Abstract
Numerous studies have confirmed that microRNAs (miRNAs or miRs) have important roles in cancer biogenesis and development including multiple myeloma (MM). MicroRNA-25-3p (miR-25-3p) has been proven to promote cancer progression, whereas its functions in MM has not yet been reported, at least to the best of our knowledge. Therefore, the present study aimed to investigate the function of miR-25-3p in MM and to identify the potential underlying mechanistic pathway. Herein, it was found that miR-25-3p expression was significantly increased in MM tissues and cell lines. The upregulation of miR-25-3p was closely associated with anemia, renal function impairment international staging system (ISS) staging and Durie-Salmon (D-S) staging. A high level of miR-25-3p was predictive of a poor prognosis of patients with MM. In vitro, the knockdown of miR-25-3p suppressed the proliferation and promoted the apoptosis of RPMI-8226 and U266 cells, while the overexpression of miR-25-3p exerted opposite effects. In addition, phosphatase and tensin homolog deleted on chromosome 10 (PTEN), a well-known tumor suppressor, was confirmed as a target of miR-25-3p in MM cells. Moreover, it was found that the PTEN expression levels were decreased, and inversely correlated with miR-25-3p expression levels in MM tissues. Further analyses revealed that the overexpression of PTEN exerted effects similar to those of miR-25-3p knockdown, whereas the knockdown of PTEN partially abolished the effects of miR-25-3p inhibitor on MM cells. Accompanied by PTEN induction, miR-25-3p promoted PI3K/AKT signaling pathway activation in MM cells. Collectively, these findings demonstrate critical roles for miR-25-3p in the pathogenesis of MM, and suggest that miR-25-3p may serve as a novel prognostic biomarker and therapeutic target of MM.
Keywords: microRNA-25-3p, multiple myeloma, phosphatase and tensin homolog deleted on chromosome 10, PI3K/AKT signaling pathway
Introduction
Multiple myeloma (MM) is a malignant plasma cell disorder, accounting for the leading cause of mortality in patients with hematological cancer (1). Despite recent and extensive progress being made in the treatment of MM, novel strategies for the more effective management of MM are urgently required (2-5). Thus, it is necessary to identify novel prognostic biomarkers and therapeutic regimens for patients with MM.
MicroRNAs (miRNAs or miRs), endogenous small (approximately 22 nucleotides in length) non-coding RNAs, act as unique regulators of gene expression by targeting mRNAs for translational repression or cleavage (6). Recently, increasing evidence has unveiled the roles of miRNAs in the development of MM (7-10). For example, Gupta et al found that circulatory levels of miR-203 were decreased in the serum of patients with MM, and may be an independent predictive biomarker for the diagnosis of MM (11). In addition to diagnosis, exploiting the therapeutic potentials of miRNAs has been reported in cancer research. Zhang et al demonstrated the tumor suppressive ability of miR-29b through the downregulation of Mcl-1, functioning as a potential therapeutic target for MM (12). Jia et al reported the antitumor role of miR-26b-5p by targeting JAG1 (13). These data suggest the crucial role of miRNAs in the progression of MM.
As one of the most well-studied miRNAs, miR-25-3p is aberrantly expressed and principally functions as an oncogenic miRNA in the development and progression of human cancers. For example, Zhang et al found that miR-25-3p promoted tumorigenesis by regulating CDKN1C expression in glioma (14). In liver cancer, Sanchez-Mejias et al also reported the oncogenic role of miR-25-3p with the inhibition of the SOCS5/mammalian target of rapamycin (mTOR) signaling axis (15). Notably, in MM, miR-25-3p has been found to be significantly upregulated in the serum of patients with MM, functioning as a potential diagnostic target in MM (16,17). Although miR-25-3p has been reported to be highly expressed in MM, the roles and molecular mechanisms of miR-25-3p in MM remain unknown.
Thus, the present study aimed to elucidate the potential involvement of miR-25-3p in MM and to explored the possible underlying mechanisms. The findings suggest that miR-25-3p may be a possible biomarker and effective therapeutic target for MM.
Materials and methods
Human tissue samples
Bone marrow samples from 50 patients with MM were obtained from the Department of Hematology, the First Affiliated Hospital of Xinxiang Medical University between June, 2017 and June, 2018. All patients were diagnosed based on the World Health Organization diagnostic criteria for MM (18,19). The inclusion criteria were as follows: i) Patients newly diagnosed with MM according to the World Health Organization diagnostic criteria for MM; ii) an age >18 years old; iii) patients had to be able to be regularly followed-up. The exclusion criteria were as follows: i) Patients with relapsed/refractory MM; ii) patients that had received chemotherapy, radiotherapy or other systematic treatments prior to enrollment; iii) patients with a history of solid tumors or hematological malignancies other than MM. At the same time, 10 healthy volunteers undergoing bone marrow transplant were recruited as the controls. All patient characteristics are presented in Table I. Written informed consents were acquired from all patients and healthy volunteers and the present study was approved by the Ethics Committee of the First Affiliated Hospital of Xinxiang Medical University. The samples were rapidly stored at -80°C until use.
Table I.
Association between miR-25-3p and the pathological characteristics of patients with MM.
