Abstract
Biosensors are important devices in clinical diagnostics, food processing, and environmental monitoring for detecting various analytes, especially viruses. These biosensors provide rapid and effective instruments for qualitative and quantitative detection of infectious diseases in real-time. Here, we report the development of biosensors based on various techniques. Additionally, we will explain the mechanisms, advantages, and disadvantages of the most common biosensors that are currently used for viral detection, which could be optical (e.g., surface-enhanced Raman scattering (SERS), Surface plasmon resonance (SPR)) and electrochemical biosensors. Based on that, this review recommends methods for efficient, simple, low-cost, and rapid detection of SARS-CoV-2 (the causative agent of COVID-19) that employ the two types of biosensors depending on attaching hemoglobin β-chain and binding of specific antibodies with SARS-CoV-2 antigens, respectively.
Keywords: COVID-19, Rapid detection, Viral biosensor, Bioreceptors, Electrochemical, SPR, SARS-CoV-2, Spike proteins, ACE2 receptors
Graphical abstract
1. Introduction
Viruses are obligatory intracellular parasites that require a suitable host for replication [1,2]. It is established that viruses constantly changing their genetic makeup as a mechanism to evade the host immune system and potentially could cause major diseases and even death [3]. Viral pathogens can be detected by traditional methods, for example; Cell culture [4], hemagglutination inhibition test, i [5] and complement fixation test [6]. Electron microscopy, can also be used for viral imaging. Electron microscope, on the contrary to other viral detection techniques, requires no organism-specific reagents [7]. Shell vial technique, is also used to detect viruses in various bodily fluids. Cell monolayers of different specimens such as cerebrospinal fluid, stools, urine, genital fluids, etc. are precipitated in flat-bottom tubes by centrifugation, and then incubated for viral detection by immuno-detection methods using specific antibody and others [8].
The development in the field of diagnostics led to using other immunological assays for instance; Radioimmunoassay (RIA) and enzyme-linked immunoassay (EIA). These methods measures viral specific immunoglobulins levels in patients' serum [9]. Others are based on nucleic acid detection as viral nucleic acids (DNA or RNA) are amplified by Polymerase chain reaction (PCR) technique as quantitative and qualitative nucleic acid detection technique [10]. In viral diagnosis, virus isolation is considered the gold standard and the most sensitive method, however it is laborious and work taking 3–7 days. The serological investigations for antibodies against viral antigens are less sensitive and could be non-specific. The high sensitivity and selectivity of RT-qPCR require expensive laboratory appliances and technical experiences, which are employ RNA extraction steps that limit their applications in this field [11]. Biosensors are currently important devices in clinical diagnostics, food processing, and environmental monitoring to detect various analytes, such as specific proteins, cancer biomarkers, nucleic acids, bacteria, viruses, and toxins [3].
1.1. Biosensors
Biosensors are analytical techniques that could be used as simple, real-time and effective devises for the detection of various infectious diseases.
The biosensors research field began in 1962 with the designing of glucose oxidase biosensor, which was introduced by Clark and Lyons. After that, numerous applications of sensors and biosensors have been described [12]. Biosensors have been invented over decades ago by biotechnologists to detect bacteria and viruses by recognizing biomarkers or characteristics of the targets.
Bio-receptors act as sensing elements Due to their biochemical properties making them sensitive and selective for biomarkers detection with minimum interference with other microorganisms or molecules present in the tested sample.
Biosensors comprise three main elements: the bio-receptor, the transducer and the signal processing system [13]. The Bio-receptors component of biosensors may be monoclonal antibody, nucleic acids, glycan, lectin, enzyme, tissue or whole- cell interact specifically with a biomarker. The transductor convert these interactions to a measurable signal, then the qualitative and quantitative identification of pathogen are viewed/reported by recording and displaying the signals [14,15]. Fig. 1 illustrates the main principles of biosensors.
Fig. 1.
