Abstract
Heart failure (HF) is a serious threat to human health. Long noncoding RNAs (lncRNAs) are critical regulators of HF. The aim of the study was to investigate the molecular mechanism of MALAT1 in HF rats. MALAT1 expression was detected in serum of normal volunteers and HF patients, HF rats and isoproterenol (ISO)-induced H9C2 cells, and its diagnostic value was evaluated in HF patients. Indexes related to cardiac functions and hemodynamics, myocardial injury, lipid metabolism, lipid oxidation, and inflammation were detected. Moreover, the downstream mechanism of MALAT1 was predicted and verified and in vivo experiments were further performed in ISO-induced H9C2 cells to verify the effects of MALAT1 in HF. MALAT1 was highly expressed in serum of HF patients, HF rats and ISO-induced H9C2 cells and was valuable in predicting HF. Inhibition of MALAT1 increased cardiac function and anti-inflammation and alleviated myocardial injury, lipid metabolism, lipid oxidation and apoptosis rates. Inhibition of MALAT1 reduced H9C2 cell injury. MALAT1 competitively bound to microRNA (miR)-532-3p to upregulate LDLR protein. Inhibition of miR-532-3p weakened the protective effect of downregulated MALAT1 against H9C2 cell injury. We concluded that MALAT1 upregulated LDLR expression by competitively binding to miR-532-3p, thereby increasing pathological injury in HF.
Keywords: heart failure, long non-coding RNA MALAT1, microRNA-532-3p, lipid metabolism, lipid oxidation, inflammatory responses
Introduction
Heart failure (HF), as a heterogeneous clinical syndrome, mainly develops as a consequence of cardiac overload and injury (1). HF results in low quality of life given its high mortality and incidence rate (2). Elderly individuals over 65 years of age and women are more likely to develop HF (3,4). In addition, HF patients may experience breathing problems, fatigue, poor endurance during exercise and fluid retention (5). Chronic inflammatory processes and lipid metabolism disorders are implicated in HF (6). Risk factors, including diseases (ischemic heart disease, hypertension and diabetes) and unhealthy living habits (tobacco use and high-fat diets), function as predictors for HF incidence and severity (7). Although progress has been made in the development of new drugs for the treatment of HF, the efficacy is not satisfactory (8). Therefore, given the significant health risks caused by HF, it is crucial to identify new and more effective methods to treat HF. Recently, some long noncoding RNAs (lncRNAs) have been demonstrated to be involved in the HF-related lncRNA-mRNA network, demonstrating potent diagnostic value for HF (9). Circulating lncRNAs are also useful biomarkers for myocardial infarction and other adult heart diseases (10). Therefore, the aim of the present study was to identify a novel lncRNA-based therapy for HF from the aspects of lipid metabolism and inflammatory responses.
LncRNAs, which are RNA molecules greater than 200 nucleotides in length (11), regulate cholesterol [(mostly oxidized low-density lipoprotein (oxLDL)] accumulation and inflammation in macrophages in the progression of atherosclerosis (AS) (12) which is closely related to HF (13). LncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is highly conserved and widely expressed in mammalian tissue cells (14). MALAT1 is involved in the pathogenesis of AS (15) and regulates oxLDL-induced endothelial inflammation during AS (16). However, studies on the regulatory mechanism of MALAT1 in HF are limited. In addition, it has been shown that lncRNAs interacting with microRNAs (miRNAs) to regulate mRNA are strongly linked to the development of cardiovascular diseases, including HF (17). miRNAs, which are recognized as small noncoding RNAs, modulate gene expression, and potentially serve as biomarkers for HF (18). miR-532-3p is involved in atherothrombotic events that may lead to HF (19,20). miR-532-3p is of great value in the regulation of diverse metabolic disorders, including inflammation and obesity (21). Nevertheless, the direct role of miR-532-3p in HF has not been reported.
Therefore, it was reasonable to hypothesize that MALAT1 played an underlying role in regulating lipid metabolism and inflammatory responses in HF through the miRNA-mRNA system. Consequently, we performed a series of histological and molecular experiments to identify the lncRNA-miRNA-mRNA network of MALAT1 and study the underlying molecular machinery with the aim of providing some novel therapies against HF.
Materials and methods
Ethics approval
This study was approved and supervised by the human ethics committee of Yantai Yuhuangding Hospital. All the subjects signed the informed consent. All experiments were approved by the animal ethics committee of Yantai Yuhuangding Hospital.
Clinical samples
From May 2018 to June 2019, 57 HF patients (39 males and 18 females, aged 48-73 years) diagnosed via coronary angiography were enrolled in Yantai Yuhuangding Hospital. In addition, 48 healthy controls (28 males and 20 females, aged 50-73 years) were included during the same period. Venous blood was extracted from the subjects under fasting conditions and stored at -80°C, and relevant physiological and biochemical indexes were detected in the patients and controls. Inclusion criteria for patients with HF were as follows: i) At least one major coronary artery exhibited >60% stenosis; ii) informed consent was signed. Exclusion criteria were: i) Unstable angina pectoris or myocardial infarction; ii) complicated with another organic heart disease; iii) presence of severe hypertension, diabetes, shock, arrhythmia, liver and kidney dysfunction, familial hypercholesterolemia, and/or malignant tumor or inflammatory diseases.
Animal treatment and grouping
A total of 40 healthy Sprague-Dawley (SD) male rats [220-260 g, SYXK (Zhejiang) 2019-0021] purchased from Hangzhou Qingda Kerui Biotechnology Co., Ltd. (Hangzhou, Zhejiang, China) were housed under standard conditions for one week before the experiments. All experimental animals were kept in a specific pathogen-free animal room (22-24°C, light/dark=12/12 h, humidity 50%), and could freely obtain standard food and water.
After rats were anesthetized using intraperitoneal injection of 60 mg/kg pentobarbital sodium, the left anterior descending coronary artery was ligated to construct a rat model of HF (22). Rats in the sham group were only threaded without ligation. The rat model was identified using an electrocardiogram (ECG) 3 days after the operation. HF model rats were assigned to 3 groups: HF group (injection of the same amount of normal saline via the tail vein), HF + small hairpin-negative control (sh-NC) group (injection of 50 nM sh-NC via the tail vein), and HF + sh-MALAT1 group (injection of 50 nM sh-MALAT1 via the tail vein). The normal saline, sh-NC and sh-MALAT1 groups were injected equally twice a week for 4 weeks. The above adenovirus vectors were purchased from Guangzhou RiboBio Co., Ltd.
Cardiac function and hemodynamics
Cardiac function and hemodynamics were determined as described previously (22). Briefly, the rats (6 rats in each group) were anesthetized by inhalation of 1.5-2% isoflurane prior to scanning using the isoflurane anesthesia system (JD Medical Dist Co Inc.). Heart rate (HR) was measured by inserting a right ventricular catheter into the right ventricle via the right jugular vein. Left ventricular ejection fraction (LVEF) and fraction shortening (FS) were detected dynamically using a cardiac color Doppler ultrasonic cardiogram (ACSON512, Philips). Then, the BL-420S biological function experimental system (Taimeng Technology Co., Ltd.) was used to monitor the cardiac function of rats. Left ventricular systolic pressure (LVSP), left ventricular end-diastolic pressure (LVEDP), and left ventricular maximum rates of rise and fall (±dp/dtmax) of rats in each group were recorded.
Sample collection
After the detection of cardiac function and hemodynamics, rats were euthanized by intraperitoneal injection of excessive pentobarbital sodium (800 mg/kg) (23), and serum was collected and stored. Some rats in each group were used to generate homogenate, and the myocardial tissues of the remaining rats were fixed in 4% paraformaldehyde buffer. Paraffin-embedded tissues were routinely sectioned at 4 µM for tissue staining.
