed] [Google Scholar]
- 17.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: The Rosetta stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Y, Huang A, Li T, Su X, Ding H, Li H, Qin X, Hou L, Zhao Q, Ge X, et al. miR-152 reduces human umbilical vein endothelial cell proliferation and migration by targeting ADAM17. FEBS Lett. 2014;588:2063–2069. doi: 10.1016/j.febslet.2014.04.037. [DOI] [PubMed] [Google Scholar]
- 19.Dang YW, Zeng J, He RQ, Rong MH, Luo DZ, Chen G. Effects of miR-152 on cell growth inhibition, motility suppression and apoptosis induction in hepatocellular carcinoma cells. Asian Pac J Cancer Prev. 2014;15:4969–4976. doi: 10.7314/APJCP.2014.15.12.4969. [DOI] [PubMed] [Google Scholar]
- 20.Huang H, Hu M, Li P, Lu C, Li M. miR-152 inhibits cell proliferation and colony formation of CD133(+) liver cancer stem cells by targeting KIT. Tumour Biol. 2015;36:921–928. doi: 10.1007/s13277-014-2719-x. [DOI] [PubMed] [Google Scholar]
- 21.Miao CG, Qin D, Du CL, Ye H, Shi WJ, Xiong YY, Zhang XL, Yu H, Dou JF, Ma ST, et al. DNMT1 activates the canonical Wnt signaling in rheumatoid arthritis model rats via a crucial functional crosstalk between miR-152 and the DNMT1, MeCP2. Int Immunopharmacol. 2015;28:344–353. doi: 10.1016/j.intimp.2015.06.013. [DOI] [PubMed] [Google Scholar]
- 22.Jiang H, Liu J, Wang T, Gao JR, Sun Y, Huang CB, Meng M, Qin XJ. Urinary metabolite profiling provides potential differentiation to explore the mechanisms of adjuvant-induced arthritis in rats. Biomed Chromatogr. 2016;30:1397–1405. doi: 10.1002/bmc.3697. [DOI] [PubMed] [Google Scholar]
- 23.Zhu L, Li J, Guo L, Yu X, Wu D, Luo L, Zhu L, Chen W, Chen C, Ye C, Zhang D. Activation of NALP1 inflammasomes in rats with adjuvant arthritis; a novel therapeutic target of carboxyami-dotriazole in a model of rheumatoid arthritis. Br J Pharmacol. 2015;172:3446–3459. doi: 10.1111/bph.13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang Y, Jia X, Sun X, Xu S, Wu Y, Zhang L, Wei W. APRIL promotes proliferation, secretion and invasion of fibroblast-like synoviocyte from rats with adjuvant induced arthritis. Mol Immunol. 2015;64:90–98. doi: 10.1016/j.molimm.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 25.Luo C, Liang JS, Gong J, Zhang HL, Feng ZJ, Yang HT, Zhang HB, Kong QH. miRNA-31 over-expression improve synovial cells apoptosis induced by RA. Bratisl Lek Listy. 2018;119:355–360. doi: 10.4149/BLL_2018_066. [DOI] [PubMed] [Google Scholar]
- 26.Kragstrup TW, Sohn DH, Lepus CM, Onuma K, Wang Q, Robinson WH, Sokolove J. Fibroblast-like synovial cell production of extra domain A fibronectin associates with inflammation in osteoarthritis. BMC Rheumatol. 2019;3:46. doi: 10.1186/s41927-019-0093-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen X, Wang Z, Tong F, Dong X, Wu G, Zhang R. lncRNA UCA1 promotes gefitinib resistance as a ceRNA to target FOSL2 by sponging miR-143 in non-small cell lung cancer. Mol Ther Nucleic Acids. 2020;19:643–653. doi: 10.1016/j.omtn.2019.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Präbst K, Engelhardt H, Ringgeler S, Hübner H. Basic colorimetric proliferation assays: MTT, WST, and Resazurin. Methods Mol Biol. 2017;1601:1–17. doi: 10.1007/978-1-4939-6960-9_1. [DOI] [PubMed] [Google Scholar]