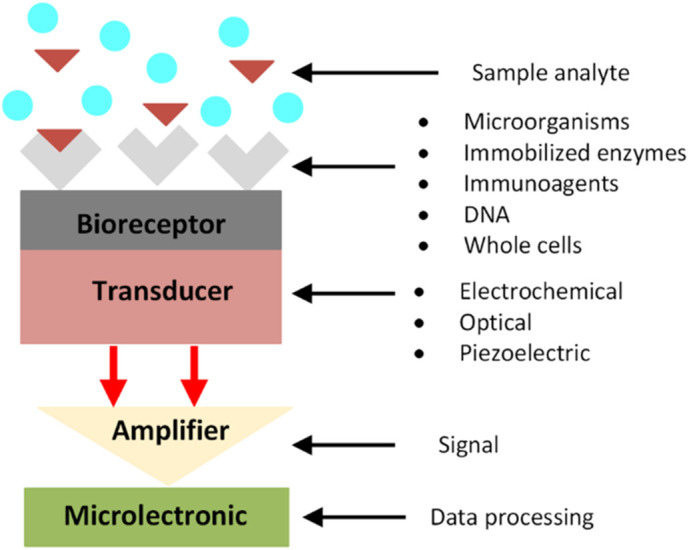
Principle of biosensor, where the analyte bind specifically to bioreceptor that lead to generating a signal (electrochemical, optical or piezoelectric) which can be amplified to be read by data processing.
1.2. Viral bioreceptors
Bioreceptors are highly specific biomolecules that are selected for viral analyte immobilized onto transducers to act as functional sensors.
The target analyte involves viral antigenic part that may be whole virus, viral proteins (capsid proteins), viral nucleic acids (RNA or DNA genomes) or viral-specific antibodies. In viral bioreceptors, sensing constituents are whole cells, peptides, nucleic acids, aptamers and antibodies which are the most common bioreceptors [16]. Peptides are called viral fusion proteins (VFPs). These peptides are oligomeric glycoproteins, the hydrophobic transmembrane sequences of these peptides in the C-terminal region anchored in the viral membrane [17]. Viral genome may have composed of ssRNA with interaction with capsid proteins due to their icosahedral or helical particles forming crystals or fibers-like; Human respiratory syncytial virus (RSV), or composed of dsDNA like; dsDNA bacteriophages and dsDNA animal viruses [18]. Aptamers are short functional biomolecules like oligonucleotides or peptides that bind specifically to targets with extremely high affinity and selectivity depending on their structural conformations. Aptamers are selected in vitro in 1990s by systematic evolution of ligands by exponential enrichment (SELEX) and other methods which have been reported RNA and DNA aptamers selection efficiently. Nucleic acid aptamers are RNA and single-stranded (ss) DNA oligonucleotides ranging from 15 to 70 mers length [19,20]. RNA aptamers need adding extra chemical modifications for improving their chemical stability due to their chemical instability, because in RNA nucleotides a reactive hydroxyl group (−OH) present at the 2׀ position of the ribose sugar. Deprotonation of (−OH) group in solution, especially in alkaline solutions resulting formation of anionic 2 ׀-O¯ which may attack the phosphorus atom of the phosphodiester linkage nucleophilically, and finally RNA molecules hydrolyzed. On the other hand, DNA aptamers have much more stability than natural RNA aptamers in 10% fetal bovine serum (FBS) and human serum, because of the C—H bonds at the 2 ׀ position of the deoxyribose sugar of DNA nucleotides [21]. Since high affinity of aptamers to fold upon binding with their target molecules they are termed as “chemical antibodies” due to analogical or even better attributes to antibodies. The in vitro artificial synthesis of aptamers based on (SELEX) includes the fabrication of aptamers to bind specifically to non-immunogenic and toxic targets unlike natural antibodies produced by animal immune system induction. (SELEX) synthesized aptamers to specific regions of targets that may be difficult discovered by antibodies. Broad range of targets can be discovered by aptamers including metal ions like (K+, Hg+2 and Pb+2), amino acids, drugs, nucleotides, larger molecules like antibiotics or even whole cell like bacteria and viruses [[22], [23], [24]]. Antibody bioreceptors are most popular due to their specificity against diverse analytes like whole virus or viral proteins which can bind by high affinity. Production of polyclonal and monoclonal antibodies occurs by the host in response to artificial infection with virus. Antibodies in biosensors can interact tightly with their antigens (analytes) forming complex mixture by non-covalent bonds with their targets [16]. Although their specificity and high affinity against analyte, antibodies have some disadvantages like instability comparing with peptide-based probes and recognition of different epitopes on the same pathogen by polyclonal antibodies, whereas monoclonal antibodies are more selective to analytes than polyclonal antibodies [16,25].