Hematoxylin and eosin (H&E) staining
After euthanasia, the rats were perfused with phosphate-buffered saline (PBS) from the left ventricle. Myocardial tissues were isolated, fixed with 4% paraformaldehyde at 25°C for 4 h and embedded in paraffin. Paraffin-embedded sections were deparaffinized and dehydrated. Then, sections at 4 µm underwent treatment with xylene I, xylene II and gradient alcohol successively for 5 min each. Following 2-min washing with water, the sections were stained with H&E for 8 min at 25°C, washed with water, differentiated using 1% hydrochloric acid alcohol for 5 sec, and then washed 3 times with water. The samples were washed with 0.25% ammonia water for 1 min followed by 3 washes with water for 2 min each. Next, the sections were stained with 1% eosin for 30 sec, washed with water three times, and immersed in gradient alcohol, xylene I and xylene II for 3 min separately. After the xylene on the slide was completely dried, the slide was sealed with neutral gum, covered by a clean cover slide, and observed under an optical microscope (BX 51, Olympus Optical Co., Ltd.).
Terminal deoxynucleotidyl transferase (TdT)-mediated dUTP nick end labeling (TUNEL) staining
After paraffin-embedded sections were deparaffinized, surgery was performed according to the instructions of TUNEL apoptosis detection kit (Beyotime Biotechnology Co., Ltd.). TUNEL-positive cells were observed under the optical microscope after development using 3,3′-diaminobenzidine (DAB).
After dewaxing and hydration, the paraffin sections were added with proteinase K working solution at 37°C for 20 min. After PBS washing, the sections were immersed in sealing solution (3% H2O2 dissolved in methanol) at room temperature (25°C) for 10 min. Then the sections were incubated with TUNEL reaction solution at 37°C for 60 min, and then 2,4-diaminobutyric acid (DAB) color-developing solution was added. After color development, 4′,6-diamidino-2-phenylindole (DAPI) (100 ng/ml, at 25°C) was added for nuclear staining for 10 min, and then observed under fluorescence microscope (Olympus, BX51). At least three sections were observed in each tissue. Three visual fields (×400) were randomly selected for statistical analysis.
Myocardial cell culture and grouping
H9C2 myocardial cells of rats (Cell Bank of Chinese Academy of Sciences) were cultured in Dulbecco's modified Eagle's medium (DMEM) (Beijing Solarbio) containing 10% fetal bovine serum (FBS) at 37°C with 5% CO2. When cells grew to 80% confluency, the cells were allocated into the control group (H9C2 myocardial cells were cultured normally) and isoproterenol (ISO) group (H9C2 myocardial cells were treated with 80 µM ISO for 48 h) (24). In addition, cells were transfected with si-NC, si-MALAT1, inhibitor NC, miR-532-3p inhibitor, miR-532-3p inhibitor + si-NC and miR-532-3p inhibitor + si-MALAT1 (all from Guangzhou RiboBio) using Lipofectamine 2000 (Thermo Fisher Scientific) as per the manufacturer's instructions. After transfection for 24 h, the cells were treated with 80 µM ISO for 48 h and collected for subsequent experiments.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
TRIzol reagent was used to extract total RNA from myocardial tissues and cells of rats in each group, and RNA was reverse transcribed into cDNA using the PrimeScript RT reagent kit (RR047A, Takara Bio Inc.) or NcodeTM miRNA First-Strand cDNA Synthesis (Thermo Scientific Fisher). The synthesized cDNA was detected by RT-qPCR using the Fast SYBR-Green PCR kit (Applied Biosystems, Inc.) and the ABI PRISM 7300 RT-PCR system (Applied Biosystems). The reaction system (20 µl) included 2X SYBR Green (10 µl), upstream and downstream primers (20 µM; 0.3 µl for each), cDNA (1.0 µl) and RNase-free dH2O (8.4 µl). The reaction procedure was 95°C for 5 min followed by 40 cycles of 95°C for 30 sec and 60°C for 1 min. Each sample was assessed in triplicate. U6 or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as internal references. The relative expression of related genes was analyzed using the 2−ΔΔCq method as follows: ΔΔCq=(Cq target gene in the experimental group-Cq internal reference in the experimental group)-(Cq target gene in the control group-Cq internal reference in the control group) (25). Primers are presented in Table I.
Table I.
RT-qPCR primer sequences.
| Gene | Primer sequence |
|---|---|
| hsa-MALAT1 | F: 5′-AAAGCAAGGTCTCCCCACAAG-3′ |
| R: 5′-GGTCTGTGCTAGATCAAAAGGCA-3′ | |
| rno-MALAT1 | F: 5′-AGCGGAAGAACGAATGTAAC-3′ |
| R: 5′-GAACAGAAGGAAGAGCCAAG-3′ | |
| rno-miR-532-3p | F: 5′-ACGGCTTTTCTCTTCCATGCCT-3′ |
| R: 5′-CAGTGCAGGGTCCGAGGTAT-3′ | |
| rno-U6 | F: 5′-CTCGCTTCGGCAGCACA-3′ |
| R: 5′-AACGCTTCACGAATTTGCGT-3′ | |
| rno-LDLR | F: 5′-GTGGCAGTAGTGAGTGTATC-3′ |
| R: 5′-ATCCCAAAGACGAGAAGT-3′ | |
| hsa-GAPDH | F: 5′-GCAACTAGGATGGTGTGGCT-3′ |
| R: 5′-TCCCATTCCCCAGCTCTCATA-3′ | |
| rno-GAPDH | F: 5′-CTCCTCGAAGTACCCTGTGC-3′ |
| R: 5′-CATGGTGCAGCGATGCTTTA-3′ |
Western blot analysis
Total protein in myocardial tissues and cells of rats in each group was extracted, and the protein concentration was measured using a bicinchoninic acid (BCA) protein quantification kit (Boster Biological Technology Co., Ltd.). Protein (30 µg) was separated by 10% SDS-PAGE, transferred to polyvinylidene fluoride membranes and blocked with 5% bovine serum albumin at room temperature for 2 h to block non-specific binding. Membranes were then incubated with a primary antibody against low-density lipoprotein receptor (LDLR) (1/200, no. ab30532) at 4°C overnight and a secondary immunoglobulin G (IgG) antibody (1 µg/ml, no. ab8226) (Abcam Inc.) at room temperature for 1 h. Then, the membranes were washed with 1X PBS containing 0.5% Triton X-100 (PBST), developed and visualized using enhanced chemiluminescence solution (EMD Millipore Corporation). Each band in the western blot images was analyzed for gray value quantification using ImageJ (National Institute of Health). β-actin (1/2000, ab205718) was used as an internal reference.
Nuclear and cytoplasmic fractionation assay
According to the instructions of NE-PER™ nuclear and cytoplasmic fractionation kit (Thermo Scientific Fisher), MALAT1 expression in the isolated cytoplasm and nuclear extract was detected using RT-qPCR.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay
After transfection, when the cell confluency reached 80%, H9C2 cells were washed twice with PBS solution and detached with 0.25% trypsin to prepare a single cell suspension. Then, 3×103-6×103 cells/well were seeded into 96-well plates (0.2 ml/well; repeated for 6 wells) and incubated in incubators at 37°C containing 5% CO2. Next, the plates were removed at 24, 48 and 72 h, and the medium was replaced with medium containing 10% MTT solution (5 g/l) (GD-Y1317, Guduo Biotechnology Company) for further culture for 4 h at 37°C. Then, the supernatant was absorbed, and 100 µl dimethyl sulfoxide (D5879-100ML, Sigma-Aldrich) was added to each well followed by 10 min of gentle agitation and mixing to fully dissolve the formazan crystal produced by living cells. The optical density (OD) at 490 nm of each well was detected using a microplate reader (Nanjing Detie Experimental Equipment Co., Ltd.), and the cell survival rate was calculated. The cell survival rate (%) was calculated as: OD value in the experimental group/OD value in the control group ×100%. The cell survival rate of the control group was set as 100%.
Flow cytometry
Briefly, cells were collected by centrifugation at 4,500 × g for 5 min at 4°C, and the culture medium was discarded. Next, the cells were washed twice with cold PBS and suspended with 400 µl 1X binding buffer. Then, 5 µl Annexin V-fluorescein isothiocyanate (FITC) was added to the cell suspension, and the suspension was gently mixed and incubated at 4°C for 15 min in the dark. Next, 10 µl propidium iodide (PI) was added to the cells, and the suspension was gently mixed for 5 min at 4°C. Cells were assessed within 1 h using a flow cytometer (FACS Calibur, BD Biosciences). Each experiment was repeated three times.