| Clinical feature | Patients (total) n=50 | miR-25-3p expression
|
P-value | |
|---|---|---|---|---|
| High | Low | |||
| Sex | 0.9453 | |||
| Male | 22 | 10 | 12 | |
| Female | 28 | 13 | 15 | |
| Age (years) | 0.6233 | |||
| <50 | 17 | 7 | 10 | |
| ≥50 | 33 | 16 | 17 | |
| Anemia | 0.0150a | |||
| Mild | 20 | 5 | 15 | |
| Moderate/severe | 30 | 18 | 12 | |
| Impairment of renal function | 0.0208a | |||
| Renal inadequacy | 38 | 24 | 14 | |
| No impairment | 12 | 3 | 9 | |
| ISS staging | 0.0354a | |||
| Stage I | 21 | 6 | 15 | |
| Stage II-III | 29 | 17 | 12 | |
| D-S staging | 0.0100a | |||
| Stage I | 13 | 2 | 11 | |
| Stage II-III | 37 | 21 | 16 | |
P<0.05. MM, multiple myeloma; ISS, International Staging System; D-S, Durie-Salmon.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from the MM tissues and cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). For the detection of miR-25-3p, total RNA was reverse transcribed into cDNA using the TaqMan™ MicroRNA Reverse Transcription kit (cat no. 4366596, Thermo Fisher Scientific, Inc.). qPCR was conducted using the miScript SYBR-Green PCR kit (cat no. 218073, Qiagen GmbH) in a Light Cycler instrument (Bio-Rad Laboratories, Inc.). For the detection of phosphatase and tensin homolog deleted on chromosome 10 (PTEN) mRNA, total RNA was reverse transcribed into cDNA using a Reverse Transcription kit (cat no. RR047A, Takara Bio, Inc.). SYBR Premix Ex Taq II (Takara Bio, Inc., Tokyo, Japan) was then used for PCR. The thermocycling conditions were as follows: 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 10 min. Primers used for miR-25-3p and PTEN were as follows: miR-25-3p forward, 5′-CAT TGC ACT TGT CTC GGT CTG A-3′ and reverse, 5′-GCT GTC AAC GAT ACG CTA CGT AAC G-3′; U6 forward, 5′-GCT TCG GCA GCA CAT ATA CTA AAA T-3′ and reverse, 5′-CGC TTC ACG AAT TTG CGT GTC AT-3′; PTEN forward, 5′-CCA GGA CCA GAG GAA ACC T-3′ and reverse, 5′-GCT AGC CTC TGG ATT TGA-3′; and GAPDH forward, 5′-GTG GTG AAG ACG CCA GTG GA-3′ and reverse, 5′-CGA GCC ACA TCG CTC AGA CA-3′. U6 and GAPDH were used as internal controls for detecting miR-25-3p and PTEN expression, respectively, and fold changes were calculated through the 2−∆∆Cq method (20).
Cells and cell culture
The NCI-H929 (ATCC® CRL-9068), RPMI-8226 (ATCC® CCL-155) and U266 (ATCC® TIB-196) cell lines were purchased from the American Type Culture Collection (ATCC). The MM.1R cell line was purchased from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (cat. no. SCSP-505). Normal plasma cells (nPCs) were obtained from Procell Life Science & Technology Co., Ltd. All cells were cultured in RPMI-1640 medium (Cambrex) supplemented with 10% fetal bovine serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C and 5% CO2.
Cell transfection
The miR-25-3p mimics, mimics negative control (mimics NC), miR-25-3p inhibitor and inhibitor NC were purchased from RiboBio Co., Ltd. PTEN overexpression vector pcDNA-PTEN and pcDNA vector were purchased from GenePharma, Co. Ltd. In addition, PTEN siRNA (si-PTEN, 5′-CAU CAG CAU AUG GAA AGC UUC AUU U-3′) and si-Scramble (5′-CAU ACG UAU AGG GAA UUC ACC AUU U-3′) were also obtained from RiboBio Co., Ltd. RPMI-8226 and U266 cells (8.0×105/well) in 6-well plates were grown to approximately 80% confluency, and then respectively transfected with miR-25-3p inhibitor (20 nM), inhibitor NC (20 nM), si-PTEN (50 nM), or 2 µg pcDNA-PTEN using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). After 24 h, the cells were collected, and RT-qPCR was performed to ensure silencing was achieved.
Cell proliferation
For the detection of cell proliferation, RPMI-8226 and U266 cells (5×103/well) were seeded in 96-well plates for 24 h, and then transfected with 20 nM miRNA inhibitor and 50 nM si-PTEN, or 2 µg pcDNA-PTEN as described above. At 1, 2 and 3 days post-transfection, 10 µl CCK-8 solution (Dojindo Molecular Technologies, Inc.) were added and the cells were cultured at 37°C for an additional 2 h. The OD absorbance at 450 nm was then detected using an iMark microplate reader (Bio-Rad Laboratories, Inc.).
Cell apoptosis assay
For the detection of cell apoptosis, the Annexin V-FITC/PI apoptosis detection kit (cat no. ab14085, Abcam) was applied according to the manufacturer's instructions. Briefly, 48 h following transfection, cells were centrifuged at 300 × g at 4°C for 10 min and washed with PBS, and stained with Annexin V and propidium iodide (PI) for 15 min at room temperature in the dark. Apoptosis was then measured on a FACScan flow cytometer (Beckman Coulter, Inc.) and then analyzed using FlowJo 8.7.1 software (FlowJo, LLC). The results revealed healthy viable cells in the lower left quadrant (Q4) on the scatter plot as (FITC−/PI−); the lower right quadrant (Q3) represented early-stage apoptotic cells (FITC+/PI−); the upper right quadrant (Q2) represented necrotic cells and late-stage apoptotic cells (FITC+/PI+). The rate of apoptosis was calculated as follows: Apoptotic rate=percentage of early stage apoptotic cells (Q3) + percentage of late stage apoptotic cells (Q2). The experiment was repeated 3 times independently.
Luciferase assay
PicTar (http://pictar.mdc-berlin.de/) and TargetScan (http://www.targetscan.org) were used to search for the putative targets of miR-25-3p. The luciferase reporter plasmids (wt-PTEN-UTR-pGL3 or mut-PTEN-UTR-pGL3) were synthesized by GenePharma, Co., Ltd. 293T cells (8×104) (ATCC® CRL-11268) were co-transfected with the luciferase reporter along with miR-25-3p mimics/inhibitor using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). At 48 h post-transfection, the luciferase activity was measured using the dual-luciferase reporter assay system (Promega Corporation). Renilla luciferase was used to normalize the cell number at 48 h following transfection.
Western blot analysis
Proteins from the transfected cells were extracted using RIPA lysis buffer (EMD Millipore) containing protease inhibitors and phosphatase inhibitors. The protein concentration was determined using a bicinchoninic acid (BCA) assay (cat no. P6651, Beyotime Institute of Biotechnology). Briefly, the protein samples (40 µg/lane) were separated by 12% SDS-PAGE gel and transferred to polyvinylidene difluoride membranes (EMD Millipore). The membranes were then blocked with 5% skim milk for 2 h at room temperature, followed by incubation with PTEN anti-body (cat no. 9559; 1:2,000), p-AKT antibody (cat no. 4060; 1:1,000), AKT antibody (cat no. 4685; 1:1,000), p-mTOR antibody (cat no. 5536; 1:1,000 dilution), mTOR antibody (cat no. 2983; 1:2,000), PCNA (cat no. 13110; 1:2,000) and β-actin antibody (cat no. 3700, 1:2,000) at 4°C overnight. Anti-mouse IgG (H + L; cat no. 8887, 1:2,000) were used as the secondary antibodies for 1 h at room temperature. All antibodies were obtained from Cell Signaling Technology, Inc. Detection was performed using an enhanced chemiluminescence kit (cat no. 34096; GE Healthcare; Cytiva) and the densities of protein bands was analyzed using ImageJ software (version 1.46; National Institutes of Health).
