Abstract
Emerging studies have indicated that the dysregulation of microRNAs (miRNAs or miRs) plays a vital role in the development and metastasis of tumors. However, the role of miR-93-5p in esophageal carcinoma (EC) has not been extensively reported. The present study thus focused on the role of miR-93-5p and its downstream target in the occurrence and development of EC. Firstly, miRNA expression profiles associated with EC were accessed from the TCGA_ESCA dataset and analyzed. Subsequently, the expression patterns of miR-93-5p and TGFβR2 were characterized in the human esophageal cell line, Het-1A, and the human EC cell lines, TE-1, Eca-109 and EC9706, by RT-qPCR and western blot analysis. WST-1 assay, flow cytometry, Transwell assay, wound healing assay and bioinformatics analysis were used to explore their functions in EC cells. Finally, a dual-luciferase reporter assay was employed to determine the targeted association between miR-93-5p and TGFβR2. The results revealed that the expression of miR-93-5p was markedly higher in EC cell lines compared with that in the normal cell line. The overexpression of miR-93-5p facilitated cell proliferation, migration and invasion, and inhibited cell apoptosis. Additionally, TGFβR2 was identified as a functional target of miR-93-5p in EC cells, as judged by a series of in vitro experiments. Furthermore, it was found that the simultaneous overexpression of miR-93-5p and TGFβR2 almost had no effect on the biological behaviors of EC cells. On the whole, the present study demonstrates that miR-93-5p promotes the proliferation, migration and invasion, and inhibits the apoptosis of EC cells by targeting TGFβR2.
Keywords: miR-93-5p, transforming growth factor-β receptor 2, esophageal carcinoma, proliferation, migration and invasion, apoptosis
Introduction
Esophageal carcinoma (EC) is one of the most common gastrointestinal tumors with two main histological subtypes: Esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC). In East Asia, the incidence of EC is known to be the highest worldwide (1). Although much progress has been made recently towards EC treatment with respect to targeted therapy, surgery and neoadjuvant chemotherapy, the 5-year survival rate of patients with locally advanced tumors remains <55% (2). Thus, optimal and effective therapeutic strategies are required.
MicroRNAs (miRNAs or miRs) are highly conserved non-coding small RNAs that are capable of regulating gene expression via the translational repression and degradation induction of mRNAs in cells (3), and have been shown to be involved in the major signaling pathways of histogenesis and cell apoptosis (4). Moreover, miRNAs can serve as biomarkers for tumor staging and prognosis. miR-93-5p is a paralog (miR-106b-25) derived from the miR-17-92 cluster. It is implicated in the occurrence and development of various human solid cancers, including breast cancer, colorectal cancer, liver cancer, lung cancer, ovarian cancer and pancreatic cancer, etc. (5). Some studies have demonstrated the abnormally high expression of miR-93-5p in liver (6), breast (7) and lung cancer (8), and it is able to promote cell proliferation and migration by binding to various target genes. Moreover, it has been reported that miR-93-5p may be a potential biomarker for the detection of the presence of cancer (9).
The transforming growth factor-β (TGF-β) pathway is a pivotal player in cell carcinogenesis and metastasis. TGF-β receptor 2 (TGFβR2) is a key molecule that regulates the TGF-β pathway and its expression is often downregulated or lost in several cancer (10). The decrease in TGFβR2 expression can result in a variety changes in tumor behavior, such as poor tumor differentiation, higher tumor staging and an increase in the lymph node metastasis rate (11). In tumor cells, TGFβR2 signaling can regulate a variety of activities, such as epithelial-mesenchymal transition (EMT), cell migration and invasion, angiogenesis, immune regulation and cytokine secretion (12). The present study explored the regulatory effects of miR-93-5p on TGFβR2 in EC cells, with an aim to provide novel targeted diagnostic and prognostic approaches for EC.
Materials and methods
Bioinformatics analysis
miRNA expression profiles, including 13 normal samples and 176 EC tissue samples, and mRNA expression profiles, including 11 normal samples and 160 EC tissue samples, were obtained from the TCGA-ESCA database (https://portal.gdc.cancer.gov/). The R package 'edgeR' was employed to identify the differentially expressed miRNAs (DEmiRNAs) with the criteria of |logFC|>2 and adj. P-value <0.05. Target genes for miR-93-5p were predicted through bioinformatics analysis and 3 target prediction databases, miRDB (http://mirdb.org/miRDB/index.html), miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php) and TargetScan (http://www.targetscan.org/vert_71/). Survival analysis was then conducted to identify the mRNA of interest. Based on the downloaded data, the t-test was used to determine the significance of the expression of these genes in normal tissues and tumor tissues. The samples were divided into the high and low expression groups based on the median expression of each gene, and survival analysis was performed using the 'survival' package, respectively, and genes that were significantly related to the prognosis and targeted downregulation were selected as the target gene. Finally, the targeted binding sites between miR-93-5p and its target gene were predicted.
Cells and cell culture
The human normal esophageal cell line, Het-1A (BNCC337688), and EC cell lines [TE-1 (BNCC100151), Eca-109 (BNCC337687) and EC9706 (BNCC339892)] were obtained from Bena Culture Collection (BNCC, Beijing, China). The Het-1A, Eca-109 and EC9706 cell lines were grown in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin. The TE-1 cell lines were cultured in RPMI-1640 medium containing 10% FBS. All of the cells were placed in a wet incubator with 5% CO2 at 37°C.