1.3. Viral transducers
Functional sensing platforms are formed by immobilization of solid phase (transducers) physical absorption onto a conducting polymer surface like; polypyrrole or polyaniline or by covalent coupling to a linker molecule such as; an mSAM through amino, carboxyl, maleimido or thiol groups which bind to transducer surface. The transducer surface may be gold, carbon, silicon or hydrogels by direct attachment, streptavidin/ biotin affinity or silanisation [16]. The reaction of analyte with bioreceptor performs chemical changes like new chemical production, heat release, pH or mass change or electrons flow. These biochemical signals are converted by transducer into electrical signal. Ultimately, the electrical signal is amplified by amplifier element and sent to (microelectronics and data processor) producing a measurable signal, such as (digital display) which exhibit optical change or print-out [26]. Different types of transducers are used for medical diagnosis like optical, electrochemical, piezoelectric, magnetic, micromechanical, and thermal. The most common biosensor transducers that used for viral detection are optical (e.g. surface-enhanced Raman scattering (SERS), Surface plasmon resonance (SPR)) and electrochemical [10,12].
2. Optical transducer
Optical biosensors are the most common analytical techniques which depend on visual phenomena for detection of biological element and the target analyte interaction that employ absorption, fluorescence, phosphorescence, Raman, refraction, and surface plasmon resonance (SPR). Optical biosensors detection techniques can be achieved by two ways, indirect and direct optical biosensors. Indirect optical biosensors depend on binding with fluorophores or chromophores as labels for detection process and amplifying the signal. The indirect method generates a high signal but suffers from non-specific binding. The direct optical biosensors method depends on affecting of analyte with optical properties of the sensing environment by measuring the change in the refractive index (RI) at the analyte-sensor interface as in SPR biosensor [26,27].
3. Surface plasmon resonance biosensors
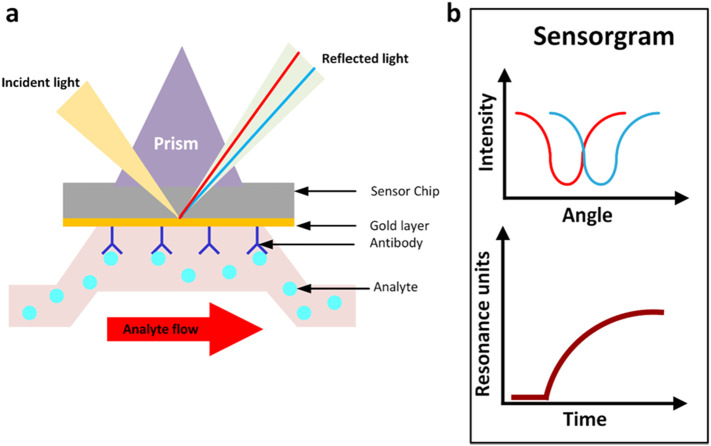
Surface plasmon resonance is an optical detection method that uses conjugation of prisms that allow biomolecular interactions in real- time [26].
The interaction between biomolecules can be analyzed by measuring the change in refractive index in real time. Change in refractive index is generated from the interaction between the immobilized molecule (ligand) on the platform and the analyte. The analyte is injected continuously into the buffer solution through the flow cell and accumulates on the platform leading to increasing the refractive index [28]. In this method, a photon of incident light strikes a metal surface (usually gold surface) at a given angle of incidence. Then, a part of the light energy pairs via the coated metal with the electrons in the layer of metal surface leading to excitation due to electrons movement. This state is called plasmon which transmitted parallel to the surface of metal (platform) [29]. Novel nanomaterials such as Ag NPs, Au NPs and quantum dots are the most commonly highlighted in optical transducers because of their plasmonic properties [29,30] plasmonic nanomaterials applications can be divided into two systems; plasmonic and non-plasmonic system. According to plasmonic systems, metal nanoparticles considered as a plasmonic probes that have a proper inter-particle distance, smaller in diameter than the particles to create particles plasmonic coupling. That produce a visible color from red to blue diversely with colorimetric detectability. The direct aggregation of plasmonic nanomaterials is the advanced example without specific ligands between (a single-stranded primer DNA). In addition, indirect aggregation can be used for virus detection by modifying targeting molecules on the surface of virus. This technology is dedicated in order to control particle aggregation in (a reproducible manner), which can be improved in a protein-glycan pairing utilizing depending on the glycan multivalence advantages for improving weak protein detection of surface proteins of viruses. Whereas, in non-plasmonic systems, nanomaterials must be functionalized with fluorescent labels [30]. Optical sensing is illustrated in Fig. 2 .