Statistical analysis
The SPSS 19.0 software package (SPSS, Inc.) was applied to analyze the data. All data are presented as the means ± S.D. Differences between 2 groups were analyzed using an unpaired Student's t-test. Multiple comparisons were performed using one-way analysis of variance followed by Tukey's post hoc test. When only 2 groups were compared, a Student's t-test was conducted. The Chi-squared test was used to evaluate the difference of categorical variables in Table I. The differences in overall survival were assessed by Kaplan-Meier survival analysis and the log rank test. Patients were divided into a low expression group and high expression group by the cut-off of the median expression value of miR-25-3p. Pearson's correlation coefficient was used for correlation analysis between miRNA and mRNA levels. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of miR-25-3p is upregulated in MM and is associated with patient clinicopathologic parameters
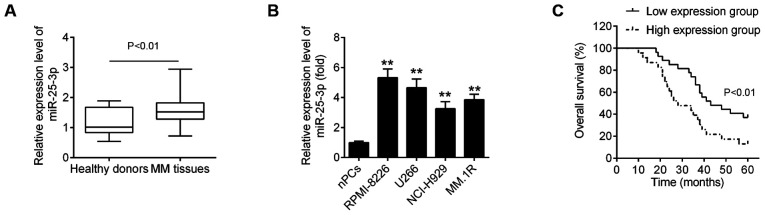
To evaluate the expression status of miR-25-3p in MM, the relative expression of miR-25-3p was first assessed in 50 bone marrow samples from patients with MM and in 10 bone marrow samples from healthy donors. As shown in Fig. 1A, miR-25-3p expression was significantly upregulated in MM tissues compared with healthy donors. In addition, the levels of miR-25-3p expression were measured in 4 MM cell lines relative to the nPCs. Compared with the nPCs, miR-25-3p expression was upregulated, with differential expression levels, in the 4 MM cell lines (Fig. 1B), particularly in the RPMI-8226 and U266 cells.
Figure 1.
miR-25-3p expression is significantly upregulated in MM tissues and MM cell lines. (A) Expression of miR-25-3p was detected in 50 bone marrow tissue samples from MM patients and 10 normal bone marrow tissue samples from healthy donors by RT-qPCR. (B) Expression of miR-25-3p in MM cell lines was determined by RT-qPCR. nPCs cells were used as control cells. Data are presented as the means ± SD of 3 independent experiments. **P<0.01 vs. nPCs. (C) Prognostic value of miR-25-3p for MM patients assessed by Kaplan-Meier analysis. MM, multiple myeloma; nPCs, normal plasma cells.
Subsequently, to explore the clinical and pathological role of miR-25-3p in MM, the association between miR-25-3p expression and the clinicopathological characteristics of the patients with MM we analyzed. As shown in Table I, a high expression of miR-25-3p was closely associated with anemia, renal function impairment, the international staging system (ISS) staging and Durie-Salmon (D-S) staging (Table I). However, other clinicopathological parameters, such as sex and age exhibited no significant association with miR-25-3p. It was also found that the overall survival (OS) rate of patients with MM was longer in the miR-25-3p low expression group compared with the miR-25-3p high expression group (Fig. 1C). Collectively, these data indicate that miR-25-3p expression was indeed upregulated in MM and was mainly associated with disease progression.
Knockdown of miR-25-3p suppresses cell proliferation and induces cell apoptosis
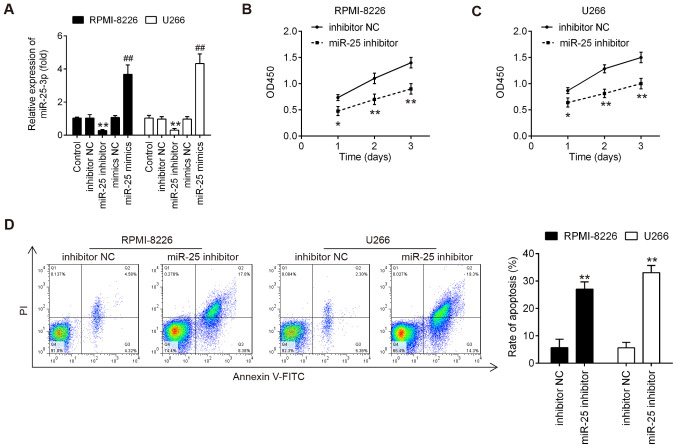
To determine the role of miR-25-3p in MM cells, both the RPMI-8226 and U266 cells were transfected with miR-25-3p inhibitor/mimics or mimics/inhibitor NC. As was expected, miR-25-3p expression was notably increased or decreased following transfection with mimics or inhibitor, respectively (Fig. 2A), which confirmed the transfection efficiency. According to the results of CCK-8 assay, miR-25-3p knockdown markedly inhibited cell proliferation at varying degrees in both the RPM-I8226 and U266 cells (Fig. 2B and C). The present study then determined whether the effects of miR-25-3p on cell proliferation may be associated with apoptosis. To determine this, these treated cells were subjected to FITC-Annexin V/PI staining followed by flow cytometric analysis. As shown in Fig. 2D, miR-25-3p knockdown resulted in a significant increase in the apoptotic portion of both the RPMI-8226 and U266 cells compared to the inhibitor NC. These results suggested that miR-25-3p knockdown inhibited cell proliferation by promoting cell apoptosis in vitro.
Figure 2.
Knockdown of miR-25-3p suppresses cell proliferation and induces cell apoptosis. RPMI-8226 and U266 cells were transfected with the miR-25-3p inhibitor or miR-25-3p mimics for 48 h, and the cells were then used for analysis. (A) Transfection efficiency was assessed by RT-qPCR. (B and C) Cell proliferation was measured by CCK-8 assay at the indicated time points. (D) Cell apoptosis was measured by flow cytometry. Data are presented as the means ± SD of 3 independent experiments. *P<0.05, **P<0.01 vs. inhibitor NC group; ##P<0.01 vs. mimics NC group.