Cell transfection and vector construction
The EC cells in logarithmic growth phase were collected for transfection with miR-93-5p mimic, miR-93-5p inhibitor and miR-93-5p mimic + oe-TGFβR2 as well as their corresponding controls (obtained from Guangzhou RiboBio Co., Ltd.) using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.), respectively. The transfection concentration was 50 nM. Following 6 h of incubation with 5% CO2 at 37°C, the cells were then continuously cultured in fresh medium for 48 h for use in subsequent experiments. The lentiviral vector was used to construct the TGFβR2 overexpression vector: TGFβR2 cDNA with restriction enzyme sites of KpnI and XhoI, as well as a corresponding control sequence (designed by BLOCK-iT™ RNAi Deshgner website) was synthesized, and then ligated into the lentiviral expression vector pLVX-IRES-neo (Clontech Laboratories, Inc.) by T4 ligase. Following 24 h of culture at 37°C, the expression vector was extracted and sequenced. Finally, the packaged vector and viral particle were used to infect EC cells (2×104 cells) cultured in 5 µg/ml Polybrene cultured in a 12-well plate with a multiplicity of infection (MOI) of 50. At 24 h following transfection, cells were harvested for analysis.
RT-qPCR
Total RNA was isolated from the cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). miR-93-5p cDNA was synthesized using the qScript microRNA cDNA synthesis kit (Quantabio), and TGFβR2 cDNA was synthesized using cDNA synthesis kit (Thermo Fisher Scientific, Inc.). qPCR was performed using the miScript SYBR-Green PCR kit (Qiagen GmbH) under the following thermal cycling conditions: 95°C for 2 min; 95°C for 5 sec and 60°C for 30 sec, with a total of 40 cycles. U6 and GAPDH were used as loading controls for miR-93-5p and TGFβR2, respectively. The primer sequences are listed in Table I. The quantitative expression values were calculated using the 2−ΔΔCq method (13).
Table I.
Sequences of primers used for RT-qPCR.
| Gene | Forward | Reverse |
|---|---|---|
| U6 | CAGCACATATACTAAAATTGGAACG | ACGAATTTGCGTGTCATCC |
| hsa-miR-93-5p | GCCGCCAAAGTGCTGTTC | CAGAGCAGGGTCCGAGGTA |
| TGFβR2 | GTAGCTCTGATGAGTGCAATGAC | GGGGTCATTGATGGCAACAATA |
| GAPDH | AAGGTGAAGGTCGGAGTCAAC | GGGGTCATTGATGGCAACAATA |
Western blot analysis
Cells (following 48 h of transfection) of each group were lysed on ice for 10 min with RIPA lysis buffer (Sigma-Aldrich; Merck KGaA). The BCA protein assay kit (Thermo Fisher Scientific, Inc.) was employed to detect the concentration of the protein samples. The protein samples (30 mg per lane) were then separated using sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with 10% gel, and then transferred onto nitrocellulose membranes (ZY-160FP, Zeye Bio Co., Ltd.). The membranes were then blocked with 5% BSA/TBST for 2 h at room temperature and washed 3 times with 1X TBST. The membranes were incubated with primary antibodies, including rabbit polyclonal antibody TGFβR2 (ab184948; 1:1,000) and rabbit polyclonal antibody GAPDH (ab181602; 1:2,500) at 4°C overnight. The membranes were then washed with 1X TBST and incubated with horseradish peroxidase-labeled goat anti-rabbit IgG (ab205718; 1:1,000) for hybridization at room temperature for 2 h. Finally, protein bands were visualized using electrochemiluminescence (ECL) solution and then observed and analyzed. All the antibodies mentioned above were purchased from Abcam.
Detection of cell proliferation by WST-1 assay
The WST-1 kit (Roche Diagnostics) was used to determine cell proliferation. EC cells were plated into 96-well plates at a density of 2.5×105 cell/well, and incubated with WST-1 at 37°C for 3 h. The absorbance at 450 nm was measured using a microplate reader (SpectraMax i3, Molecular Devices, LLC) to determine the cell proliferation rate.
Detection of cell apoptosis by flow cytometry
Cell apoptosis was detected by flow cytometry using an Apoptosis kit with Annexin V FITC/propidium iodide (PI) (V13242, Thermo Fisher Scientific, Inc.) in accordance with the manufacturer's guidelines. Cells were harvested and rinsed following 48 h of incubation at 37°C. The cell suspension was mixed with 10 µl of Annexin V and 5 µl of PI at room temperature for 10 min. Subsequently, fluorescence activated cell sorting (FACS) flow cytometry (FACSCalibur, 342976, BD Biosciences) was used to detect cell apoptosis.
Wound healing assay
After 24 h of cell transfection, when the cells reached 70-80% confluency, a wound was scraped into the cells using a tip of 200 µl pipette across the hole center and the cells were then washed with PBS twice to remove the floating cells. Subsequently, cells were cultured in fresh serum-free DMEM for a further 24 h at 37°C. The relative distance of the scratches was observed under a microscope (Axioskop 40, Carl Zeiss AG). The photographs were analyzed using an image analysis system (Wound Healing ACAS, ibidi). For each field of view, 2 straight lines were drawn on the front of both sides of the gap, and the average distance was calculated as the average of the distances of the 2 straight lines at the left, center and right points of the view.