Fluorescence in situ hybridization assay
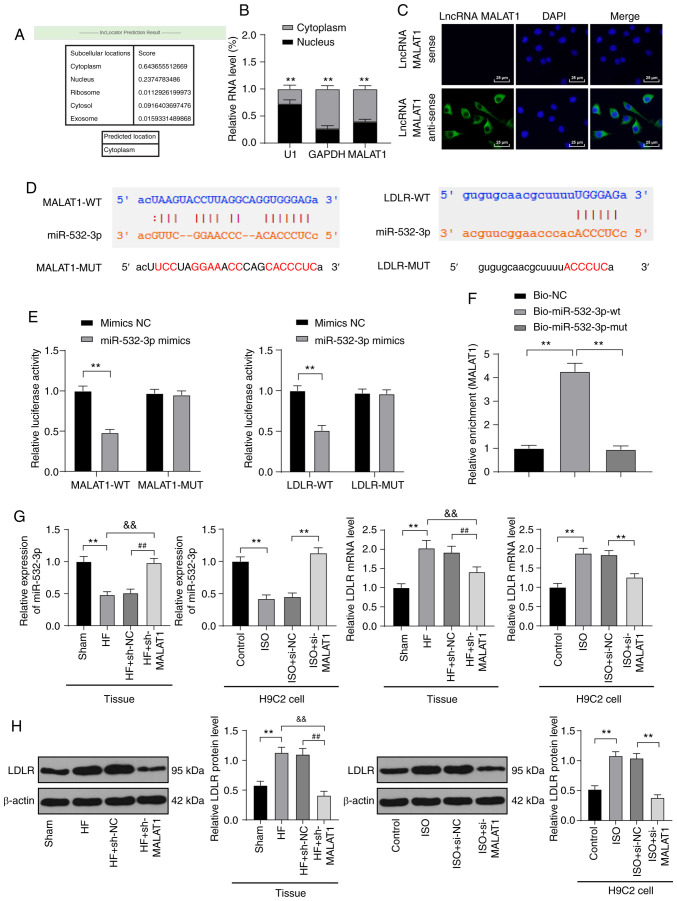
The localization of MALAT1 in cells was predicted by online analysis website (http://www.csbio.sjtu.edu.cn/cgi-bin/lncLocator.py). According to the instructions of the FISH kit (Guangzhou Ribobio), cells were fixed with 4% paraformaldehyde. PBST was added, and the samples were blocked with prehybridization solution at 37°C. Cells were hybridized with lncRNA FISH probe (DNA) overnight at 37°C, washed with the hybridizing solution at 42°C in the dark and stained using 4′,6-diamidino-2-phenylindole (DAPI) for 10 min. Next, the cells were fixed on the glass slide with a sealing agent in the dark and observed under a fluorescence microscope (BX53, Olympus).
Dual-luciferase reporter gene assay
Starbase (http://starbase.sysu.edu.cn/index.php) predicted the binding relationship between MALAT1 and miR-532-3p as well as miR-532-3p and LDLR. Briefly, 293T cells were seeded into the 6-well plates (2×105 cells/well). After the cells adhered to the wall, MALAT1-wild type (WT), MALAT1-mutant (MUT), LDLR-WT, and LDLR-MUT were co-transfected with NC mimic and miR-552-3p mimic, separately, into 293T cells using Lipofectamine 2000 (Invitrogen) as per the manufacturer's instructions. After successful transfection, the cells were cultured for 48 h and collected. Changes in luciferase activity in the cells were detected according to the protocol of the dual-luciferase detection kit (D0010, Beijing Solarbio Science & Technology Co., Ltd.). The fluorescence intensity was detected using a GloMax 20/20 luminometer (Promega). Each experiment was repeated three times.
RNA pull-down
RNA pull-down was carried out as previously described (26,27). H9C2 myocardial cells were transfected with 50 nM of biotinylated Bio-miR-542-3p-WT, Bio-miR-542-3p-MUT and the corresponding Bio-NC. After 48 h, the cells were collected, washed with PBS, and vortexed. Next, the cells were incubated with the specific lysis buffer (Ambion, Inc.) for 10 min and then underwent a 3-h incubation with M-280 streptavidin magnetic beads precoated with RNase-free and yeast tRNA (Sigma-Aldrich) at 4°C followed by 2 washes with precooled lysis buffer, 3 washes with low-salt buffer and one wash with high-salt buffer. Finally, RNA was purified using TRIzol, and MALAT1 expression was detected using RT-qPCR.
Detection of biochemical indexes
The levels of creatine kinase isoenzyme (CK-MB), cardiac troponin I (cTn-I), reactive oxygen species (ROS), superoxide dismutase (SOD), malondialdehyde (MDA), lipid metabolism-related indexes [endothelin (ET), total cholesterol (TC), triglyceride (TG), LDL cholesterol (LDL-C), and high-density lipoprotein cholesterol (HDL-C)], and inflammatory-related cytokines [tumor necrosis factor alpha (TNF-α), interleukin-1 beta (IL-1β), IL-6, transforming growth factor beta (TGF-β), IL-4 and IL-10] in serum of clinical patients and normal volunteers and in serum and cells of rats in each group were determined, respectively, according to the instructions of the ELISA kits (Nanjing Jiancheng Bioengineering Institute). In addition, ROS in cells was detected using the DCFDA/H2DCFDA-Cellular ROS assay kit (cat. no. ab113851, Abcam). The samples were incubated with DCFDA/H2DCFDA for 20 min. After PBS washing, DAPI (100 ng/ml) was added for nuclear staining at 25°C for 10 min and the samples were observed under fluorescence microscope.
Statistical analysis
All data were analyzed by SPSS 21.0 statistical software (IBM Corp.). The data exhibited a normal distribution as demonstrated by the Kolmogorov-Smirnov test. The results were expressed by mean ± standard deviation. One-way or two-way analysis of variance (ANOVA) was used for the comparison among multiple groups followed by Tukey's multiple comparison test. In addition, receiver operating characteristic (ROC) curves were used to evaluate the diagnostic value of MALAT1. The Chi-square test was used for gender data analysis, and t-test was used for other clinical data analysis. The P-value was calculated using a bilateral test, and P<0.05 indicates that the difference was statistically significant.
Results
LncRNA MALAT1 exhibited important value in predicting HF patients
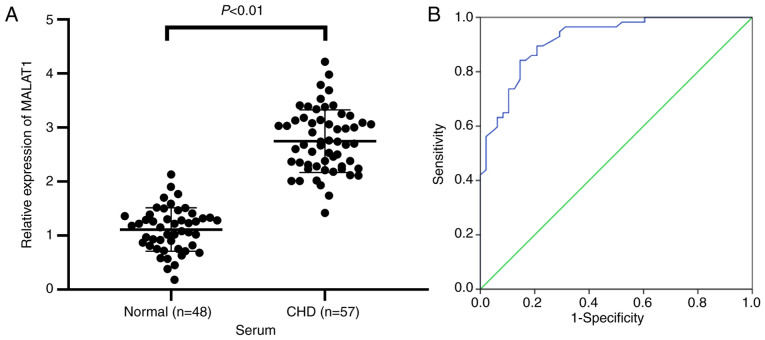
To investigate the diagnostic value of serum lncRNA MALAT1 levels in HF, we enrolled 48 healthy volunteers and 57 HF patients and compared their general characteristics (Table II). No significant differences in sex, age, body mass index (BMI), smoking, alcohol and hypertension, as well as indexes related to lipid metabolism (TC, TG, and LDL-C) were identified between the two groups. However, HDL-C in HF patient serum was reduced compared with that in normal volunteers, and myocardial injury-related indexes (cTn-I and CK-MB) in HF patient serum were increased compared with those in normal volunteers (all P<0.01). To examine the relationship between MALAT1 and HF, MALAT1 expression in serum of normal volunteers and HF patients was measured. The results showed that MALAT1 expression in serum of HF patients was markedly upregulated compared with that in the normal controls (P<0.01, Fig. 1A). The diagnostic value of MALAT1 levels in HF patients was analyzed using ROC curves. The area under the ROC curve was 0.918, the 95% confidence interval was 0.868-0.969. The sensitivity and specificity were 84.2 and 85.4%, respectively (Fig. 1B). The results showed that lncRNA MALAT1 exhibited great value in the prediction of HF patients.
Table II.