Overexpression of miR-25-3p promotes the proliferation and inhibits the apoptosis of MM cells
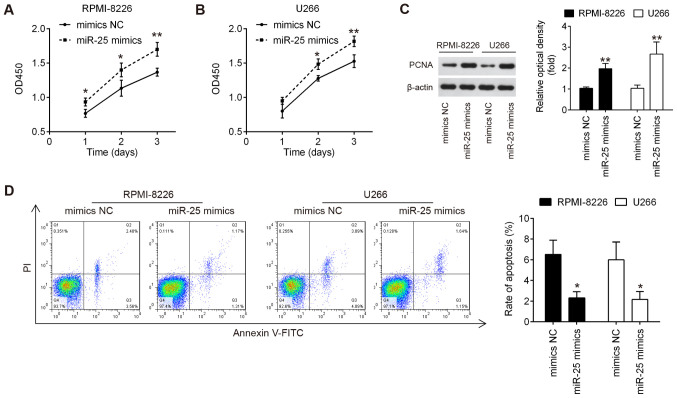
The effects of miR-25-3p overexpression on cell proliferation and apoptosis in vitro were then evaluated. The RPMI-8226 and U266 cells were transfected with miR-25-3p mimics or mimics NC. As shown in Fig. 3A and B, miR-25-3p upregulation markedly promoted the proliferation of both RPMI-8226 and U266 cells. The results of western blot analysis revealed that compared with the mimics NC group, the expression levels of PCNA in both the RPMI-8226 and U266 cells were significantly increased in the miR-25-3p mimics group (Fig. 3C). In addition, compared with the mimics NC group, the apoptotic portion of cells in the miR-25-3p mimics group was significantly decreased (Fig. 3D). All results suggest that miR-25-3p overexpression promoted cell proliferation by inhibiting apoptosis in vitro.
Figure 3.
Overexpression of miR-25-3p promotes cell proliferation and inhibits cell apoptosis. RPMI-8226 and U266 cells were transfected with the miR-25-3p mimics or mimics NC for 48 h, and then cells were used for analysis. (A and B) Cell proliferation was measured by CCK-8 assay at the indicated time points. (C) Expression of PCNA protein was measured by western blot analysis. (D) Cell apoptosis was measured by flow cytometry. Data are presented as the means ± SD of 3 independent experiments. *P<0.05, **P<0.01 vs. inhibitor NC group.
PTEN was a direct target of miR-25-3p
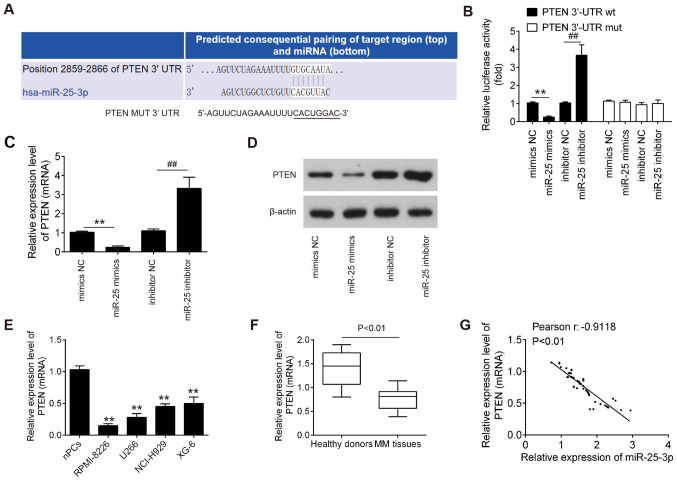
To examine the molecular mechanism by which miR-25-3p functions in MM, candidate target genes of miR-25-3p were computationally screened using the online database PicTar and TargetScan. Among several predicted target genes, PTEN was identified as a potential target of miR-25-3p (Fig. 4A). To verify whether miR-25-3p directly binds to PTEN, a dual luciferase reporter assay was conducted. It was found that miR-25-3p upregulation markedly inhibited, whereas miR-25-3p inhibition promoted the luciferase activity of the PTEN-2-3′UTR wt reporter; however, co-transfection with mutant vector did not lead to any changes in luciferase activity (Fig. 4B). To further confirm whether PTEN is regulated by miR-25-3p, the mRNA and protein expression levels of PTEN in the RPMI-8226 and U266 cells were measured by RT-qPCR and western blot analysis, respectively. It was found that PTEN expression was significantly decreased following transfection with miR-25-3p mimics, while it was increased following miR-25-3p inhibitor transfection (Fig. 4C and D). Subsequently, the expression levels of PTEN in MM cell lines and the above-mentioned clinical samples were detected by RT-qPCR. As shown in Fig. 4E and F, PTEN expression was significantly downregulated in MM cell lines and MM tissues, compared with the nPCs and healthy donor tissues. In addition, an evident inverse correlation was observed between the PTEN and miR-25-3p expression levels in tumor tissues (Fig. 4G, r=−0.9118; P<0.01). These results indicate that PTEN may be a downstream target of miR-25-3p.
Figure 4.
PTEN is a direct target of miR-25-3p. (A) Schematic of the PTEN1 3′UTR containing the miR-25-3p binding sites. (B) Luciferase assay of 293T cells co-transfected with Firefly luciferase constructs containing the PTEN wt or mut 3′UTRs and miR-25-3p mimics, mimics NC, miR-25-3p inhibitor or inhibitor NC, as indicated (n=3). Data are presented as the means ± SD of 3 independent experiments. (C and D) RPMI-8226 and U266 cells were transfected with miR-25-3p mimics, mimics NC, miR-25-3p inhibitor or inhibitor NC for 48 h, and the expression levels of PTEN mRNA and protein were determined by RT-qPCR and western blot analysis, respectively. Data are presented as the means ± SD of 3 independent experiments. **P<0.01 vs. mimics NC; ##P<0.01 vs. inhibitor NC. (E) Expression of miR-25-3p in MM cell lines was determined by RT-qPCR. nPCs cells were used as control cells. Data are presented as the means ± SD of 3 independent experiments. **P<0.01 vs. nPCs group. (F) PTEN expression was measured by RT-qPCR in 50 bone marrow tissue samples from MM patients and 10 normal bone marrow tissue samples from healthy donors. P<0.01 vs. healthy donor group. (G) Pearson's correlation analysis was used to analyze the correlation between the expression of PTEN and the expression of miR-25-3p expression in multiple myeloma tissues (r=−0.9118, P<0.01).