Transwell assay
For the invasion assay, 24-well Transwell chambers (8 µm pores, BD Biosciences) were used. Approximately 2×104 cells were seeded into the upper chamber coated with Matrigel (Corning, Inc.), and DMEM medium containing 10% FBS was added to the lower chamber. The cells in the upper chamber were wiped off with a cotton swab following incubation at 37°C for 24 h, while the cells invading the lower chamber were stained with crystal violet (Sigma-Aldrich; Merck KGaA) at room temperature for 20 min. Finally, cells were counted in randomly selected fields under a microscope (Axioskop 40, Carl Zeiss AG,) at ×100 magnification.
Dual-luciferase reporter assay
Dual-luciferase reporter gene assay was utilized to validate whether TGFβR2 is the direct target gene of miR-93-5p. DNA fragments of TGFβR2 3′-UTR containing the putative binding sites with miR-93-5p (or mutated binding sites) were amplified by PCR, digested with XhoI and BamHI, and then ligated into the Firefly luciferase vector pGL3 (Promega Corporation). The constructs were named wild-type (TGFβR2-wt) and mutated-type (TGFβR2-mut) TGFβR2, respectively. TGFβR2-wt or TGFβR2-mut reporter was transfected into cells along with miR-93-5p mimic or mimic NC. Following 48 h of transfection, cells were harvested and lysed. Luciferase activity was determined using a dual luciferase assay system (Promega Corporation). Relative Firefly luciferase activity was normalized to Renilla luciferase activity as a control for transfection efficiency.
Statistical analysis
All data were processed using SPSS 22.0 software (IBM, Corp.). Measurement data are expressed as the means ± standard deviation. All experiments were repeated at least 3 times. A Student's t-test was used for comparisons between 2 groups, while one-way ANOVA and Tukey's post hoc test were adopted in the case of comparisons among ≥3 groups. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-93-5p is highly expressed in EC tissues and cells
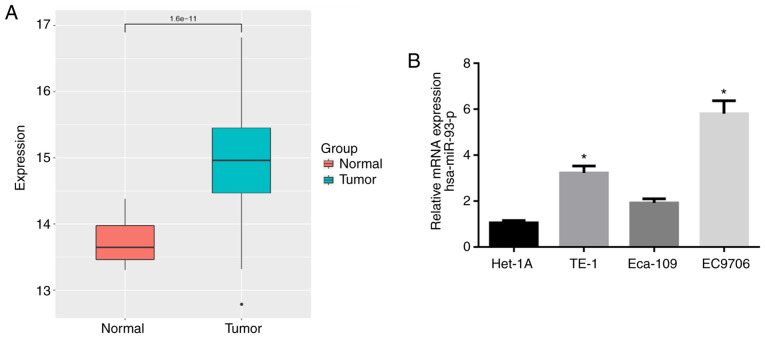
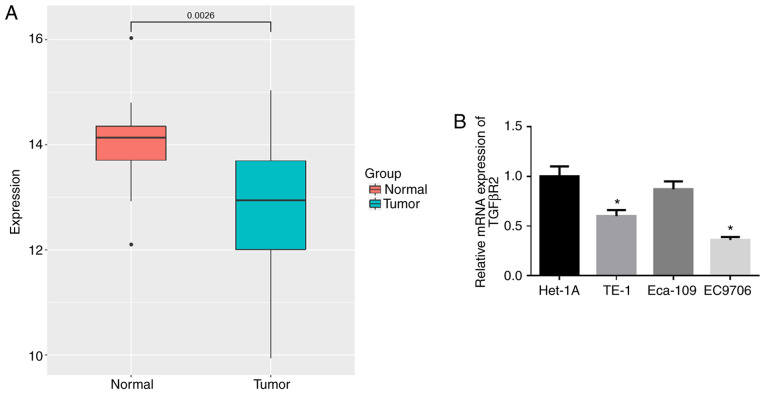
The expression data of miR-93-5p in 176 EC tissue samples and 13 normal samples were retrieved from the TCGA database. As shown in Fig. 1A, the expression of miR-93-5p was significantly higher in EC tissues compared with normal tissues. RT-qPCR was then performed to verify that miR-93-5p was highly expressed in the EC cell lines (TE-1, Eca-109 and EC9706) compared with that in the human normal esophageal cell line Het-1A (Fig. 1B). The EC9706 cell line with the most differential expression of miR-93-5p (P<0.05) was thereby selected for use in subsequent experiments.
Figure 1.
miR-93-5p is highly expressed in EC tissues and cells. (A) miR-93-5p expression data were retrieved from the TCGA database. (B) RT-qPCR was performed to detect the expression of miR-93-5p in normal esophageal cell line and EC cell lines. *P<0.05, compared to normal Het-1A cell line. EC, esophageal carcinoma.