Basic clinical characteristics of the patient.
| Characteristic | Normal (n=48) | HF (n=57) | P-value |
|---|---|---|---|
| Sex | 0.46 | ||
| Male | 28 (58.33%) | 39 (68.42%) | |
| Female | 20 (41.67%) | 18 (31.58%) | |
| Age | 60.21±13.32 | 61.95±12.16 | 0.49 |
| BMI | 24.21±3.14 | 25.33±3.26 | 0.08 |
| Smoking | 29/19 | 39/18 | 0.22 |
| Alcohol | 25/23 | 33/24 | 0.36 |
| Hypertension | 27/21 | 38/19 | 0.31 |
| TC (mmol/l) | 1.68±0.56 | 1.76±0.61 | 0.49 |
| TG (mmol/l) | 4.21±1.05 | 4.38±1.12 | 0.43 |
| LDL-C (mmol/l) | 3.21±0.86 | 3.56±0.92 | 0.16 |
| HDL-C (mmol/l) | 1.15±0.21 | 0.81±0.16 | <0.01 |
| cTn-I (µg/l) | 0.17±0.06 | 1.55±0.26 | <0.01 |
| CK-MB (U/l) | 15.37±6.18 | 48.51±6.62 | <0.01 |
HF, heart failure; BMI, body mass index; TC, total cholesterol; TG, triglyceride; LDL-C, low-density lipoprotein cholesterol; HDL-C, high-density lipoprotein cholesterol; cTn-I, cardiac troponin I; CK-MB, creatine kinase isoenzyme. Chi-square test was used for gender data analysis, and t-test was used for other data analysis.
Figure 1.
LncRNA MALAT1 expression could predict HF diagnosis. (A) The expression of MALAT1 in the serum of normal volunteers (n=48) and HF patients (n=57) was detected by RT-qPCR. (B) ROC curve analysis of the diagnostic value of MALAT1 levels in HF patients. Independent t-test and ROC analysis were used for data analysis. P<0.01.
Inhibition of lncRNA MALAT1 reduced myocardial injury in HF rats
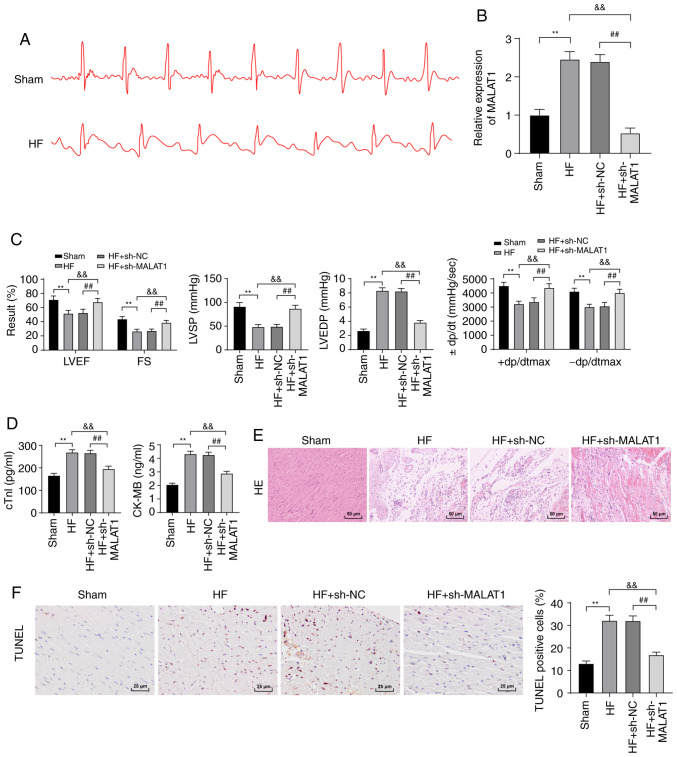
To examine the molecular mechanism of MALAT1 in HF, we first constructed an HF rat model. ECG detection revealed that compared with the sham-operated rats, HF rats exhibited an increased ST segment in ECG (Fig. 2A), and MALAT1 expression was clearly upregulated in the tissues of HF rats (P<0.01, Fig. 2B). To further study the effect of MALAT1 on HF, HF rats were injected with sh-NC or sh-MALAT1 via the tail vein, and the relevant indexes were detected. Compared with sh-NC-treated HF rats, sh-MALAT1-treated HF rats exhibited significantly decreased expression of MALAT1 (P<0.01, Fig. 2B). After the establishment of the HF model, HR results revealed no significant difference in HR between the experimental group and the control group (Fig. S1). LVEF, FS, LVSP and ±dp/dtmax levels were reduced, and LVEDP levels were obviously increased in HF rats (Fig. 2C). However, MALAT1 inhibition yielded opposite results (all P<0.01, Fig. 2C). ELISA showed that the serum levels of cTn-I and CK-MB in HF rats were significantly increased compared with sham-operated rats, and these levels were reversed after inhibition of MALAT1 (P<0.01, Fig. 2D). H&E staining showed that the myocardial cells of sham-operated rats exhibited a normal shape and an ordered arrangement, whereas those of HF rats were swollen, disordered and extremely damaged. After MALAT1 inhibition, the degree of myocardial injury in the rats was reduced (Fig. 2E). TUNEL staining showed that the number of TUNEL-positive cells in HF rats was increased compared with sham-operated rats; the above results were reversed after MALAT1 knockdown (P<0.01, Fig. 2F). The above results showed that HF rats exhibited severe myocardial injury, which was relieved by MALAT1 inhibition.
Figure 2.
Inhibition of lncRNA MALAT1 reduced myocardial injury in HF rats. (A) ECG detection of rats, n=10. (B) Expression of MALAT1 in rat tissues was detected by RT-qPCR, n=4. (C) Detection of cardiac function and left ventricular hemodynamics in rats, n=10. (D) ELISA detection of myocardial injury-related indexes in rats, n=4. (E) Observation of the pathological changes of rat tissues using H&E staining. Magnification, ×200, n=6. (F) Detection of myocardial cell apoptosis in rats using TUNEL staining, Magnification, ×200, n=6; scale, 25 µm. Three independent repeated tests were conducted. The data were expressed as the mean ± standard deviation and analyzed using one-way or two-way ANOVA followed by Tukey's multiple comparison test. **P<0.01, compared with the sham group; &&P<0.01, compared with the HF group; ##P<0.01, compared with the HF + sh-NC group.
Inhibition of MALAT1 improved lipid metabolism disorders and relieved inflammation in HF rats
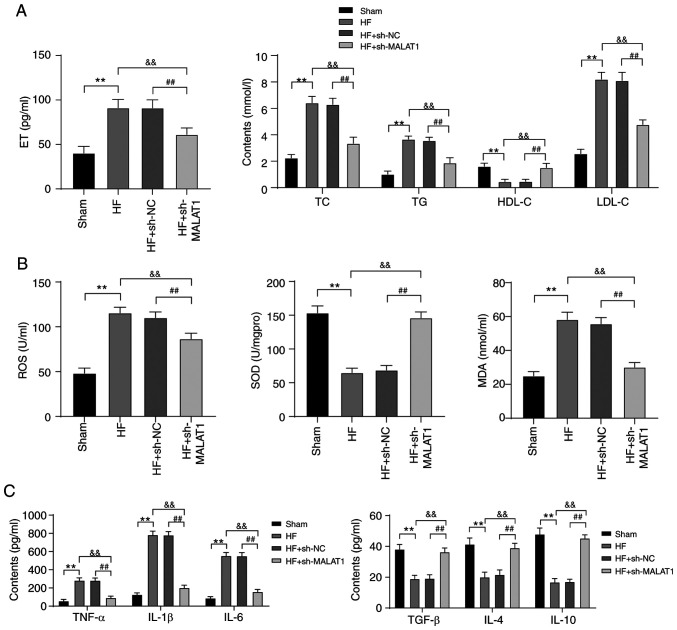
Lipid metabolism disorders and inflammatory responses are important pathogenic conditions of HF (28,29). Therefore, lipid metabolism and inflammatory responses in HF rats were detected. Compared with rats in the sham group, HF rats exhibited lipid metabolism disorders. Specifically, ET, TC, TG and LDL-C levels were clearly increased, whereas the level of HDL-C was significantly reduced. However, inhibition of MALAT1 reversed the above results (all P<0.01) (Fig. 3A). In addition, in HF rats, ROS and MDA levels were increased, while SOD levels were decreased. In addition, the levels of anti-inflammatory cytokines (TGF-β, IL-4 and IL-10) were reduced, whereas the levels of pro-inflammatory cytokines (TNF-α, IL-1β and IL-6) were increased. MALAT1 inhibition yielded the completely opposite results (all P<0.01, Fig. 3B and C). The above results showed that HF rats displayed lipid metabolism disorders and increased levels of lipid oxidation and inflammation responses, which were alleviated after MALAT1 downregulation.