Fig. 2.
(a) Immune complex bounded to sensor chip that is coated with suitable material (such as gold metal) which reflects the incident light in a given angle appropriate for detection of analyte, (b) measurement of sensing angle over time. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.1. Advantages of SPR biosensor
It allows label-free measurement with high reliability, sensitivity and in a real-time [28].
SPR sensor device design is trending toward miniaturization, low cost and user friendliness. [31].
3.2. Disadvantages of SPR biosensors
The SPR is not appropriate for analyzing small analytes. Because the SPR measures the material mass which bind to sensor surface, the quite small analytes give quite small responses [30].
4. Electrochemical sensors
Electrochemical sensors can be used as quantitative or semi-quantitative analysis of oxidation and reduction reactions with high specificity and sensitivity of electroactive species. It works by potentiometric, amperometric, conductometric, polarographic, capacitive or piezoelectric ways [32,33]. The effective physical transduce in electrochemical transducer is the working electrode, the sensitive layer is the interface between the electrode and the analyzed environment [34]. The produced current is directly related to the electroactive species concentration (present/produced), the transducer of electrochemical biosensors must be a conductor and helpful for the bio-recognition element attachment after appropriate surface modification steps [32].
It is essential to choose a suitable material when designing the electrochemical biosensor. This material must be inert at the potential when the electrochemical reaction takes place. Solid electrodes have been recently made from metals such as, gold, silver, nickel, copper, platinum, mercury and heterogeneous carbon electrodes which contain carbon as an electrically conductive material [33]. Electrode transducers are often used for viral detection due to ease of modification of the surface and compatibility of electrochemical transducer electrode. Presently, many researchers construct biosensors based on glycan (glycocalyx) forming a compact layer on the surface of the measurement reach to 100 mM concentration, these modified biosensors considered as natural receptors for viruses that have selectivity for subtypes of pathogens [35]. The modification occurs by immobilization of bio-recognition element which represented by a receptor on the electrode surface, hybridized electrochemical biosensor as a probe or affinity EB (targeting molecule) are commonly used as bioreceptors.
Using of simple electrodes Modification method could significantly reduce the testing time without the need for antibodies and labelling.
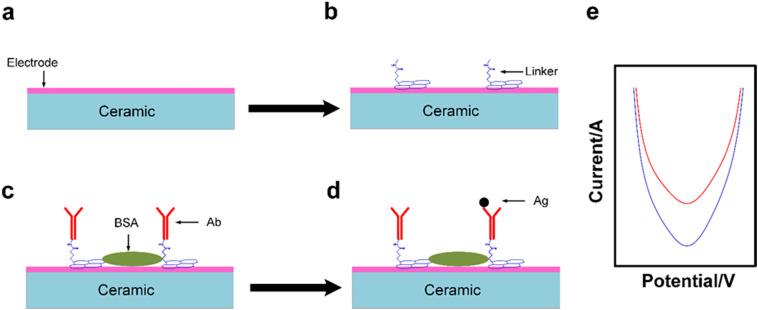
By applying AC electric field on electrode, a positive di-electrophoresis will be induced leading to attract viral particles to the sensor. This is followed by the detection of simple signal or signal amplification by amplifier which then converted into a quantitative amperometric, potentiometric or impedimetric signal [12,35]. Fig. 3 illustrates the working mechanism of electrochemical biosensor.
Fig. 3.
(a) a schematic example of platform of the sensor (b) linkers used to linking the bioreceptors components with biosensor platform (c) bioreceptors (Abs) bind to linker, (BSA) as a blocking agent (d) the analyte (Ag) attached with bioreceptors (Abs) (e) the immune complex formation generates a quantitative electric signal.
4.1. Advantages of electrochemical biosensor
Due to the easily modification of surface and compatibility of electrochemical transducer electrode they used for viral detection [35].
Capability of fast providing specific (quantitative or semi-quantitative) analytical information using biochemical receptor due to direct contact with an electrochemical transduction element [33].