Overexpression of miR-93-5p facilitates the proliferation, migration and invasion, and inhibits the apoptosis of EC cells
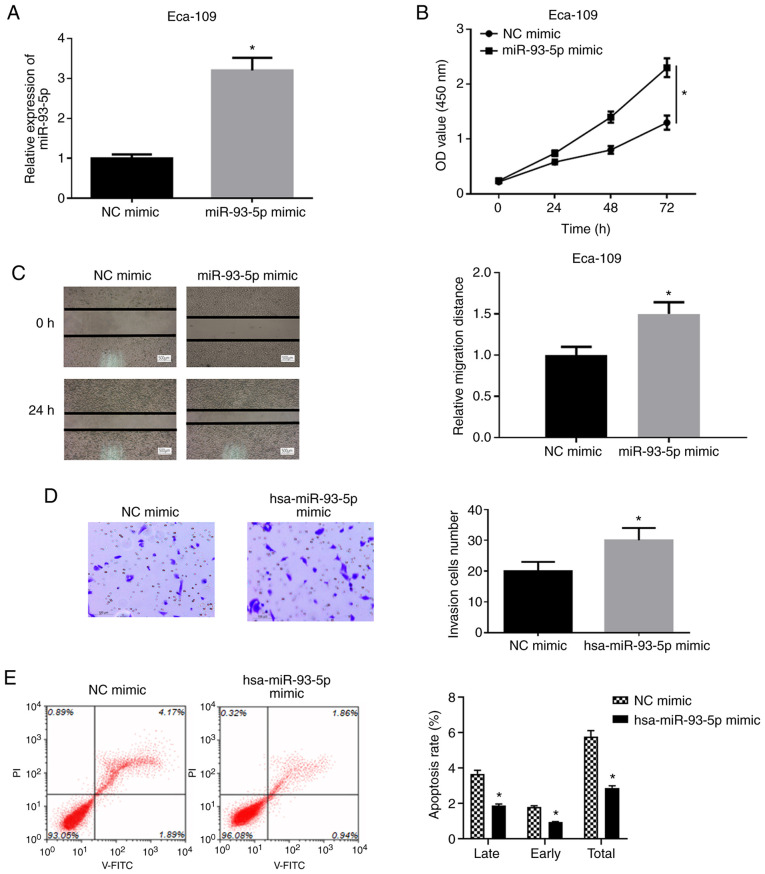
To determine the effects of miR-93-5p on the biological function of EC cells, hsa-miR-93-5p mimic or NC mimic were transfected into EC9706 cells and it was found that the expression of hsa-miR-93-5p was significantly upregulated in the cells transfected with hsa-miR-93-5p mimic (Fig. 2A). A series of experiments were then performed using the EC9706 cell line. WST-1 assay was applied to examine the viability of the cells in the NC mimic group and miR-93-5p mimic group. As shown in Fig. 2B, the viability of the EC9706 cells in the miR-93-5p mimic group was significantly increased by comparison with that in the NC mimic group (P<0.05). Wound healing assay was implemented to detect the migratory ability of the EC9706 cells (Fig. 2C). The results revealed that the migration rate of the EC9706 cells in miR-93-5p mimic group was significantly increased relative to that in the NC mimic group (P<0.05). Moreover, Transwell invasion assay was conducted to measure the cell invasive ability (Fig. 2D), and it was found that the invasive ability of the cells was increased in the miR-93-5p mimic group (P<0.05). Flow cytometry was also performed for the detection of cell apoptosis following 48 h of transfection (Fig. 2E). The apoptotic rate of the cells in the miR-93-5p mimic group was significantly decreased relative to that in the NC mimic group (P<0.05). The above-mentioned results indicated that the overexpression of miR-93-5p facilitated the viability, migration and invasion, and decreased the apoptosis of EC cells.
Figure 2.
miR-93-5p promotes the proliferation, migration and invasion, and inhibits the apoptosis of EC cells. The EC cell line, EC9706, was transfected with miR-93-5p mimic and its control. (A) RT-qPCR detection of the expression of hsa-miR-93-5p following transfection with hsa-miR-93-5p mimic or NC mimic; (B) WST-1 assay was performed to detect cell viability. (C) Wound healing assay was applied to detect the migratory ability (500 µm). (D) Transwell assay was carried out to detect the invasive ability (100 µm). (E) Flow cytometry was used to determine cell apoptotic rate. *P<0.05, compared to NC mimic. EC, esophageal carcinoma.