- 29.Deng H, Zheng M, Hu Z, Zeng X, Kuang N, Fu Y. Effects of daphnetin on the autophagy signaling pathway of fibroblast-like synoviocytes in rats with collagen-induced arthritis (CIA) induced by TNF-α. Cytokine. 2020;127:154952. doi: 10.1016/j.cyto.2019.154952. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, Feng T, Duan S, Shi Y, Li S, Zhang X, Zhang L. miR-155 promotes fibroblast-like synoviocyte proliferation and inflammatory cytokine secretion in rheumatoid arthritis by targeting FOXO3a. Exp Ther Med. 2020;19:1288–1296. doi: 10.3892/etm.2019.8330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019;47(D1):D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, Liu S. LncRNA GAS5 suppresses inflammatory responses and apoptosis of alveolar epithelial cells by targeting miR-429/DUSP1. Exp Mol Pathol. 2020;113:104357. doi: 10.1016/j.yexmp.2019.104357. [DOI] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Zhu W, Xu J, Jiang C, Wang B, Geng M, Wu X, Hussain N, Gao N, Han Y, Li D, et al. Pristane induces autophagy in macrophages, promoting a STAT1-IRF1-TLR3 pathway and arthritis. Clin Immunol. 2017;175:56–68. doi: 10.1016/j.clim.2016.11.017. [DOI] [PubMed] [Google Scholar]
- 36.Kikuta S, Nakamura Y, Yamamura Y, Tamura A, Homma N, Yanagawa Y, Tamura H, Kasahara J, Osanai M. Quantitative activation-induced manganese-enhanced MRI reveals severity of Parkinson's disease in mice. Sci Rep. 2015;5:12800. doi: 10.1038/srep12800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye Y, Gao X, Yang N. lncRNA ZFAS1 promotes cell migration and invasion of fibroblast-like synoviocytes by suppression of miR-27a in rheumatoid arthritis. Hum Cell. 2018;31:14–21. doi: 10.1007/s13577-017-0179-5. [DOI] [PubMed] [Google Scholar]
- 38.Javanmard AR, Dokanehiifard S, Bohlooli M, Soltani BM. LOC646329 long non-coding RNA sponges miR-29b-1 and regulates TGFβ signaling in colorectal cancer. J Cancer Res Clin Oncol. 2020;146:1205–1215. doi: 10.1007/s00432-020-03145-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao X, Sun S, Xu J, Luo Y, Xin Y, Wang Y. MicroRNA-152 inhibits cell proliferation of osteosarcoma by directly targeting Wnt/β-catenin signaling pathway in a DKK1-dependent manner. Oncol Rep. 2018;40:767–774. doi: 10.3892/or.2018.6456. [DOI] [PubMed] [Google Scholar]
- 40.Sun S, Xie F, Xu X, Cai Q, Zhang Q, Cui Z, Zheng Y, Zhou J. Advanced oxidation protein products induce S-phase arrest of hepatocytes via the ROS-dependent, β-catenin-CDK2-mediated pathway. Redox Biol. 2018;14:338–353. doi: 10.1016/j.redox.2017.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tögel L, Nightingale R, Chueh AC, Jayachandran A, Tran H, Phesse T, Wu R, Sieber OM, Arango D, Dhillon AS, et al. Dual targeting of bromodomain and extraterminal domain proteins, and WNT or MAPK signaling, inhibits c-MYC expression and proliferation of colorectal cancer cells. Mol Cancer Ther. 2016;15:1217–1226. doi: 10.1158/1535-7163.MCT-15-0724. [DOI] [PubMed] [Google Scholar]
- 42.Tetsuo F, Arioka M, Miura K, Kai M, Kubo M, Igawa K, Tomooka K, Takahashi-Yanaga F, Nishimura F, Sasaguri T. Differentiation-inducing factor-1 suppresses cyclin D1-induced cell proliferation of MCF-7 breast cancer cells by inhibiting S6K-mediated signal transducer and activator of transcription 3 synthesis. Cancer Sci. 2019;110:3761–3772. doi: 10.1111/cas.14204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dong ZC, Zhang D, Wang SB, Lin ZQ. Target inhibition on GSK-3beta by miR-9 to modulate proliferation and apoptosis of bladder cancer cells. Differentiation-inducing factor-1 suppresses cyclin D1-induced cell proliferation of MCF-7 breast cancer cells by inhibiting S6K-mediated signal transducer and activator of transcription 3 synthesis. Eur Rev Med Pharmacol Sci. 2018;22:3018–3026. doi: 10.26355/eurrev_201805_15059. [DOI] [PubMed] [Google Scholar]