4.2. Nanotechnology and biosensors
Nanotechnology has various applications in several areas, like coatings, sensors, optical communications, agriculture, food, electromechanical systems, electronics, and biomedical applications [36]. Nanomaterials have physical and chemical surface properties like solubility, diffusivity, optical, toxicity, thermodynamic, color and magnetic properties compared to bulk material depending on their size [36,37]. Metal oxides are mostly used in microelectronic circuits, sensors, piezoelectric devices, and as catalysts because of the electronic structure difference. Oxygen vacancies in an oxide nanoparticle produces atomic arrangements are different from that in the bulk material which enhances the chemical activities of metal oxides [38].
Sizes of Nanomaterials ranging between 1 and 100 nm, this offers a large surface area to volume ratios. To enhance the properties of biosensors that could be used for viral detection, nanomaterials are used in designing biosensors to achieve large biocompatible areas with the analyte (antibodies, enzymes, DNA, cells, and proteins) and improve their applicability and sensitivity [39]. By using nanotechnology strategies in viral biosensors, we can overcome the disadvantages of present techniques for viral detection by minimizing cost and detection time. Nanomaterials used in biomedical sensing have functional electrical and mechanical characteristics which participate in enhancing electrochemical, optical, and magnetic properties of biosensors [39,40]. Several types of nanomaterials used for diagnosis and biosensing such as nanoparticles (NPs), nanocomposites, carbon nanotubes (CNTs), quantum dots (QDs) and graphene or graphene-based nanomaterials [41]. Table 1 lists the type of viral transducers, their material, type of signal generation and the type of analyte.
Table 1.
non-transducers (electrochemical and optical) with type of nanoparticles and viral analytes.
| Type of viral transducer | Nanomaterial mostly used | Generation of signal | Type of analyte |
|---|---|---|---|
| Electrochemical transducer | Carbon nanomaterial allotrops like Carbon nanotube(CNT), Graphene and Graphene based materials [42] Au and magnetic NPs [43] Nanosized semiconductor crystals and Nanowires [44] |
Combination of nanomaterials with analyte for construction of electrode surface fabrication to enhance electrochemical signal [43]. | Antibodies, ssDNA [41] and double-stranded DNA (dsDNA) [40]. |
| Optical transducers (SPR biosensors) | Plasmonic nanorod materials [45] Quantum dots (QDs) Graphene oxide (GO) and reduced graphene oxide (RGO) [39] Ag NPs [46] Spherical AuNPs [47] |
Metal nanoparticles (NPs) act as plasmonic probes due to increasin Raman scattering signals make them as raman probes [30] | single-stranded primer DNA [48] Antibodies and Aptamers [43] Specific oligonucleotide from RNA-α [48] |
4.3. Recommendations about COVID-19 biosensors
Since (December 2019), a novel coronavirus [COVID-19] which causes severe acute respiratory syndrome (SARS-CoV-2), firstly reported in Wuhan city in central China leading to an explosion in the number of cases reported globaly. SARS-CoV-2 is considered the new type of beta-coronavirus that belongs to the coronavirus family of viruses. The main viruses of the corona virus family that can infect human are.
MERS-CoV (Middle East Respiratory Syndrome, or MERS), SARS-CoV (acute respiratory syndrome, or SARS) and SARS-CoV-2 (coronavirus disease 2019, or COVID-19) according to CDC (https://www.cdc.gov/coronavirus/types.html).