Figure 3.
Inhibition of MALAT1 improved lipid metabolism disorders and relieved inflammation in HF rats. ELISA was used to detect (A) lipid metabolism, (B) lipid oxidation and (C) inflammatory responses in rats, n=10. Three independent repeated tests were conducted. The data were expressed as the mean ± standard deviation and analyzed using one-way or two-way ANOVA followed by Tukey's multiple comparison test. **P<0.01, compared with the sham group; &&P<0.01, compared with the HF group; ##P<0.01, compared with the HF + sh-NC group.
Inhibition of MALAT1 reduced ISO-induced H9C2 myocardial cell injury
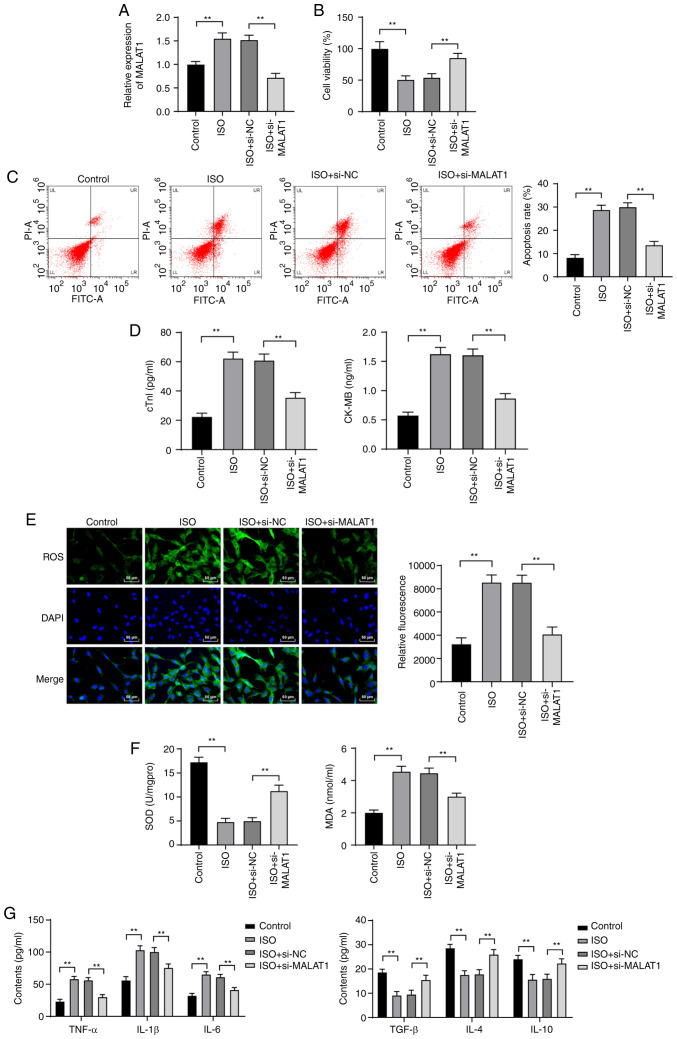
To further investigate the effect of MALAT1 on HF, si-NC or si-MALAT1 was transfected into ISO-induced H9C2 cells. MALAT1 expression was distinctly increased in H9C2 cells after ISO treatment. Compared with that in H9C2 cells treated with si-NC, MALAT1 expression was significantly decreased in H9C2 cells after MALAT1 inhibition (P<0.01, Fig. 4A). The viability of ISO-induced H9C2 cells was obviously decreased. The apoptosis rate cTn-I and CK-MB levels were significantly increased. However, silencing MALAT1 expression reversed the above results (all P<0.01, Fig. 4B-D). ISO treatment increased ROS, MDA, TNF-α, IL-1β and IL-6 levels and reduced SOD, TGF-β, IL-4 and IL-10 levels; however, MALAT1 knockdown exhibited the opposite trend (all P<0.01, Fig. 4E-G).
Figure 4.
Inhibition of MALAT1 reduced ISO-induced H9C2 myocardial cell injury. (A) MALAT1 expression in H9C2 cells was detected by RT-qPCR. (B) H9C2 cell viability was detected by MTT assays. (C) The apoptosis rate of H9C2 cells was detected using flow cytometry. (D) ELISA kits were used to detect the levels of cTn-I and CK-MB in H9C2 cells. (E-G) ELISA kits were used to detect the levels of lipid oxidation-related factors (ROS, MDA and SOD) and inflammatory factors (TNF-α, IL-1β, IL-6, TGF-β, IL-4 and IL-10) in H9C2 cells; scale in panel E, 50 µm. Three independent repeated tests were conducted. The data were expressed as the mean ± standard deviation and analyzed using one-way or two-way ANOVA followed by Tukey's multiple comparison test. **P<0.01, compared with the control group or the ISO + si-NC group.
LncRNA MALAT1 competitively bound to miR-532-3p to upregulate LDLR
In the above in vitro and in vivo experimental results, we verified that inhibition of MALAT1 reduced injury in HF rats and ISO-induced H9C2 cells, but the downstream action mechanism of MALAT1 was still unclear. The mechanism of action of lncRNA depended on its subcellular location. The online website (http://www.csbio.sjtu.edu.cn/cgi-bin/lncLocator.py) (30) predicted that MALAT1 is located in the cytoplasm of myocardial cells (Fig. 5A). Nuclear and cytoplasmic fractionation assays (P<0.01, Fig. 5B) and FISH assays (Fig. 5C) verified that MALAT1 is mainly located in the cytoplasm of H9C2 cells, indicating that MALAT1 plays a regulatory role in myocardial cells through the mechanism of ceRNA. Subsequently, Starbase (http://starbase.sysu.edu.cn/index.php) predicted that MALAT1 and miR-532-3p as well as miR-532-3p and LDLR had targeted binding sites (Fig. 5D), which were subsequently confirmed by dual-luciferase reporter gene assays and RNA pull-down assays (P<0.01, Fig. 5E-F). In myocardial tissues of HF rats, miR-532-3p expression was obviously decreased, and LDLR mRNA and protein levels were obviously increased. After MALAT1 inhibition, these results were reversed. The detection results were consistent with that in H9C2 cells (all P<0.01, Fig. 5G-H).
Figure 5.
LncRNA MALAT1 competitively bound to miR-532-3p to upregulate LDLR. (A) An online website predicted the sublocation of lncRNA MALAT1. (B) Localization expression of lncRNA MALAT1 was detected using nuclear and cytoplasmic fractionation assays. (C) The localization of lncRNA MALAT1 was observed using FISH assay; scale, 25 µm; (D). An online software predicted the binding sites between MALAT1 and miR-532-3p as well as miR-532-3p and LDLR. (E) In 293T cells, binding interaction between MALAT1 and miR-532-3p as well as miR-532-3p and LDLR was verified using dual-luciferase reporter gene assays. (F) Binding relationship between MALAT1 and miR-532-3p was verified using RNA pull-down assays. (G) miR-532-3p expression and LDLR mRNA levels in rat tissues and H9C2 cells were detected using RT-qPCR, n=4. (H) LDLR protein levels in rat tissues and H9C2 cells were detected using western blot analysis, n=4. Three independent repeated tests were conducted. The data were expressed as the mean ± standard deviation and analyzed using one-way or two-way ANOVA followed by Tukey's multiple comparison test. **P<0.01, compared with the sham group or the control group or the ISO + si-NC group; &&P<0.01, compared with the HF group; ##P<0.01, compared with the HF + sh-NC group.