- 44.Perumal V, Dharmarajan AM, Fox SA. The Wnt regulator SFRP4 inhibits mesothelioma cell proliferation, migration, and antagonizes Wnt3a via its netrin-like domain. Int J Oncol. 2017;51:362–368. doi: 10.3892/ijo.2017.4011. [DOI] [PubMed] [Google Scholar]
- 45.Madav Y, Barve K, Prabhakar B. Current trends in theranostics for rheumatoid arthritis. Eur J Pharm Sci. 2020;145:105240. doi: 10.1016/j.ejps.2020.105240. [DOI] [PubMed] [Google Scholar]
- 46.DeMizio DJ, Geraldino-Pardilla LB. Autoimmunity and inflammation link to cardiovascular disease risk in rheumatoid arthritis. Rheumatol Ther. 2020;7:19–33. doi: 10.1007/s40744-019-00189-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang CA, Li JP, Yen JC, Lai IL, Ho YC, Chen YC, Lan JL, Chang JG. lncRNA NTT/PBOV1 axis promotes monocyte differentiation and is elevated in rheumatoid arthritis. Int J Mol Sci. 2018;19:2806. doi: 10.3390/ijms19092806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Samarpita S, Ganesan R, Rasool M. Cyanidin prevents the hyperproliferative potential of fibroblast-like synoviocytes and disease progression via targeting IL-17A cytokine signalling in rheumatoid arthritis. Toxicol Appl Pharmacol. 2020;391:114917. doi: 10.1016/j.taap.2020.114917. [DOI] [PubMed] [Google Scholar]
- 49.Xu Q, Yin S, Yao Y, Li X, Song B, Yang Y, Liu Y, Chen R, Li J, Ma T, et al. MAST3 modulates the inflammatory response and proliferation of fibroblast-like synoviocytes in rheumatoid arthritis. Int Immunopharmacol. 2019;77:105900. doi: 10.1016/j.intimp.2019.105900. [DOI] [PubMed] [Google Scholar]
- 50.Wang Y, Jiao T, Fu W, Zhao S, Yang L, Xu N, Zhang N. miR-410-3p regulates proliferation and apoptosis of fibroblast-like synoviocytes by targeting YY1 in rheumatoid arthritis. Biomed Pharmacother. 2019;119:109426. doi: 10.1016/j.biopha.2019.109426. [DOI] [PubMed] [Google Scholar]
- 51.Zhang X, Wang W, Zhu W, Dong J, Cheng Y, Yin Z, Shen F. Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int J Mol Sci. 2019;20:5573. doi: 10.3390/ijms20225573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang Z, Li L, Hou Z, Liu W, Wang H, Zhou T, Li Y, Chen S. lncRNA HAND2-AS1 inhibits 5-fluorouracil resistance by modulating miR-20a/PDCD4 axis in colorectal cancer. Cell Signal. 2020;66:109483. doi: 10.1016/j.cellsig.2019.109483. [DOI] [PubMed] [Google Scholar]
- 53.Zhao CC, Jiao Y, Zhang YY, Ning J, Zhang YR, Xu J, Wei W, Kang-Sheng G. lnc SMAD5-AS1 as ceRNA inhibit proliferation of diffuse large B cell lymphoma via Wnt/β-catenin pathway by sponging miR-135b-5p to elevate expression of APC. Cell Death Dis. 2019;10:252. doi: 10.1038/s41419-019-1479-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang H, Wang G, Gao Y, Zhao C, Li X, Zhang F, Jiang C, Wu B. lnc-SNHG1 activates the TGFBR2/SMAD3 and RAB11A/Wnt/β-catenin pathway by sponging MiR-302/372/373/520 in invasive pituitary tumors. Cell Physiol Biochem. 2018;48:1291–1303. doi: 10.1159/000492089. [DOI] [PubMed] [Google Scholar]