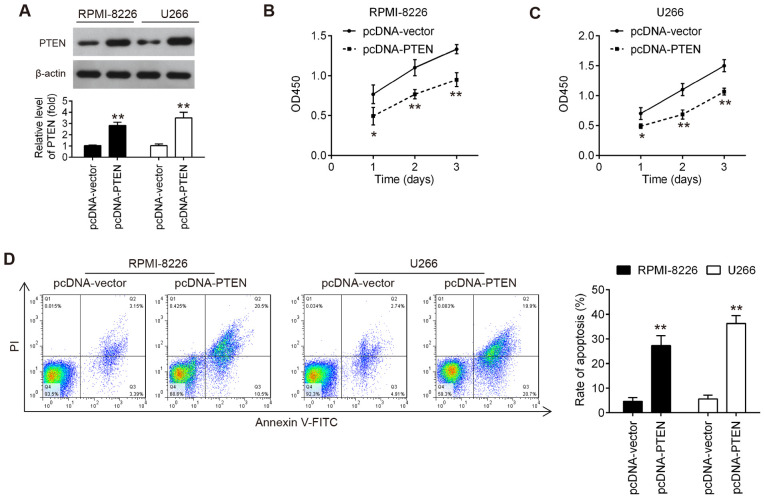
Overexpression of PTEN inhibits the proliferation and promotes the apoptosis of MM cells
The results mentioned above demonstrated that miR-25-3p inhibited cell proliferation in vitro and miR-25-3p directly targeted PTEN. The present study then wished to determine whether the upregulation of PTEN exerted a similar effect to that of miR-20a knockdown. To address this question, pcDNA-PTEN plasmid was transfected into RPMI-8226 and U266 cells. As shown in Fig. 5A, the PTEN protein level was notably increased in the RPMI-8226 and U266 cells transfected with the PTEN overexpression plasmid. Cell proliferation and apoptosis were then estimated. As shown by the results of CCK-8 assay, PTEN upregulation significantly suppressed the proliferation of RPMI-8226 and U266 cells compared with the pcDNA-vector group (Fig. 5B and C). It was also found that PTEN upregulation markedly promoted the apoptosis of RPMI-8226 and U266 cells, compared with the pcDNA-vector group (Fig. 5D). Consequently, PTEN upregulation exerted similar effects to those of miR-25-3p knockdown in MM cells.
Figure 5.
Overexpression of PTEN inhibits the proliferation and promotes the apoptosis of MM cells. RPMI-8226 and U266 cells were transfected with pcDNA-PTEN or pcDNA-vector for 48 h, and the cells were then used for analysis. (A) Transfection efficiency was assessed by western blot analysis. (B and C) Cell proliferation was measured by CCK-8 assay at the indicated time points. (D) Cell apoptosis was measured by flow cytometry. Data are presented as the means ± SD of 3 independent experiments. *P<0.05, **P<0.01 vs. pcDNA-vector group.
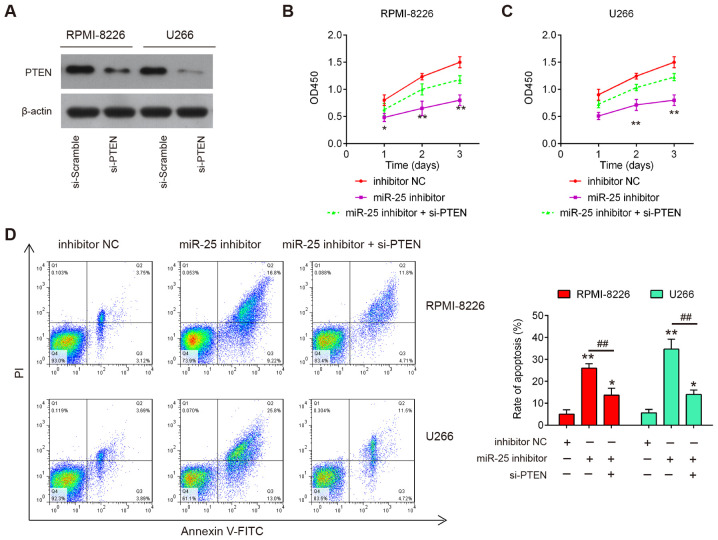
PTEN inhibition reverses the effects of miR-25-3p on the proliferation and apoptosis of MM cells
To verify whether PTEN mediates the inhibitory effects of miR-25-3p on cell proliferation and cell apoptosis, rescue experiments were performed by transfecting si-PTEN with miR-25-3p inhibitor into RPMI-8226 and U266 cells. As shown in Fig. 6A, the protein expression levels of PTEN were notably decreased in the RPMI-8226 and U266 cells following si-PTEN transfection. The results of CCK8 assay revealed that PTEN knockdown partially reversed the inhibitory effects of miR-25-3p inhibitor on cell proliferation (Fig. 6B and C). Moreover, the increased apoptotic portion induced by miR-25-3p inhibitor was significantly reduced by PTEN knockdown in the RPMI-8226 and U266 cells (Fig. 6D). Taken together, these findings demonstrated that miR-25-3p inhibited cell proliferation and promotes apoptosis by directly targeting PTEN expression in MM cells.
Figure 6.
Knockdown of PTEN markedly reverses the effects of miR-25-3p inhibition on the proliferation and apoptosis of MM cells. (A) Transfection efficiency was assessed by western blot analysis following si-PTEN transfection in RPMI-8226 and U266 cells. RPMI-8226 and U266 cells were co-transfected with the si-PTEN and miR-25-3p inhibitor for 48 h, and the cells were then used for analysis. (B and C) Cell proliferation was measured by CCK-8 assay at the indicated time points. (D) Cell apoptosis was measured by flow cytometry. Data are represented as the means ± SD of 3 independent experiments. *P<0.05, **P<0.01 vs. inhibitor NC group; ##P<0.01 vs. miR-25-3p inhibitor.