Suppression of miR-93-5p inhibits the proliferation, migration and invasion, and promotes the apoptosis of EC cells
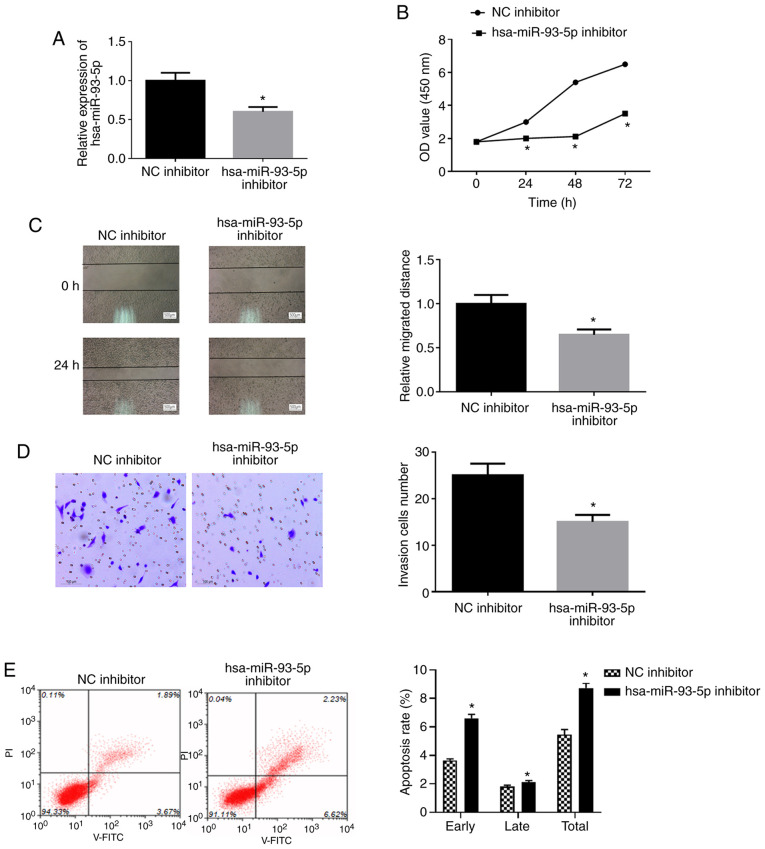
To further examine the effects of miR-93-5p on EC, EC9706 cells were transfected with miR-93-5p inhibitor or NC inhibitor (Fig. 3A). WST-1 assay, wound healing assay and Transwell assay revealed that the viability (Fig. 3B), migration (Fig. 3C) and invasion (Fig. 3D) of the EC9706 transfected with miR-93-5p inhibitor were significantly decreased compared with those in the control group. Flow cytometry was employed for examination of cell apoptosis. As shown in Fig. 3E, the apoptotic rate of the cells was significantly increased in the miR-93-5p inhibitor group compared with the NC inhibitor group (P<0.05). Overall, these findings indicate that the suppression of miR-93-5p inhibits the proliferation, migration and invasion, and promotes the apoptosis in EC cells.
Figure 3.
Suppression of miR-93-5p inhibits proliferation, migration and invasion, and promotes the apoptosis of EC cells. The EC cell line, EC9706, was transfected with miR-93-5p inhibitor and its control. (A) RT-qPCR to detect the expression of hsa-miR-93-5p following transfection with hsa-miR-93-5p inhibitor or NC inhibitor. (B) WST-1 assay was performed to detect cell viability. (C) Wound healing assay was applied to detect the migratory ability (500 µm). (D) Transwell assay was carried out to detect the cell invasive ability (100 µm). (E) flow cytometry was used to determine cell apoptotic rate. *P<0.05, compared to NC inhibitor. EC, esophageal carcinoma.
miR-93-5p targets and downregulates TGFβR2 expression
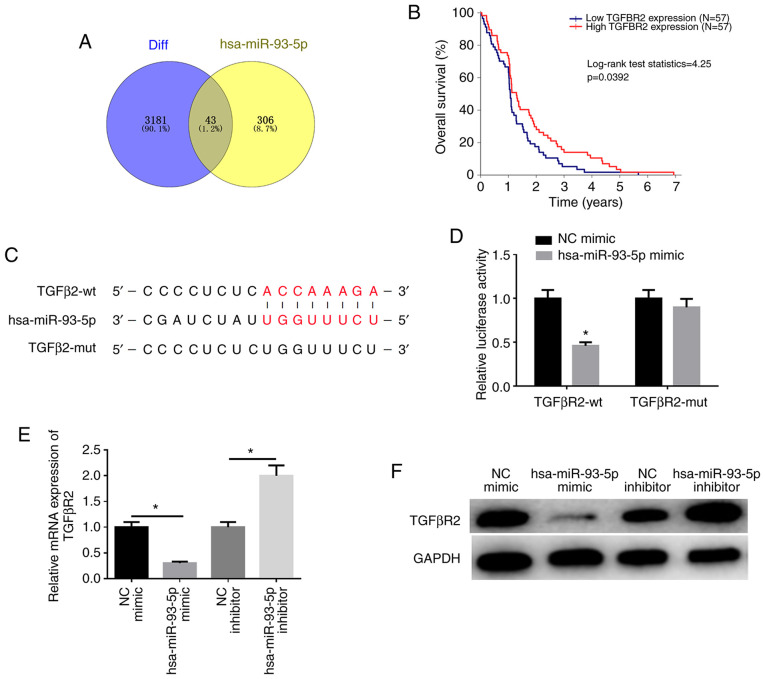
miRDB (http://mirdb.org/), miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php) and TargetScan (http://www.targetscan.org/vert_72/) databases were employed to predict the potential mRNAs binding to miR-93-5p and it was found that there were 43 overlapping mRNAs between the predicted mRNAs and downregulated DEmRNAs (Fig. 4A). Among these genes, TGFβR2 was found to be significantly associated with EC prognosis (Fig. 4B). The biological website, TargetScan (http://www.targetscan.org/vert_72/), was used and it was found that there was a potential binding site of miR-93-5p on the TGFβR2 3′-UTR (Fig. 4C), which was further verified by dual-luciferase reporter gene assay (Fig. 4D). Compared with the control group, the luciferase activity of TGFβR2-wt was suppressed by miR-93-5p overexpression, while that of TGFβR2-mut was unaffected (P<0.05). These findings demonstrate that miR-93-5p can target TGFβR2. In addition, the results of western blot analysis and RT-qPCR revealed that the expression of TGFβR2 was markedly downregulated in the miR-93-5p mimic group compared with the NC mimic group, while the expression of TGFβR2 was considerably upregulated in the miR-93-5p inhibitor group than that in the NC inhibitor group (P<0.05; Fig. 4E and F). These results indicate that hsa-miR-93-5p targets TGFβR2 expression.