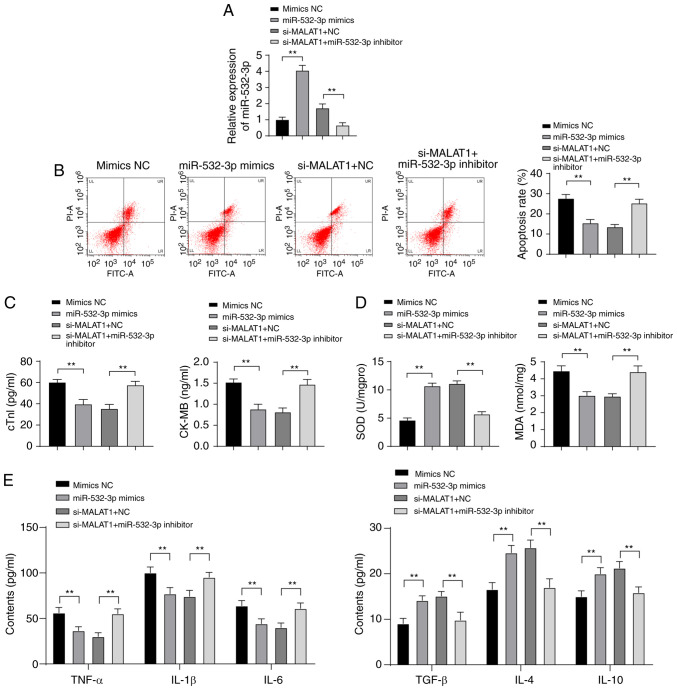
Inhibition of miR-532-3p weakened the protective effect of downregulated MALAT1 against H9C2 myocardial cell injury
To further investigate the effect of miR-532-3p on H9C2 myocardial cell injury and the mechanism by which MALAT1 regulates myocardial injury through miR-532-3p, mimic NC or miR-532-3p mimic was transfected into ISO-treated H9C2 myocardial cells, and a functional rescue experiment was established via the combined treatment of si-MALAT1 + NC and si-MALAT1 + miR-532-3p inhibitor. After overexpression of miR-532-3p, miR-532-3p expression was significantly increased in H9C2 myocardial cells. Compared with cells treated with si-MALAT1 + NC, the expression of miR-532-3p in H9C2 myocardial cells was clearly decreased in the si-MALAT1 + miR-532-3p inhibitor group (P<0.01, Fig. 6A). miR-532-3p overexpression reduced the apoptosis rate and levels of cTn-I, CK-MB, MDA, TNF-α, IL-1β and IL-6 in H9C2 myocardial cells and increased levels of SOD, TGF-β, IL-4 and IL-10. Compared with cells treated with miR-532-3p inhibitor + si-NC, ISO-treated H9C2 myocardial cells in the miR-532-3p inhibitor + si-MALAT1 group were more damaged (all P<0.01, Fig. 6B-E). Overexpression of miR-532-3p reduced injury of ISO-treated H9C2 myocardial cells, and inhibition of miR-532-3p weakened the protective effect of downregulated MALAT1 against ISO-treated H9C2 myocardial cell injury.
Figure 6.
Inhibition of miR-532-3p weakened the protective effect of downregulated MALAT1 against H9C2 myocardial cell injury. (A) The expression of miR-532-3p in H9C2 cells was detected using RT-qPCR. (B) The apoptosis rate was detected using flow cytometry. (C) The injury index of H9C2 cells was detected using ELISA. (D and E) Lipid oxidation and inflammatory responses in rats were detected using ELISA. Three independent repeated tests were conducted. The data were expressed as the mean ± standard deviation and analyzed using one-way or two-way ANOVA followed by Tukey's multiple comparison test. **P<0.01.
Discussion
HF is a chronic disease, and its rate of incidence is likely to continue to increase in the next few years (31). Previous findings have shown that lncRNAs play an important role in regulating mammalian cardiogenesis (32). A study has revealed the association of MALAT1 expression profiles in peripheral blood of coronary artery disease patients with previous cardiac events (33). In this study, we demonstrated that lncRNA MALAT1 was highly expressed in HF patients and exhibited great value for HF diagnosis. MALAT1 targets miR-532-3p to upregulate LDLR in HF, and inhibition of MALAT1 alleviated myocardial injury, lipid metabolism disorders and inflammation responses in HF rats.
As evidenced, HF may develop as a result of myocardial injury (34). MALAT1 is involved in HF (35). Both cTn-T and CK-MB are biomarkers of myocardial injury, and upregulation of these biomarkers is indicative of aggravated myocardial injury (36). It is well established that LVEF, FS, LVSP, LVEDP and ±dp/dtmax can be used to evaluate myocardial injury (37,38). Our study revealed that MALAT1 was highly expressed in HF patients and rats, and suppression of MALAT1 reduced the myocardial injury of HF rats. These conclusions were confirmed by the significantly reduced serum levels of cTn-I, CK-MB and LVEDP; increased LVEF, FS, LVSP, and ±dp/dtmax levels, and decreased TUNEL-positive cells. Consistently, MALAT1 knockdown helps to improve the cardiac function and decrease myocardial injury and apoptosis (39).
Lipid metabolism disorders are critical in the pathogenesis of HF (40). Assessment of the changes of TC, HDL and LDL may provide a better understanding of lipid metabolism (41). The current results showed that inhibition of MALAT1 decreased levels of ET, TC, TG, and LDL-C and increased the level of HDL-C in HF rats, indicating improvements in lipid metabolism disorders. Consistent with our study, decreased ET, TC, TG, and LDL-C levels and increased HDL-C levels signified reduced blood lipid levels (42). MALAT1 was highly expressed in hepatocytes of obese mice, and downregulated MALAT1 significantly inhibited lipid accumulation (43). Moreover, inflammation is widely perceived as a dominating risk factor for HF (44). In the current study, downregulated MALAT1 decreased levels of pro-inflammatory cytokines (TNF-α, IL-1β and IL-6) and increased levels of anti-inflammatory cytokines (TGF-β, IL-4 and IL-10) in HF rats, indicating mitigation of inflammation. Numerous studies have reported results similar to this study. For example, suppressed MALAT1 helps to relieve inflammation caused by hypoxia reoxygenation stimulation by decreasing transcripts and production of pro-inflammatory cytokines (45). MALAT1 knockdown alleviates inflammation after brain ischemia (46). Additionally, lipid oxidation is also strongly associated with the progression of HF (47). Results of the present study showed that MDA and ROS levels were increased, and SOD levels were reduced in HF rats. These effects were reversed after MALAT1 inhibition. Consistently, silencing MALAT1 contributes to downregulation of ROS and MDA levels and elevated SOD levels in hypoxia-induced cell injury (48). The results indicated that MALAT1 inhibition alleviated lipid oxidation in HF rats.
The above analyses revealed that inhibition of MALAT1 reduced myocardial injury and relieved lipid metabolism disorders, inflammation and lipid oxidation in HF rats. These results were further verified in ISO-induced H9C2 cells in vitro. However, the downstream regulatory mechanism of MALAT1 was still unclear. MALAT1 functions as a ceRNA and plays an essential role in cardiac progenitor cell growth via miR-mediated regulation, providing a potential method for HF therapy (49). miR-532 is enriched in mitochondria in HF (50). In this study, targeted binding relationships between MALAT1 and miR-532-3p as well as miR-532-3p and LDLR were verified. miR-532-3p was decreased and LDLR was upregulated in HF rats and ISO-induced H9C2 cells, and inhibition of miR-532-3p weakened the protective effect of downregulated MALAT1 against H9C2 myocardial cell injury. Consistent with this study, miR-532 exerts cardioprotective effects on myocardial infarction and subsequent HF (51). LDLR can serve as a risk predictor in HF (52). In addition, miR-532-3p participates in the regulation of atherosclerotic plaque formation, significantly contributing to atherothrombotic events, including heart attack, via the involvement of LDLR (20). However, the interactions between MALAT1 and miR-532-3p have not been elucidated, demonstrating the innovation of this study. Briefly, the results indicated that MALAT1 acts as a ceRNA to sponges miR-532-3p to upregulate LDLR, thereby aggravating myocardial injury in HF.
Altogether, our study revealed that suppression of MALAT1 relieved myocardial injury, lipid metabolism disorders and inflammation in HF. These results identified a novel lncRNA-based therapy for HF, and inhibition of the MALAT1/miR-532-3p/LDLR axis could potentially be developed as a promising therapeutic approach for HF treatment. Considering the limitation of animal ethics and experimental funding, no dose gradient was established, and no double-labeling method for identification of specific TUNEL-positive cells was used in this study. In addition, predisposing mutations associated with MALAT1 gene, vascularity, fibrosis and hypertrophy were not explored. The above questions are to be addressed in future research. Although the present study provides information on the therapeutic value of MALAT1 in the treatment of HF, the experimental results and clinical applications need to be further verified.