According to the current epidemic, SARS-COV-2 is more infectious than SARS-CoV [49]. SARS-CoV-2 RNA is a positive-strand including (large ssRNA) genome of approximately (29700) nucleotides. The genome is expected to consist of fourteen functional ORFs at least which encode for three classes of proteins. Where, eight auxiliary proteins that are thought provide (selective advantage) in the infected host, two large polyproteins (pp1a and pp1ab) that cleaved into (sixteen non-structural proteins (nsps)). This is important in viral RNA synthesis, four structural proteins (the S, E, M and N proteins) that are essential for cytoplasmic viral assembly which are spike (S) protein, nucleo-capsid (N) protein, membrane (M) protein and the envelope (E) protein [50]. Liu et al., (2020) and Walls et al., (2020) studied the SARS-CoV-2 spike proteins and the host ACE2 receptors. The structural glycoprotein (S) is located on the outer-envelope of the virion, it binds to the angiotensin converting enzyme-2 (ACE2) receptor of the host. The glycoprotein (S) of SARS-CoV, MERS-CoV, and SARSCoV-2 has (1104 to 1273) amino acids. In addition, it contains S1 subunit with amino (N)-terminal and S2 subunit with carboxyl (C)-terminal. The receptor-binding domain (RBD) in the S1 subunit is spanning about (200 residues) consisting of two subdomains (the core and the external subdomains). The core subdomain of RBD is responsible for the formation of trimer particles of S glycoprotein. Furthermore, the external subdomain binds with ACE2 since it contains (two exposed loops) on the surface. By investigating the RBD sequence, evolutionary relationship in (S) protein is important in realizing the virus origin tendencies. The interaction of spike RBD-receptor is the key factor that determine coronaviruses host range [51,52]. Liu and Li, 2020 found that the non-structure proteins ORF8 and E2 surface glycoproteins of SARS-CoV-2 could bind to the porphyrin of 1-Beta Chain of Hemoglobin to form a complex. That combination leads to the dissociation of iron to form the porphyrin. That attack will lead to lower hemoglobin to carry oxygen and carbon dioxide and finally results in inability for oxygen and carbon dioxide exchanging causing lung cells inflammation [53].
According to the above, we recommend designing biosensors for direct, simple, low-cost and rapid detecting of patients that are infected with SARS-CoV-2 form saliva sample; here we suggest two types of biosensors
-
1.
Electrochemical biosensor: The non-structural proteins ORF8 and E2 surface glycoproteins of SARS-CoV-2 could bind to the porphyrin of 1-Beta Chain of Hemoglobin and releasing the heme. The biosensor bioreceptors composed of 1-Beta Chain of Hemoglobin, and so if the specimen is positive, SARS-CoV-2 proteins will bind to hemoglobin molecules on the transducer releasing heme part generating electrical signal, taken in consideration measuring the heme concentration before and after the investigation.
-
2.
Optical biosensor: depends on binding of the Anti-Spike-RBD mAb (Human-IgG1) specifically with analyte Spike RBD, then measuring the change in refraction index.
5. Conclusion
In this review, we reported techniques commonly used for viral detection. Furthermore; nanotechnology role in biosensors development by binding nanomaterials with biosensors depending on nanomaterials characteristics, in order to enhance the detection processes of biosensors by increasing the surface area for most contact with analytes and increasing the electrical or optical properties of transducers. Additionally, two methods have been recommended for the detection of the novel SARS-CoV-2, depending on the two biosensor types for direct and early detection of COVID-19.
Funding
This research received no external funding.
CRediT authorship contribution statement
“Conceptualization, X.X. and Y.Y.; methodology, X.X.; software, X.X.; validation, X.X., Y.Y. and Z.Z.; formal analysis, X.X.; investigation, X.X.; resources, X.X.; data curation, X.X.; writing—original draft preparation, X.X.; writing—review and editing, X.X.; visualization, X.X.; supervision, X.X.; project administration, X.X.; funding acquisition, Y.Y. All authors have read and agreed to the published version of the manuscript.
Declaration of competing interest
The authors declare no conflict of interest.
References
- 1.La Scola B., et al. The virophage as a unique parasite of the giant mimivirus. Nature. 2008;455(7209):100–104. doi: 10.1038/nature07218. [DOI] [PubMed] [Google Scholar]
- 2.Mahmood Z., et al. Investigating virological, immunological, and pathological avenues to identify potential targets for developing covid-19 treatment and prevention strategies. Vaccines. 2020;8(3):443. doi: 10.3390/vaccines8030443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saylan Y., et al. An alternative medical diagnosis method: biosensors for virus detection. Biosensors. 2019;9(2):65. doi: 10.3390/bios9020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bibby K. Metagenomic identification of viral pathogens. Trends Biotechnol. 2013;31(5):275–279. doi: 10.1016/j.tibtech.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 5.Pedersen, J.C., Hemagglutination-inhibition assay for influenza virus subtype identification and the detection and quantitation of serum antibodies to influenza virus, in Animal Influenza Virus. 2014, Springer. p. 11–25. [DOI] [PubMed]
- 6.Actor, J.K., Introductory immunology, 2nd: Basic Concepts for Interdisciplinary Applications. 2019: Academic Press.