- 55.Li Y, Zou L, Yang X, Chu L, Ni J, Chu X, Guo T, Zhu Z. Identification of lncRNA, microRNA, and mRNA-associated CeRNA network of radiation-induced lung injury in a mice model. Dose-Response. 2019;17:1559325819891012. doi: 10.1177/1559325819891012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu J, Li H, Zheng B, Sun L, Yuan Y, Xing C. Competitive endogenous RNA (ceRNA) regulation network of lncRNA-miRNA-mRNA in colorectal carcinogenesis. Dig Dis Sci. 2019;64:1868–1877. doi: 10.1007/s10620-019-05506-9. [DOI] [PubMed] [Google Scholar]
- 57.Wang J, Yin J, Wang X, Liu H, Hu Y, Yan X, Zhuang B, Yu Z, Han S. Changing expression profiles of mRNA, lncRNA, circRNA, and miRNA in lung tissue reveal the pathophysiological of bronchopulmonary dysplasia (BPD) in mouse model. J Cell Biochem. 2019;120:9369–9380. doi: 10.1002/jcb.28212. [DOI] [PubMed] [Google Scholar]
- 58.Friedrich M, Pracht K, Mashreghi MF, Jäck HM, Radbruch A, Seliger B. The role of the miR-148/-152 family in physiology and disease. Eur J Immunol. 2017;47:2026–2038. doi: 10.1002/eji.201747132. [DOI] [PubMed] [Google Scholar]
- 59.Miao CG, Yang YY, He X, Huang C, Huang Y, Qin D, Du CL, Li J. MicroRNA-152 modulates the canonical Wnt pathway activation by targeting DNA methyltransferase 1 in arthritic rat model. Biochimie. 2014;106:149–156. doi: 10.1016/j.biochi.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 60.Sun LB, Zhao SF, Zhu JJ, Han Y, Shan TD. Long noncoding RNA UCID sponges miR-152-3p to promote colorectal cancer cell migration and invasion via the Wnt/β-catenin signaling pathway. Oncol Rep. 2020;44:1194–1205. doi: 10.3892/or.2020.7670. [DOI] [PubMed] [Google Scholar]
- 61.Miao CG, Yang YY, He X, Li XF, Huang C, Huang Y, Zhang L, Lv XW, Jin Y, Li J. Wnt signaling pathway in rheumatoid arthritis, with special emphasis on the different roles in synovial inflammation and bone remodeling. Cell Signal. 2013;25:2069–2078. doi: 10.1016/j.cellsig.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 62.Wu J, Fan W, Ma L, Geng X. miR-708-5p promotes fibroblast-like synoviocytes' cell apoptosis and ameliorates rheumatoid arthritis by the inhibition of Wnt3a/β-catenin pathway. Drug Des Devel Ther. 2018;12:3439–3447. doi: 10.2147/DDDT.S177128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang LH, Wang Y, Fan QQ, Liu YK, Li LH, Qi XW, Mao Y, Hua D. Up-regulated Wnt1-inducible signaling pathway protein 1 correlates with poor prognosis and drug resistance by reducing DNA repair in gastric cancer. World J Gastroenterol. 2019;25:5814–5825. doi: 10.3748/wjg.v25.i38.5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Malysheva K, de Rooij K, Lowik CW, Baeten DL, Rose-John S, Stoika R, Korchynskyi O. Interleukin 6/Wnt interactions in rheumatoid arthritis: Interleukin 6 inhibits Wnt signaling in synovial fibroblasts and osteoblasts. Croat Med J. 2016;57:89–98. doi: 10.3325/cmj.2016.57.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sen M, Reifert J, Lauterbach K, Wolf V, Rubin JS, Corr M, Carson DA. Regulation of fibronectin and metalloproteinase expression by Wnt signaling in rheumatoid arthritis synoviocytes. Arthritis Rheum. 2002;46:2867–2877. doi: 10.1002/art.10593. [DOI] [PubMed] [Google Scholar]