Supplementary Data
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
All the data generated or analyzed during this study are included in this published article.
Authors' contributions
PZ and YKW are the guarantors of integrity of the entire study. PZ and YKW contributed to the study concepts, study design, and definition of intellectual content. LPZ and JHZ contributed to the literature research and data acquisition. YKW contributed to manuscript editing and review. HQW contributed to the clinical studies. NL contributed to the experimental studies and data acquisition. LPZ contributed to the data analysis and statistical analysis. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved and supervised by the human ethics committee of Yantai Yuhuangding Hospital. All subjects signed the informed consent. All experiments were approved by the animal ethics committee of Yantai Yuhuangding Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors have no conflicts of interest to declare.
References
- 1.Snipelisky D, Chaudhry SP, Stewart GC. The many faces of heart failure. Card Electrophysiol Clin. 2019;11:11–20. doi: 10.1016/j.ccep.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Tomasoni D, Adamo M, Lombardi CM, Metra M. Highlights in heart failure. ESC Heart Fail. 2019;6:1105–1127. doi: 10.1002/ehf2.12555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bozkurt B, Khalaf S. Heart failure in women. Methodist Debakey Cardiovasc J. 2017;13:216–223. doi: 10.14797/mdcj-13-4-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Melander S, Miller S. Heart failure: Overcoming the physiologic dilemma through evidence-based practice. Nurs Clin North Am. 2016;51:13–27. doi: 10.1016/j.cnur.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Ziaeian B, Fonarow GC. Epidemiology and aetiology of heart failure. Nat Rev Cardiol. 2016;13:368–378. doi: 10.1038/nrcardio.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinelli AEM, Maranhao RC, Carvalho PO, Freitas FR, Silva BMO, Curiati MNC, Kalil Filho R, Pereira-Barretto AC. Cholesteryl ester transfer protein (CETP), HDL capacity of receiving cholesterol and status of inflammatory cytokines in patients with severe heart failure. Lipids Health Dis. 2018;17:242. doi: 10.1186/s12944-018-0888-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bui AL, Horwich TB, Fonarow GC. Epidemiology and risk profile of heart failure. Nat Rev Cardiol. 2011;8:30–41. doi: 10.1038/nrcardio.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosik J, Szostak B, Machaj F, Pawlik A. Potential targets of gene therapy in the treatment of heart failure. Expert Opin Ther Targets. 2018;22:811–816. doi: 10.1080/14728222.2018.1514012. [DOI] [PubMed] [Google Scholar]
- 9.Fan Z, Gao S, Chen Y, Xu B, Yu C, Yue M, Tan X. Integrative analysis of competing endogenous RNA networks reveals the functional lncRNAs in heart failure. J Cell Mol Med. 2018;22:4818–4829. doi: 10.1111/jcmm.13739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turton N, Swan R, Mahenthiralingam T, Pitts D, Dykes IM. The functions of long non-coding RNA during embryonic cardiovascular development and its potential for diagnosis and treatment of congenital heart disease. J Cardiovasc Dev Dis. 2019;6:21. doi: 10.3390/jcdd6020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Y, Wu J, Liangpunsakul S, Wang L. Long non-coding RNA in liver metabolism and disease: Current status. Liver Res. 2017;1:163–167. doi: 10.1016/j.livres.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan Y, Song D, Wu J, Wang J. Long non-coding RNAs link oxidized low-density lipoprotein with the inflammatory response of macrophages in atherogenesis. Front Immunol. 2020;11:24. doi: 10.3389/fimmu.2020.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sirtori CR, Ruscica M, Calabresi L, Chiesa G, Giovannoni R, Badimon JJ. HDL therapy today: From atherosclerosis, to stent compatibility to heart failure. Ann Med. 2019;51:345–359. doi: 10.1080/07853890.2019.1694695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Q, Yi X. Down-regulation of long noncoding RNA MALAT1 protects hippocampal neurons against excessive autophagy and apoptosis via the PI3K/Akt signaling pathway in rats with epilepsy. J Mol Neurosci. 2018;65:234–245. doi: 10.1007/s12031-018-1093-3. [DOI] [PubMed] [Google Scholar]
- 15.Liu L, Tan L, Yao J, Yang L. Long non-coding RNA MALAT1 regulates cholesterol accumulation in ox-LDL-induced macrophages via the microRNA-17-5p/ABCA1 axis. Mol Med Rep. 2020;21:1761–1770. doi: 10.3892/mmr.2020.10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L, Qi Y, Wang Y, Tang H, Li Z, Wang Y, Tang S, Zhu H. LncRNA MALAT1 suppression protects endothelium against oxLDL-induced inflammation via inhibiting expression of MiR-181b target gene TOX. Oxid Med Cell Longev. 2019;2019:8245810. doi: 10.1155/2019/8245810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Y. The novel regulatory role of lncRNA-miRNA-mRNA axis in cardiovascular diseases. J Cell Mol Med. 2018;22:5768–5775. doi: 10.1111/jcmm.13866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shah P, Bristow MR, Port JD. MicroRNAs in heart failure, cardiac transplantation, and myocardial recovery: Biomarkers with therapeutic potential. Curr Heart Fail Rep. 2017;14:454–464. doi: 10.1007/s11897-017-0362-8. [DOI] [PubMed] [Google Scholar]
- 19.Bauersachs R, Zeymer U, Briere JB, Marre C, Bowrin K, Huelsebeck M. Burden of coronary artery disease and peripheral artery disease: A literature review. Cardiovasc Ther. 2019;2019:8295054. doi: 10.1155/2019/8295054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang R, Cao Y, Li H, Hu Z, Zhang H, Zhang L, Su W, Xu Y, Liang L, Melgiri ND, et al. miR-532-3p-CSF2RA axis as a key regulator of vulnerable atherosclerotic plaque formation. Can J Cardiol. 2020;36:1782–1794. doi: 10.1016/j.cjca.2019.12.018. [DOI] [PubMed] [Google Scholar]
- 21.Arola-Arnal A, Blade C. Proanthocyanidins modulate microRNA expression in human HepG2 cells. PLoS One. 2011;6:e25982. doi: 10.1371/journal.pone.0025982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao K, Zhang J, Gao P, Wang Q, Liu Y, Liu J, Zhang Y, Li Y, Chang H, Ren P, et al. Qishen granules exerts cardioprotective effects on rats with heart failure via regulating fatty acid and glucose metabolism. Chin Med. 2020;15:21. doi: 10.1186/s13020-020-0299-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zatroch KK, Knight CG, Reimer JN, Pang DS. Refinement of intraperitoneal injection of sodium pentobarbital for euthanasia in laboratory rats (Rattus norvegicus) BMC Vet Res. 2017;13:60. doi: 10.1186/s12917-017-0982-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan C, Tang X, Ye M, Zhu G, Dai Y, Yao Z, Yao X. Qi-Li-Qiang-Xin alleviates isoproterenol-induced myocardial injury by inhibiting excessive autophagy via activating AKT/mTOR pathway. Front Pharmacol. 2019;10:1329. doi: 10.3389/fphar.2019.01329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 26.Zhao W, Geng D, Li S, Chen Z, Sun M. LncRNA HOTAIR influences cell growth, migration, invasion, and apoptosis via the miR-20a-5p/HMGA2 axis in breast cancer. Cancer Med. 2018;7:842–855. doi: 10.1002/cam4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Zhou YX, Wang C, Mao LW, Wang YL, Xia LQ, Zhao W, Shen J, Chen J. Long noncoding RNA HOTAIR mediates the estrogen-induced metastasis of endometrial cancer cells via the miR-646/NPM1 axis. Am J Physiol Cell Physiol. 2018;314:C690–C701. doi: 10.1152/ajpcell.00222.2017. [DOI] [PubMed] [Google Scholar]