Figure 4.
miR-93-5p targets and downregulates TGFβR2 expression. (A) Venn diagram of potential mRNAs binding to miR-93-5p predicted by bioinformatics websites and downregulated DEmRNAs. (B) Survival analysis based on the differential expression of TGFβR2. (C) The targeted binding sites of miR-93-5p on TGFβR2 3′UTR predicted by TargetScan website. (D) Dual luciferase reporter assay was performed to verify the targeted association between miR-93-5p and TGFβR2. (E) RT-qPCR was used to detect the effect of hsa-miR-93-5p on TGFβR2 transcription. (F) Western blot analysis was used to detect the expression of TGFβR2 following transfection with NC mimic, miR-93-5p mimic, NC inhibitor and miR-93-5p inhibitor. *P<0.05, compared to NC mimic or inhibitor. EC, esophageal carcinoma.
TGFβR2 is expressed in low levels in EC tissues and cells
In addition, the expression of TGFβR2 in EC tissue samples in TCGA-ESCA was significantly downregulated (Fig. 5A). RT-qPCR was used to examine the expression of TGFβR2 in the human normal esophageal cell line, Het-1A, and in the EC cell lines TE-1, Eca-109 and EC9706. As illustrated in Fig. 5B, TGFβR2 was markedly downregulated in the TE-1 and EC9706 cells by comparison with that in the Het-1A cell line (P<0.05), with the lowest expression observed in the EC9706 cells (P<0.05).
Figure 5.
TGFβR2 is expressed in low levels in EC cells. (A) TGFβR2 expression data were retrieved from the TCGA database. (B) RT-qPCR was performed to detect the expression of TGFβR2 in normal esophageal cell line and EC cell lines. *P<0.05, compared to normal Het-1A cell line. EC, esophageal carcinoma.
miR-93-5p promotes the proliferation, migration and invasion, and inhibits the apoptosis of EC cells by downregulating TGFβR2 expression
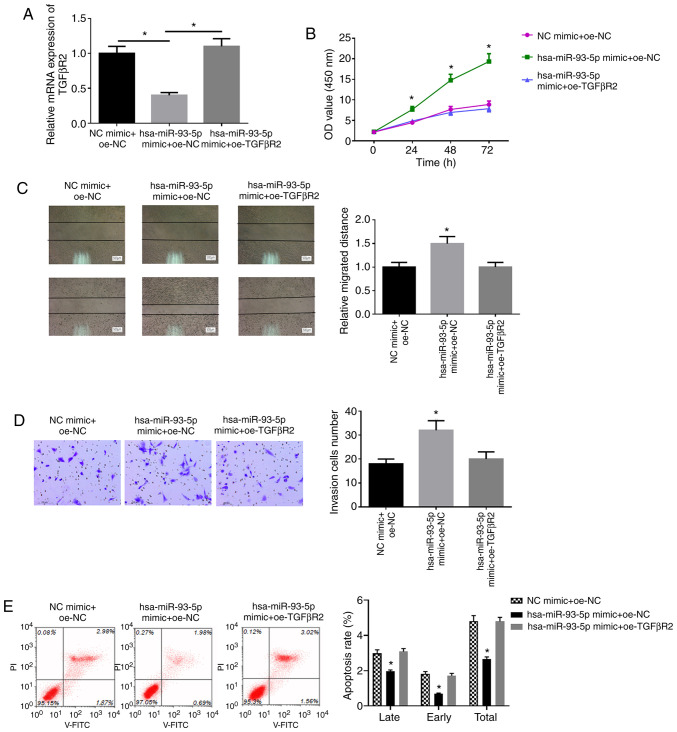
To confirm whether TGFβR2 can reverse the effects of miR-93-5p on the proliferation, migration and invasion of EC cells, EC9706 cells were transfected with miR-93-5p mimic + oe-NC, NC mimic + oe-NC, miR-93-5p mimic + oe-TGFβR2, respectively. The expression of TGFβR2 following transfection was detected by RT-qPCR. It was found that the mRNA and protein expression of TGFβR2 was successfully increased following transfection with TGFβR2 overexpression vector (Fig. S1). The results also revealed that the overexpression of hsa-miR-93-5p inhibited TGFβR2 mRNA expression, and oe-TGFβR2 reversed the inhibitory effects of hsa-miR-93-5p mimic on TGFβR2 mRNA expression (Fig. 6A). WST-1 assay was used to examine the viability of the cells in each group. As shown in Fig. 6B, the cells in the miR-93-5p mimic + oe-NC group had a higher viability compared with those in the NC mimic + oe-NC group (P<0.05), and no significant differences were observed between the NC mimic + oe-NC group and miR-93-5p mimic + oe-TGFβR2 group (P>0.05). Wound healing assay and Transwell assay were applied to detect the cell migration and invasion ability. The cells in the miR-93-5p mimic + oe-NC group also exhibited higher migratory and invasive abilities relative to those in the NC mimic + oe-NC group (P<0.05; Fig. 6C and D), and there was no marked differences between the NC mimic + oe-NC group and miR-93-5p mimic + oe-TGFβR2 group (P>0.05). Flow cytometry was performed to detect cell apoptosis. The cell apoptotic ability was significantly decreased in the miR-93-5p mimic + oe-NC group by comparison with that in the NC mimic + oe-NC group (P<0.05; Fig. 6E), while there were no obvious differences between the NC mimic + oe-NC group and miR-93-5p mimic + oe-TGFβR2 group (P>0.05). These findings confirm that overexpressing miR-93-5p can promote proliferation, migration, invasion and inhibit cell apoptosis of EC cells upon TGFβR2 suppression.