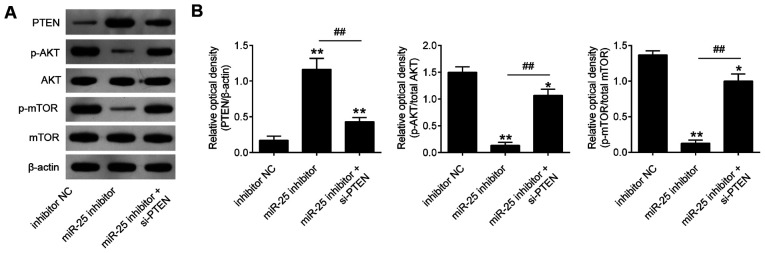
Knockdown of miR-25-3p blocks the activity of PTEN/PI3K/AKT pathway
It has been well recognized that PTEN negatively regulates the activity of the Akt pathway, and the Akt pathway is closely associated with cell growth and cell apoptosis in human cancers (21-23). To investigate whether miR-25-3p affects the activity of the PI3K/Akt signaling pathway, the expression levels of downstream proteins in the PI3K/Akt signaling pathway, namely AKT, p-AKT, mTOR and p-mTOR, were evaluated in RPMI-8226 and U266 cells. The results revealed that miR-25-3p inhibition elevated PTEN expression and reduced p-AKT and p-mTOR expression levels relative to the control group (Fig. 7). These data suggest that miR-25-3p knockdown blocked the activity of PTEN/PI3K/AKT pathway in MM cells.
Figure 7.
Knockdown of miR-25-3p suppresses the activation of the PTEN/PI3K/AKT pathway. RPMI-8226 and U266 cells were co-transfected with the si-PTEN and miR-25-3p inhibitor for 48 h, and the cells were then used for analysis. (A) The levels of PTEN, p-AKT and p-mTOR were measured by western blot analysis. (B) The bands were semi-quantitatively analyzed using ImageJ software. β-actin protein was used as the inner control of PTEN; the results of phosphorylated proteins were calculated as the ratio of phosphorylated to total protein. Data are presented as the means ± SD of 3 independent experiments. *P<0.05, **P<0.01 vs. inhibitor NC group; ##P<0.01 vs. miR-25-3p inhibitor.
Discussion
In the present study, it was found that miR-25-3p expression was upregulated in MM tissues and cell lines; high levels of miR-25-3p were closely associated with the clinicopathological characteristics of patients with MM. Moreover, miR-25-3p knockdown suppressed cell proliferation and promoted cell apoptosis by blocking the activity of the PTEN/PI3K/AKT pathway. All these findings suggest that miR-25-3p may prove to be a novel biomarker and therapeutic target for MM.
Previous studies have demonstrated that miR-25-3p expression is upregulated in MM tissues and functions as a non-invasive prognosis biomarker (16,17). Consistent with these studies, the present study found miR-25-3p expression was significantly upregulated in both MM tissues and MM cell lines. Taken together, these results suggest that an increased miR-25-3p expression is a frequent event in human MM tissues and may be involved in carcinogenesis as a tumor oncogene. Moreover, its high expression is predictive of a poor prognosis of patients with MM. In view of the above, it was hypothesized that miR-25-3p may participate in the progression of MM.
Previous studies have described the oncogenic role of miR-25-3p in several types of cancer (24,25). For example, in renal tumors, miR-25-3p expression is elevated in renal cancer tissues, and is associated with the growth of renal cancer cells (26). In melanoma, miR-25 directly targets DKK3, and thereby reduces its downstream signaling, the WNT/β-catenin pathway to promote melanoma cell proliferation (27). In gastric cancer, miR-25-3p knockdown has been shown to suppress tumor xenograft growth in mice (28). Liu et al found that the upregulation of miR-25-3p was closely associated with the progression and a poor prognosis of patients with cholangio-carcinoma (29). Although previous studies have demonstrated its high expression in MM tissue samples, the biological function of miR-25-3p in MM remains unknown. Herein, it was found that miR-25-3p knockdown suppressed the cell proliferation by promoting cell apoptosis, while miR-25-3p overexpression exerted an opposite effect on RPMI-8226 and U266 cells. Therefore, these data indicate that miR-25-3p may function as an oncogenic miRNA in the progression of MM.
As one of the most well-studied miRNAs, miR-25-3p is aberrantly expressed and principally functions as an oncogenic miRNA in cancer development and progression. For example, miR-25-3p has been shown to be significantly upregulated in triple-negative breast cancer (TNBC) tissues and cell lines, and may serve as a novel diagnostic and therapeutic target for TNBC (30). Wan et al also found that miR-25-3p was upregulated in retinoblastoma tissues and cell lines, and the enforced expression of miR-25-3p increased cell growth, cell migration and invasion in vitro, as well as promoted tumor xenograft growth in vivo (31). The increased expression of miR-25-3p has also been reported to be associated with liver, prostate and stomach cancers (32-35). Notably, previous studies demonstrated that miR-25-3p expression was significantly upregulated in MM tissues, suggesting that it may be considered as a potential prognostic and diagnostic marker in MM (16,17). Taken together, these results suggest that an increased miR-25-3p expression may be associated with the progression of MM.
One of the best strategies with which to understand the fundamental function of miRNAs is via the elucidation of their functional targets. In the present study, through the online data-bases, PicTar and TargetScan, PTEN was identified as a target gene of miR-25-3p. PTEN is a well-known tumor suppressor gene in various types of tumors, and plays an important role in regulating cell proliferation and cell apoptosis (36). A previous study by Jiang et al demonstrated that PTEN was expressed in low levels in MM (37). Wan et al found that PTEN was a direct target of miR-25-3p in retinoblastoma (31). However, whether PTEN is regulated by miR-25-3p in MM cells has not yet been determined, at least to the best of our knowledge. In the present study, the results revealed that miR-25-3p knockdown exhibited tumor suppressive roles in MM cells by targeting PTEN.
PTEN is a key upstream of the PI3K/Akt signaling pathway (36,38) and activated PI3K/Akt signaling contributes to cell growth and apoptosis in a number of tumors, including MM (37). In MM, PI3K/Akt is constitutively active and contributes to the the proliferation of MM cells, which suggests that the inhibition of this pathway may be a potential therapeutic approach for MM (39-42). Jiang et al demonstrated that miR-20a inhibited the proliferation, migration and apoptosis of MM cells, and that these effects were associated with PI3K/AKT signaling pathway inactivation (37). The results of the present study demonstrated that miR-25-3p knockdown markedly enhanced the expression levels of PTEN, leading to the subsequent downregulation of Akt and mTOR phosphorylation. This may explain why miR-25-3p inhibition suppressed the proliferation and promoted the apoptosis of MM cells.
In conclusion, the present study found that miR-25-3p expression was frequently increased in MM tissues and may serve as a prognostic biomarker in patients with MM. Mechanically, the data indicated that miR-25-3p knockdown suppressed cell proliferation through the PTEN/PI3K/AKT pathway. The findings may provide a potential target for the prevention and treatment of MM.