- 7.Goldsmith C.S., Miller S.E. Modern uses of electron microscopy for detection of viruses. Clin. Microbiol. Rev. 2009;22(4):552–563. doi: 10.1128/CMR.00027-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Doornum G., De Jong J. Rapid shell vial culture technique for detection of enteroviruses and adenoviruses in fecal specimens: comparison with conventional virus isolation method. J. Clin. Microbiol. 1998;36(10):2865–2868. doi: 10.1128/jcm.36.10.2865-2868.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Souf S. Recent advances in diagnostic testing for viral infections. Bioscience Horizons: The International Journal of Student Research. 2016;9 [Google Scholar]
- 10.Storch G.A. Diagnostic virology. Clin. Infect. Dis. 2000;31(3):739–751. doi: 10.1086/314015. [DOI] [PubMed] [Google Scholar]
- 11.Krishna V.D., et al. Giant magnetoresistance-based biosensor for detection of influenza A virus. Front. Microbiol. 2016;7:400. doi: 10.3389/fmicb.2016.00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krejcova L., et al. Nanoscale virus biosensors: state of the art. Nanobiosensors in Disease Diagnosis. 2015;4:47–66. [Google Scholar]
- 13.Perumal V., Hashim U. Advances in biosensors: principle, architecture and applications. J. Appl. Biomed. 2014;12(1):1–15. [Google Scholar]
- 14.Vidic J., et al. Advanced biosensors for detection of pathogens related to livestock and poultry. Vet. Res. 2017;48(1):11. doi: 10.1186/s13567-017-0418-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scott, K. and E.H. Yu, Microbial Electrochemical and Fuel Cells: Fundamentals and Applications. 2015: Woodhead Publishing.
- 16.Caygill R.L., Blair G.E., Millner P.A. A review on viral biosensors to detect human pathogens. Anal. Chim. Acta. 2010;681(1–2):8–15. doi: 10.1016/j.aca.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 17.Cross K.J., et al. Composition and functions of the influenza fusion peptide. Protein Pept. Lett. 2009;16(7):766–778. doi: 10.2174/092986609788681715. [DOI] [PubMed] [Google Scholar]
- 18.Speir J.A., Johnson J.E. Nucleic acid packaging in viruses. Curr. Opin. Struct. Biol. 2012;22(1):65–71. doi: 10.1016/j.sbi.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blind M., Blank M. Aptamer selection technology and recent advances. Molecular Therapy-Nucleic Acids. 2015;4 doi: 10.1038/mtna.2014.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bai H. 2012. Aptamer-based SPR Biosensor for Detection of Avian Influenza Virus. [Google Scholar]
- 21.Zhu Q., Liu G., Kai M. DNA aptamers in the diagnosis and treatment of human diseases. Molecules. 2015;20(12):20979–20997. doi: 10.3390/molecules201219739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song S., et al. Aptamer-based biosensors. TrAC Trends Anal. Chem. 2008;27(2):108–117. [Google Scholar]
- 23.Han K., Liang Z., Zhou N. Design strategies for aptamer-based biosensors. Sensors. 2010;10(5):4541–4557. doi: 10.3390/s100504541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zou X., et al. Application of aptamers in virus detection and antiviral therapy. Front. Microbiol. 2019;10:1462. doi: 10.3389/fmicb.2019.01462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abbas, A., A. Lichtman, and S. Pillai, Cellular and Molecular Immunology, Elsevier Saunders. Philadelphia, PA.[Google Scholar], 2015.
- 26.Frías I.A., et al. Trends in biosensors for HPV: identification and diagnosis. Journal of Sensors. 2015;2015 [Google Scholar]
- 27.Wang, R. and Y. Li, Biosensors for rapid detection of Avian Influenza. Steps Forwards in Diagnosing and Controlling Influenza, 2016: p. 61.