Figure 6.
miR-93-5p promotes the proliferation, migration and invasion, and inhibits the apoptosis of EC cells by downregulating the expression of TGFβR2. EC9706 cells were transfected with miR-93-5p mimic + oe-NC, NC mimic + oe-NC, miR-93-5p mimic + oe-TGFβR2, respectively. (A) RT-qPCR detection of TGFβR2 mRNA expression in each transfection group. (B) WST-1 assay was performed to detect cell viability in each group. (C) Wound healing assay was applied to detect the migratory ability in each group (500 µm). (D) Transwell assay was carried out to detect cell invasive ability in each group (100 µm). (E) Flow cytometry was used to detect the cell apoptotic rate in each group. *P<0.05, compared to NC mimic + oe-NC. EC, esophageal carcinoma.
Discussion
miRNAs are types of small RNAs with a length of approximately 20-24 bp, and they serve as a tumor promoter or suppressor via regulating the expression of their specific target genes (14). Therefore, the regulation of miRNA expression can be used as a novel method for cancer diagnosis and treatment (15). miR-93-5p is located in the intron of the MCM7 gene and is a part of the cluster containing two other miRNAs (miR-25 and miR-106b) (8). The present study found that miR-93-5p was highly expressed in EC cells, which was consistent with previous findings on breast cancer (7), gastric cancer (16), prostate cancer (17) and colorectal cancer (6). Studies have reported that tje knockdown of miR-93-5p inhibits cell proliferation, migration and invasion in gastric cancer tissues (18). The present study also revealed that tje inhibition of miR-93-5p expression inhibited the proliferation, migration and invasion of EC cells, and promoted apoptosis.
During tumorigenesis and development, TGFβR2, a receptor serine/threonine kinase, initiates downstream TGF-β signaling (17), and thje loss or decrease of TGFβR2 expression can inhibit the TGF-β signaling, which is beneficial for early tumor growth (19). In the present study, it was predicted that TGFβR2 was expressed in low levels in EC tissues by bioinformatics analysis, and western blot analysis was performed to verify this prediction and further explore the association between TGFβR2 and the occurrence and development of EC. RT-qPCR then revealed that TGFβR2 mRNA expression was decreased in EC cell lines relative to that in normal esophageal cell line. It has been demonstrated that TGFβR2 is the major target of miR-93-5p in nasopharyngeal carcinoma invasion (20). In the present study, target prediction websites were used, and it was found that miR-93-5p bound to the 3′-UTR of TGFβR2, which was further verified by performing dual-luciferase reporter gene assay. It was then demonstrated that the overexpression of miR-93-5p inhibited the expression of TGFβR2, thereby inhibiting apoptosis and promoting the proliferation, migration and invasion of EC cells. However, miR-93-5p inhibitor was used to downregulate miR-93-5p expression, and opposite results were observed with the inhibition of miR-93-5p expression. When miR-93-5p and TGFβR2 were simultaneously overexpressed in the cells, no marked changes were observed in the proliferation, migration and invasion of the cells. All the findings described above indicate that miR-93-5p targets and downregulates the expression of TGFβR2, ultimately promoting the occurrence and development of EC cells.
In conclusion, the present study identified the regulatory effects of miR-93-5p on TGFβR2 in EC cells and clarified its role in biological cell behaviors. The results provide a potential target by inhibiting miR-93-5p or promoting TGFβR2 expression, which may provide additional insight into the mechanisms underlying the occurrence and development of EC and may lead to the development of novel strategies for EC diagnosis and treatment.
Supplementary Data
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
The data used to support the findings of this study are included in the current article or are available from the corresponding author upon request.