Acknowledgments
Not applicable.
Funding
This study was supported by the Key Projects of Science and Technology of Henan Province (grant no. 182102311141) and the Research Project of Medical Education in Henan Province (grant no. Wjlx2017083).
Availability of data and materials
All data generated and/or analyzed during the present study are included in this published article.
Authors' contributions
YZi conceived and designed the experiments. YZh, YW, LZ, RY and YH performed the experiments and analyzed the data. YZi contributed reagents/materials/analysis tools and wrote the manuscript. All authors have read and agreed to the final version of manuscript.
Ethics approval and consent to participate
Written informed consents were acquired from all patients and healthy volunteers. The present study was approved by the Ethics Committee of the First Affiliated Hospital of Xinxiang Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Terpos E, Ntanasis-Stathopoulos I, Gavriatopoulou M, Dimopoulos MA. Pathogenesis of bone disease in multiple myeloma: From bench to bedside. Blood Cancer J. 2018;8:7. doi: 10.1038/s41408-017-0037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nahi H, Våtsveen TK, Lund J, Heeg BM, Preiss B, Alici E, Møller MB, Wader KF, Møller HE, Grøseth LA, et al. Proteasome inhibitors and IMiDs can overcome some high-risk cytogenetics in multiple myeloma but not gain 1q21. Eur J Haematol. 2016;96:46–54. doi: 10.1111/ejh.12546. [DOI] [PubMed] [Google Scholar]
- 3.Harding T, Baughn L, Kumar S, Van Ness B. The future of myeloma precision medicine: Integrating the compendium of known drug resistance mechanisms with emerging tumor profiling technologies. Leukemia. 2019;33:863–883. doi: 10.1038/s41375-018-0362-z. [DOI] [PubMed] [Google Scholar]
- 4.Velez R, Turesson I, Landgren O, Kristinsson SY, Cuzick J. Incidence of multiple myeloma in Great Britain, Sweden, and Malmö, Sweden: The impact of differences in case ascertainment on observed incidence trends. BMJ Open. 2016;6:e009584. doi: 10.1136/bmjopen-2015-009584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen D, Zhou D, Xu J, Zhou R, Ouyang J, Chen B. Prognostic value of 1q21 gain in multiple myeloma. Clin Lymphoma Myeloma Leuk. 2019;19:e159–e164. doi: 10.1016/j.clml.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Handa H, Murakami Y, Ishihara R, Kimura-Masuda K, Masuda Y. The role and function of microRNA in the pathogenesis of multiple myeloma. Cancers (Basel) 2019;11:1378. doi: 10.3390/cancers11111738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adamia S, Abiatari I, Amin SB, Fulciniti M, Minvielle S, Li C, Moreau P, Avet-Loiseau H, Munshi NC, Anderson KC. The effects of MicroRNA deregulation on pre-RNA processing network in multiple myeloma. Leukemia. 2020;34:167–179. doi: 10.1038/s41375-019-0498-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu YY, Song YQ, Huang ZM, Zhang HB, Chen M. MicroRNA-26a inhibits multiple myeloma cell growth by suppressing cyclin-dependent kinase 6 expression. Kaohsiung J Med Sci. 2019;35:277–283. doi: 10.1002/kjm2.12057. [DOI] [PubMed] [Google Scholar]
- 10.Tian F, Zhan Y, Zhu W, Li J, Tang M, Chen X, Jiang J. MicroRNA-497 inhibits multiple myeloma growth and increases susceptibility to bortezomib by targeting Bcl-2. Int J Mol Med. 2019;43:1058–1066. doi: 10.3892/ijmm.2018.4019. [DOI] [PubMed] [Google Scholar]
- 11.Gupta N, Kumar R, Seth T, Garg B, Sati HC, Sharma A. Clinical significance of circulatory microRNA-203 in serum as novel potential diagnostic marker for multiple myeloma. J Cancer Res Clin Oncol. 2019;145:1601–1611. doi: 10.1007/s00432-019-02896-1. [DOI] [PubMed] [Google Scholar]
- 12.Zhang YK, Wang H, Leng Y, Li ZL, Yang YF, Xiao FJ, Li QF, Chen XQ, Wang LS. Overexpression of microRNA-29b induces apoptosis of multiple myeloma cells through down regulating Mcl-1. Biochem Biophys Res Commun. 2011;414:233–239. doi: 10.1016/j.bbrc.2011.09.063. [DOI] [PubMed] [Google Scholar]
- 13.Jia CM, Tian YY, Quan LN, Jiang L, Liu AC. miR-26b-5p suppresses proliferation and promotes apoptosis in multiple myeloma cells by targeting JAG1. Pathol Res Pract. 2018;214:1388–1394. doi: 10.1016/j.prp.2018.07.025. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J, Gong X, Tian K, Chen D, Sun J, Wang G, Guo M. miR-25 promotes glioma cell proliferation by targeting CDKN1C. Biomed Pharmacother. 2015;71:7–14. doi: 10.1016/j.biopha.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Sanchez-Mejias A, Kwon J, Chew XH, Siemens A, Sohn HS, Jing G, Zhang B, Yang H, Tay Y. A novel SOCS5/miR-18/miR-25 axis promotes tumorigenesis in liver cancer. Int J Cancer. 2019;144:311–321. doi: 10.1002/ijc.31857. [DOI] [PubMed] [Google Scholar]
- 16.Navarro A, Diaz T, Tovar N, Pedrosa F, Tejero R, Cibeira MT, Magnano L, Rosiñol L, Monzó M, Bladé J, Fernández de Larrea C. A serum microRNA signature associated with complete remission and progression after autologous stem-cell transplantation in patients with multiple myeloma. Oncotarget. 2015;6:1874–1883. doi: 10.18632/oncotarget.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiang T, Hu AX, Sun P, Liu G, Liu G, Xiao Y. Identification of four potential predicting miRNA biomarkers for multiple myeloma from published datasets. PeerJ. 2017;5:e2831. doi: 10.7717/peerj.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palumbo A, Avet-Loiseau H, Oliva S, Lokhorst HM, Goldschmidt H, Rosinol L, Richardson P, Caltagirone S, Lahuerta JJ, Facon T, et al. Revised international staging system for multiple myeloma: A report from international myeloma working group. J Clin Oncol. 2015;33:2863–2869. doi: 10.1200/JCO.2015.61.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.International Myeloma Working Group Criteria for the classification of monoclonal gammopathies, multiple myeloma and related disorders: A report of the international myeloma working group. Br J Haematol. 2003;121:749–757. doi: 10.1046/j.1365-2141.2003.04355.x. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Franke TF. PI3K/Akt: Getting it right matters. Oncogene. 2008;27:6473–6488. doi: 10.1038/onc.2008.313. [DOI] [PubMed] [Google Scholar]