- 28.Kumar P.K. Monitoring intact viruses using aptamers. Biosensors. 2016;6(3):40. doi: 10.3390/bios6030040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen H.H., et al. Surface plasmon resonance: a versatile technique for biosensor applications. Sensors. 2015;15(5):10481–10510. doi: 10.3390/s150510481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neethirajan S., et al. Recent advances in biosensor development for foodborne virus detection. Nanotheranostics. 2017;1(3):272. doi: 10.7150/ntno.20301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao C.-Y., Fu W.-L. Biosensors for hepatitis B virus detection. World J Gastroenterol: WJG. 2014;20(35):12485. doi: 10.3748/wjg.v20.i35.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar N., et al. Emerging biosensor platforms for the assessment of water-borne pathogens. Analyst. 2018;143(2):359–373. doi: 10.1039/c7an00983f. [DOI] [PubMed] [Google Scholar]
- 33.Grabowska, I., et al., Electrochemical biosensors for detection of avian influenza virus—current status and future trends. Acta Biochim. Pol., 2014. 61(3). [PubMed]
- 34.Krejcova L., et al. Electrochemical sensors and biosensors for influenza detection. Int. J. Electrochem. Sci. 2012;7(11):10779–10801. [Google Scholar]
- 35.Dziąbowska K., Czaczyk E., Nidzworski D. Detection methods of human and animal influenza virus—current trends. Biosensors. 2018;8(4):94. doi: 10.3390/bios8040094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jeevanandam J., et al. Review on nanoparticles and nanostructured materials: history, sources, toxicity and regulations. Beilstein journal of nanotechnology. 2018;9(1):1050–1074. doi: 10.3762/bjnano.9.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh T., et al. Application of nanotechnology in food science: perception and overview. Front. Microbiol. 2017;8:1501. doi: 10.3389/fmicb.2017.01501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fierro, J.L.G., Metal OXIDES: CHEMISTRY and APPLICATIONS. 2005: CRC press.
- 39.Mokhtarzadeh A., et al. Nanomaterial-based biosensors for detection of pathogenic virus. TrAC Trends Anal. Chem. 2017;97:445–457. doi: 10.1016/j.trac.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Z., et al. Biosensing methods for the detection of highly pathogenic avian influenza H5N1 and H7N9 viruses. Anal. Methods. 2017;9(36):5238–5248. [Google Scholar]
- 41.Peña-Bahamonde J., et al. Recent advances in graphene-based biosensor technology with applications in life sciences. Journal of nanobiotechnology. 2018;16(1):75. doi: 10.1186/s12951-018-0400-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu Z. An overview of carbon nanotubes and graphene for biosensing applications. Nano Lett. 2017;9(3):25. doi: 10.1007/s40820-017-0128-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee T., et al. Recent advances in AIV biosensors composed of nanobio hybrid material. Micromachines. 2018;9(12):651. doi: 10.3390/mi9120651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malik P., et al. Nanobiosensors: concepts and variations. ISRN Nanomaterials. Article id. 2013;327435:9. [Google Scholar]
- 45.Kabashin A., et al. Plasmonic nanorod metamaterials for biosensing. Nat. Mater. 2009;8(11):867–871. doi: 10.1038/nmat2546. [DOI] [PubMed] [Google Scholar]
- 46.Shakoori Z., et al. Electrochemical DNA biosensor based on gold nanorods for detecting hepatitis B virus. Anal. Bioanal. Chem. 2015;407(2):455–461. doi: 10.1007/s00216-014-8303-9. [DOI] [PubMed] [Google Scholar]
- 47.Alim S., et al. Recent uses of carbon nanotubes & gold nanoparticles in electrochemistry with application in biosensing: a review. Biosens. Bioelectron. 2018;121:125–136. doi: 10.1016/j.bios.2018.08.051. [DOI] [PubMed] [Google Scholar]
- 48.Hong S., Lee C. The current status and future outlook of quantum dot-based biosensors for plant virus detection. The plant pathology journal. 2018;34(2):85. doi: 10.5423/PPJ.RW.08.2017.0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou, S., et al., CT features of coronavirus disease 2019 (COVID-19) pneumonia in 62 patients in Wuhan, China. Am. J. Roentgenol., 2020: p. 1–8. [DOI] [PubMed]
- 50.Bartlam M., Yang H., Rao Z. Structural insights into SARS coronavirus proteins. Curr. Opin. Struct. Biol. 2005;15(6):664–672. doi: 10.1016/j.sbi.2005.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Z., et al. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J. Med. Virol. 2020;92(6):595–601. doi: 10.1002/jmv.25726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abd Ellah N.H., et al. Nanomedicine as a promising approach for diagnosis, treatment and prophylaxis against COVID-19. Nanomedicine. 2020;15(21):2085–2102. doi: 10.2217/nnm-2020-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Read R. 2020. Flawed Methods in “COVID-19: Attacks the 1-Beta Chain of Hemoglobin and Captures the Porphyrin to Inhibit Human Heme Metabolism”. [Google Scholar]