- 28.Choi SS, Kim ES, Koh M, Lee SJ, Lim D, Yang YR, Jang HJ, Seo KA, Min SH, Lee IH, et al. A novel non-agonist peroxisome proliferator-activated receptor γ (PPARγ) ligand UHC1 blocks PPARγ phosphorylation by cyclin-dependent kinase 5 (CDK5) and improves insulin sensitivity. J Biol Chem. 2014;289:26618–26629. doi: 10.1074/jbc.M114.566794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madej-Pilarczyk A, Niezgoda A, Janus M, Wojnicz R, Marchel M, Fidziańska A, Grajek S, Hausmanowa-Petrusewicz I. Limb-girdle muscular dystrophy with severe heart failure overlapping with lipodystrophy in a patient with LMNA mutation p.Ser334del. J Appl Genet. 2017;58:87–91. doi: 10.1007/s13353-016-0365-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao Z, Pan X, Yang Y, Huang Y, Shen HB. The lncLocator: A subcellular localization predictor for long non-coding RNAs based on a stacked ensemble classifier. Bioinformatics. 2018;34:2185–2194. doi: 10.1093/bioinformatics/bty085. [DOI] [PubMed] [Google Scholar]
- 31.Rogers C, Bush N. Heart failure: Pathophysiology, diagnosis, medical treatment guidelines, and nursing management. Nurs Clin North Am. 2015;50:787–799. doi: 10.1016/j.cnur.2015.07.012. [DOI] [PubMed] [Google Scholar]
- 32.Scheuermann JC, Boyer LA. Getting to the heart of the matter: Long non-coding RNAs in cardiac development and disease. EMBO J. 2013;32:1805–1816. doi: 10.1038/emboj.2013.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toraih EA, El-Wazir A, Alghamdi SA, Alhazmi AS, El-Wazir M, Abdel-Daim MM, Fawzy MS. Association of long non-coding RNA MIAT and MALAT1 expression profiles in peripheral blood of coronary artery disease patients with previous cardiac events. Genet Mol Biol. 2019;42:509–518. doi: 10.1590/1678-4685-gmb-2018-0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hartupee J, Mann DL. Neurohormonal activation in heart failure with reduced ejection fraction. Nat Rev Cardiol. 2017;14:30–38. doi: 10.1038/nrcardio.2016.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo X, Wu X, Han Y, Tian E, Cheng J. LncRNA MALAT1 protects cardiomyocytes from isoproterenol-induced apoptosis through sponging miR-558 to enhance ULK1-mediated protective autophagy. J Cell Physiol. 2019;234:10842–10854. doi: 10.1002/jcp.27925. [DOI] [PubMed] [Google Scholar]
- 36.Sadoh WE, Eregie CO, Nwaneri DU, Sadoh AE. The diagnostic value of both troponin T and creatinine kinase isoenzyme (CK-MB) in detecting combined renal and myocardial injuries in asphyxiated infants. PLoS One. 2014;9:e91338. doi: 10.1371/journal.pone.0091338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, Wang J, Tu T, Iyan Z, Mungun D, Yang Z, Guo Y. Remote ischemic postconditioning protects against myocardial ischemia-reperfusion injury by inhibition of the RAGE-HMGB1 pathway. Biomed Res Int. 2018;2018:4565630. doi: 10.1155/2018/4565630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan B, Liu S, Li X, Zhong Y, Tong F, Yang S. Preconditioning with endoplasmic reticulum stress alleviated heart ischemia/reperfusion injury via modulating IRE1/ATF6/RACK1/PERK and PGC-1α in diabetes mellitus. Biomed Pharmacother. 2019;118:109407. doi: 10.1016/j.biopha.2019.109407. [DOI] [PubMed] [Google Scholar]
- 39.Fan YZ, Huang H, Wang S, Tan GJ, Zhang QZ. Effect of lncRNA MALAT1 on rats with myocardial infarction through regulating ERK/MAPK signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23:9041–9049. doi: 10.26355/eurrev_201910_19306. [DOI] [PubMed] [Google Scholar]
- 40.Abdalla S, Fu X, Elzahwy SS, Klaetschke K, Streichert T, Quitterer U. Up-regulation of the cardiac lipid metabolism at the onset of heart failure. Cardiovasc Hematol Agents Med Chem. 2011;9:190–206. doi: 10.2174/187152511797037583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurpińska AK, Jarosz A, Ożgo M, Skrzypczak WF. Changes in lipid metabolism during last month of pregnancy and first two months of lactation in primiparous cows-analysis of apolipoprotein expression pattern and changes in concentration of total cholesterol, HDL, LDL, triglycerides. Pol J Vet Sci. 2015;18:291–298. doi: 10.1515/pjvs-2015-0038. [DOI] [PubMed] [Google Scholar]
- 42.Li J, Lei HT, Cao L, Mi YN, Li S, Cao YX. Crocin alleviates coronary atherosclerosis via inhibiting lipid synthesis and inducing M2 macrophage polarization. Int Immunopharmacol. 2018;55:120–127. doi: 10.1016/j.intimp.2017.11.037. [DOI] [PubMed] [Google Scholar]
- 43.Yan C, Chen J, Chen N. Long noncoding RNA MALAT1 promotes hepatic steatosis and insulin resistance by increasing nuclear SREBP-1c protein stability. Sci Rep. 2016;6:22640. doi: 10.1038/srep22640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shirazi LF, Bissett J, Romeo F, Mehta JL. Role of inflammation in heart failure. Curr Atheroscler Rep. 2017;19:27. doi: 10.1007/s11883-017-0660-3. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y, Zhang H, Zhang Z, Li S, Jiang W, Li X, Lv J. LncRNA MALAT1 cessation antagonizes hypoxia/reoxygenation injury in hepatocytes by inhibiting apoptosis and inflammation via the HMGB1-TLR4 axis. Mol Immunol. 2019;112:22–29. doi: 10.1016/j.molimm.2019.04.015. [DOI] [PubMed] [Google Scholar]
- 46.Cao DW, Liu MM, Duan R, Tao YF, Zhou JS, Fang WR, Zhu JR, Niu L, Sun JG. The lncRNA Malat1 functions as a ceRNA to contribute to berberine-mediated inhibition of HMGB1 by sponging miR-181c-5p in poststroke inflammation. Acta Pharmacol Sin. 2020;41:22–33. doi: 10.1038/s41401-019-0284-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fragasso G. Deranged cardiac metabolism and the pathogenesis of heart failure. Card Fail Rev. 2016;2:8–13. doi: 10.15420/cfr.2016:5:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang L, Xu F, Zhang M, Shang XY, Xie X, Fu T, Li JP, Li HL. Role of LncRNA MALAT-1 in hypoxia-induced PC12 cell injury via regulating p38MAPK signaling pathway. Neurosci Lett. 2018;670:41–47. doi: 10.1016/j.neulet.2018.01.036. [DOI] [PubMed] [Google Scholar]
- 49.Li L, Wang Q, Yuan Z, Chen A, Liu Z, Wang Z, Li H. LncRNA-MALAT1 promotes CPC proliferation and migration in hypoxia by up-regulation of JMJD6 via sponging miR-125. Biochem Biophys Res Commun. 2018;499:711–718. doi: 10.1016/j.bbrc.2018.03.216. [DOI] [PubMed] [Google Scholar]
- 50.Wang X, Song C, Zhou X, Han X, Li J, Wang Z, Shang H, Liu Y, Cao H. Mitochondria associated MicroRNA expression profiling of heart failure. Biomed Res Int. 2017;2017:4042509. doi: 10.1155/2017/4042509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bayoumi AS, Teoh JP, Aonuma T, Yuan Z, Ruan X, Tang Y, Su H, Weintraub NL, Kim IM. MicroRNA-532 protects the heart in acute myocardial infarction, and represses prss23, a positive regulator of endothelial-to-mesenchymal transition. Cardiovasc Res. 2017;113:1603–1614. doi: 10.1093/cvr/cvx132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bayes-Genis A, Núñez J, Zannad F, Ferreira JP, Anker SD, Cleland JG, Dickstein K, Filippatos G, Lang CC, Ng LL, et al. The PCSK9-LDL receptor axis and outcomes in heart failure: BIOSTAT-CHF subanalysis. J Am Coll Cardiol. 2017;70:2128–2136. doi: 10.1016/j.jacc.2017.08.057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data generated or analyzed during this study are included in this published article.