- 22.Wang X, Jiang X. PTEN: A default gate-keeping tumor suppressor with a versatile tail. Cell Res. 2008;18:807–816. doi: 10.1038/cr.2008.83. [DOI] [PubMed] [Google Scholar]
- 23.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 24.Razumilava N, Bronk SF, Smoot RL, Fingas CD, Werneburg NW, Roberts LR, Mott JL. miR-25 targets TNF-related apoptosis inducing ligand (TRAIL) death receptor-4 and promotes apoptosis resistance in cholangiocarcinoma. Hepatology. 2012;55:465–475. doi: 10.1002/hep.24698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng S, Pan W, Jin Y, Zheng J. MiR-25 promotes ovarian cancer proliferation and motility by targeting LATS2. Tumour Biol. 2014;35:12339–12344. doi: 10.1007/s13277-014-2546-0. [DOI] [PubMed] [Google Scholar]
- 26.Boguslawska J, Rodzik K, Poplawski P, Kędzierska H, Rybicka B, Sokół E, Tański Z, Piekiełko-Witkowska A. TGF-beta1 targets a microRNA network that regulates cellular adhesion and migration in renal cancer. Cancer Lett. 2018;412:155–169. doi: 10.1016/j.canlet.2017.10.019. [DOI] [PubMed] [Google Scholar]
- 27.Huo J, Zhang Y, Li R, Wang Y, Wu J, Zhang D. Upregulated MicroRNA-25 mediates the migration of melanoma cells by targeting DKK3 through the WNT/β-catenin pathway. Int J Mol Sci. 2016;17:1124. doi: 10.3390/ijms17111124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ning L, Zhang M, Zhu Q, Hao F, Shen W, Chen D. miR-25-3p inhibition impairs tumorigenesis and invasion in gastric cancer cells in vitro and in vivo. Bioengineered. 2020;11:81–90. doi: 10.1080/21655979.2019.1710924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu H, Ma L, Wang J. Overexpression of miR-25 is associated with progression and poor prognosis of cholangiocarcinoma. Exp Ther Med. 2019;18:2687–2694. doi: 10.3892/etm.2019.7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen H, Pan H, Qian Y, Zhou W, Liu X. MiR-25-3p promotes the proliferation of triple negative breast cancer by targeting BTG2. Mol Cancer. 2018;17:4. doi: 10.1186/s12943-017-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wan W, Wan W, Long Y, Li Q, Jin X, Wan G, Zhang F, Lv Y, Zheng G, Li Z, Zhu Y. MiR-25-3p promotes malignant phenotypes of retinoblastoma by regulating PTEN/Akt pathway. Biomed Pharmacother. 2019;118:109111. doi: 10.1016/j.biopha.2019.109111. [DOI] [PubMed] [Google Scholar]
- 32.Peng G, Yang C, Liu Y, Shen C. miR-25-3p promotes glioma cell proliferation and migration by targeting FBXW7 and DKK3. Exp Ther Med. 2019;18:769–778. doi: 10.3892/etm.2019.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim YK, Yu J, Han TS, Park SY, Namkoong B, Kim DH, Hur K, Yoo MW, Lee HJ, Yang HK, Kim VN. Functional links between clustered microRNAs: Suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res. 2009;37:1672–1681. doi: 10.1093/nar/gkp002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Tan W, Neo TW, Aung MO, Wasser S, Lim SG, Tan TM. Role of the miR-106b-25 microRNA cluster in hepatocellular carcinoma. Cancer Sci. 2009;100:1234–1242. doi: 10.1111/j.1349-7006.2009.01164.x. [DOI] [PubMed] [Google Scholar]
- 35.Poliseno L, Salmena L, Riccardi L, Fornari A, Song MS, Hobbs RM, Sportoletti P, Varmeh S, Egia A, Fedele G, et al. Identification of the miR-106b~25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci Signal. 2010;3:ra29. doi: 10.1126/scisignal.2000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carnero A, Blanco-Aparicio C, Renner O, Link W, Leal JF. The PTEN/PI3K/AKT signalling pathway in cancer, therapeutic implications. Curr Cancer Drug Targets. 2008;8:187–198. doi: 10.2174/156800908784293659. [DOI] [PubMed] [Google Scholar]
- 37.Jiang Y, Chang H, Chen G. Effects of microRNA-20a on the proliferation, migration and apoptosis of multiple myeloma via the PTEN/PI3K/AKT signaling pathway. Oncol Lett. 2018;15:10001–10007. doi: 10.3892/ol.2018.8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qu L, Gao Y, Sun H, Wang H, Liu X, Sun D. Role of PTEN-Akt-CREB signaling pathway in nervous system impairment of rats with chronic arsenite exposure. Biol Trace Elem Res. 2016;170:366–372. doi: 10.1007/s12011-015-0478-1. [DOI] [PubMed] [Google Scholar]
- 39.Aziz AUR, Farid S, Qin K, Wang H, Liu B. PIM kinases and their relevance to the PI3K/AKT/mTOR pathway in the regulation of ovarian cancer. Biomolecules. 2018;8:7. doi: 10.3390/biom8010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waniczek D, Śnietura M, Lorenc Z, Nowakowska-Zajdel E, Muc-Wierzgon M. Assessment of PI3K/AKT/PTEN signaling pathway activity in colorectal cancer using quantum dot-conjugated antibodies. Oncol Lett. 2018;15:1236–1240. doi: 10.3892/ol.2017.7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu T, Li L, Liu W, Ya B, Cheng H, Xin Q. Silencing of NADPH oxidase 4 attenuates hypoxia resistance in neuroblastoma cells SH-SY5Y by inhibiting PI3K/Akt-dependent glycolysis. Oncol Res. 2019;27:525–532. doi: 10.3727/096504018X15179668157803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu H, Li J, Zhou ZG. NEAT1 promotes cell proliferation in multiple myeloma by activating PI3K/AKT pathway. Eur Rev Med Pharmacol Sci. 2018;22:6403–6411. doi: 10.26355/eurrev_201810_16053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated and/or analyzed during the present study are included in this published article.