Authors' contributions
YC, WR, JD and KZ contributed to the study design. NW, JW and HZ conducted the literature search. NM acquired the data. YC, WR and KZ wrote the article. NM, GW, WS and YL performed the data analysis. All authors gave the final approval of the version to be submitted. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Nishimura T, Tamaoki M, Komatsuzaki R, Oue N, Taniguchi H, Komatsu M, Aoyagi K, Minashi K, Chiwaki F, Shinohara H, et al. SIX1 maintains tumor basal cells via transforming growth factor-β pathway and associates with poor prognosis in esophageal cancer. Cancer Sci. 2017;108:216–225. doi: 10.1111/cas.13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu K, He J, Pu W, Peng Y. The role of exportin-5 in MicroRNA biogenesis and cancer. Genomics Proteomics Bioinformatics. 2018;16:120–126. doi: 10.1016/j.gpb.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Chen S, Chen X, Sun KX, Xiu YL, Liu BL, Feng MX, Sang XB, Zhao Y. MicroRNA-93 promotes epithelial-mesenchymal transition of endometrial carcinoma cells. PLoS One. 2016;11:e0165776. doi: 10.1371/journal.pone.0165776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paul S, Lakatos P, Hartmann A, Schneider-Stock R, Vera J. Identification of miRNA-mRNA modules in colorectal cancer using rough hypercuboid based supervised clustering. Sci Rep. 2017;7:42809. doi: 10.1038/srep42809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li N, Miao Y, Shan Y, Liu B, Li Y, Zhao L, Jia L. MiR-106b and miR-93 regulate cell progression by suppression of PTEN via PI3K/Akt pathway in breast cancer. Cell Death Dis. 2017;8:e2796. doi: 10.1038/cddis.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du L, Zhao Z, Ma X, Hsiao TH, Chen Y, Young E, Suraokar M, Wistuba I, Minna JD, Pertsemlidis A. miR-93-directed downregulation of DAB2 defines a novel oncogenic pathway in lung cancer. Oncogene. 2014;33:4307–4315. doi: 10.1038/onc.2013.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu W, He J, Chen D, Zhang B, Xu L, Ma H, Liu X, Zhang Y, Le H. Expression of miR-29c, miR-93, and miR-429 as potential biomarkers for detection of early stage non-small lung cancer. PLoS One. 2014;9:e87780. doi: 10.1371/journal.pone.0087780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kucuksayan H, Akgun S, Ozes ON, Alikanoglu AS, Yildiz M, Dal E, Akca H. TGF-β-SMAD-miR-520e axis regulates NSCLC metastasis through a TGFBR2-mediated negative-feedback loop. Carcinogenesis. 2019;40:695–705. doi: 10.1093/carcin/bgy166. [DOI] [PubMed] [Google Scholar]
- 11.Malkoski SP, Haeger SM, Cleaver TG, Rodriguez KJ, Li H, Lu SL, Feser WJ, Barón AE, Merrick D, Lighthall JG, et al. Loss of transforming growth factor beta type II receptor increases aggressive tumor behavior and reduces survival in lung adenocarcinoma and squamous cell carcinoma. Clin Cancer Res. 2012;18:2173–2183. doi: 10.1158/1078-0432.CCR-11-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fricke F, Lee J, Michalak M, Warnken U, Hausser I, Suarez-Carmona M, Halama N, Schnölzer M, Kopitz J, Gebert J. TGFBR2-dependent alterations of exosomal cargo and functions in DNA mismatch repair-deficient HCT116 colorectal cancer cells. Cell Commun Signal. 2017;15:14. doi: 10.1186/s12964-017-0169-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Liu JJ, Zhang X, Wu XH. miR-93 promotes the growth and invasion of prostate cancer by upregulating its target genes TGFBR2, ITGB8, and LATS2. Mol Ther Oncolytics. 2018;11:14–19. doi: 10.1016/j.omto.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu YF, Yu JR, Yang Z, Zhu GX, Gao P, Wang H, Chen SY, Zhang J, Liu MY, Niu Y, et al. Promoter hypomethylation mediated upregulation of MicroRNA-10b-3p targets FOXO3 to promote the progression of esophageal squamous cell carcinoma (ESCC) J Exp Clin Cancer Res. 2018;37:301. doi: 10.1186/s13046-018-0966-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma DH, Li BS, Liu JJ, Xiao YF, Yong X, Wang SM, Wu YY, Zhu HB, Wang DX, Yang SM. miR-93-5p/IFNAR1 axis promotes gastric cancer metastasis through activating the STAT3 signaling pathway. Cancer Lett. 2017;408:23–32. doi: 10.1016/j.canlet.2017.08.017. [DOI] [PubMed] [Google Scholar]
- 17.Zhou H, Wu G, Ma X, Xiao J, Yu G, Yang C, Xu N, Zhang B, Zhou J, Ye Z, Wang Z. Attenuation of TGFBR2 expression and tumour progression in prostate cancer involve diverse hypoxia-regulated pathways. J Exp Clin Cancer Res. 2018;37:89. doi: 10.1186/s13046-018-0764-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schumacher K, Dagres N, Hindricks G, Husser D, Bollmann A, Kornej J. Characteristics of PR interval as predictor for atrial fibrillation: Association with biomarkers and outcomes. Clin Res Cardiol. 2017;106:767–775. doi: 10.1007/s00392-017-1109-y. [DOI] [PubMed] [Google Scholar]
- 19.Mishra S, Deng JJ, Gowda PS, Rao MK, Lin CL, Chen CL, Huang T, Sun LZ. Androgen receptor and microRNA-21 axis downregulates transforming growth factor beta receptor II (TGFBR2) expression in prostate cancer. Oncogene. 2014;33:4097–4106. doi: 10.1038/onc.2013.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lyu X, Fang W, Cai L, Zheng H, Ye Y, Zhang L, Li J, Peng H, Cho WC, Wang E, et al. TGFβR2 is a major target of miR-93 in nasopharyngeal carcinoma aggressiveness. Mol Cancer. 2014;13:51. doi: 10.1186/1476-4598-13-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used to support the findings of this study are included in the current article or are available from the corresponding author upon request.