- 66.Nakamura Y, Nawata M, Wakitani S. Expression profiles and functional analyses of Wnt-related genes in human joint disorders. Am J Pathol. 2005;167:97–105. doi: 10.1016/S0002-9440(10)62957-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang W, Guo P, Chen M, Chen D, Cheng Y, He L. FOXM1/LINC00152 feedback loop regulates proliferation and apoptosis in rheumatoid arthritis fibroblast-like synoviocytes via Wnt/β-catenin signaling pathway. Biosci Rep. 2020;40:BSR20191900. doi: 10.1042/BSR20191900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoshioka R, Kita Y, Nagahira A, Manno A, Makita N, Tomita U, Murakawa M. Quantitative analysis of cadherin-11 and β-catenin signalling during proliferation of rheumatoid arthritis-derived synovial fibroblast cells. J Pharm Pharmacol. 2015;67:1075–1082. doi: 10.1111/jphp.12410. [DOI] [PubMed] [Google Scholar]
- 69.Ramanan M, Pilli VS, Aradhyam GK, Doble M. Transcriptional regulation of microsomal prostaglandin E synthase 1 by the proto-oncogene, c-myc, in the pathogenesis of inflammation and cancer. Biochem Biophys Res Commun. 2017;482:556–562. doi: 10.1016/j.bbrc.2016.11.073. [DOI] [PubMed] [Google Scholar]
- 70.Liu Q, Loo WT, Sze SC, Tong Y. Curcumin inhibits cell proliferation of MDA-MB-231 and BT-483 breast cancer cells mediated by down-regulation of NFkappaB, cyclinD and MMP-1 transcription. Phytomedicine. 2009;16:916–922. doi: 10.1016/j.phymed.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 71.Wang H, Jia XZ, Sui CJ, Zhao YP, Mei YF, Zheng YN, Zhang ZY. Effects of thapsigargin on the proliferation and survival of human rheumatoid arthritis synovial cells. ScientificWorldJournal. 2014;2014:605416. doi: 10.1155/2014/605416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang XY, Zhang XM, Chen FH, Zhou LL, Deng XF, Liu YJ, Li XJ. Anti-proliferative effect of recombinant human endostatin on synovial fibroblasts in rats with adjuvant arthritis. Eur J Pharmacol. 2014;723:7–14. doi: 10.1016/j.ejphar.2013.10.068. [DOI] [PubMed] [Google Scholar]
- 73.Zhu B, Zhang S, Meng N, Zhang H, Yuan S, Zhang J. Long non-coding RNA RNCR3 promotes glioma progression involving the Akt/GSK-3β pathway. Oncol Lett. 2019;18:6315–6322. doi: 10.3892/ol.2019.11002. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 74.Li S, Oh YT, Yue P, Khuri FR, Sun SY. Inhibition of mTOR complex 2 induces GSK3/FBXW7-dependent degradation of sterol regulatory element-binding protein 1 (SREBP1) and suppresses lipogenesis in cancer cells. Oncogene. 2016;35:642–650. doi: 10.1038/onc.2015.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pronobis MI, Rusan NM, Peifer M. A novel GSK3-regulated APC: Axin interaction regulates Wnt signaling by driving a catalytic cycle of efficient βcatenin destruction. Elife. 2015;4:e08022. doi: 10.7554/eLife.08022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guan H, Zhang Y, Gao S, Bai L, Zhao S, Cheng XW, Fan J, Liu E. Differential patterns of secreted frizzled-related protein 4 (SFRP4) in adipocyte differentiation: Adipose depot specificity. Cell Physiol Biochem. 2018;46:2149–2164. doi: 10.1159/000489545. [DOI] [PubMed] [Google Scholar]
- 77.Ghoshal A, Ghosh SS. Antagonizing canonical Wnt signaling pathway by recombinant human sFRP4 purified from E. coli and its implications in cancer therapy. Mol Cell Biochem. 2016;418:119–135. doi: 10.1007/s11010-016-2738-6. [DOI] [PubMed] [Google Scholar]
- 78.Haraguchi R, Kitazawa R, Mori K, Tachibana R, Kiyonari H, Imai Y, Abe T, Kitazawa S. sFRP4-dependent Wnt signal modulation is critical for bone remodeling during postnatal development and age-related bone loss. Sci Rep. 2016;6:25198. doi: 10.1038/srep25198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.