Abstract
Computed tomography (CT) and X-ray are effective methods for diagnosing COVID-19. Although several studies have demonstrated the potential of deep learning in the automatic diagnosis of COVID-19 using CT and X-ray, the generalization on unseen samples needs to be improved. To tackle this problem, we present the contrastive multi-task convolutional neural network (CMT-CNN), which is composed of two tasks. The main task is to diagnose COVID-19 from other pneumonia and normal control. The auxiliary task is to encourage local aggregation though a contrastive loss: first, each image is transformed by a series of augmentations (Poisson noise, rotation, etc.). Then, the model is optimized to embed representations of a same image similar while different images dissimilar in a latent space. In this way, CMT-CNN is capable of making transformation-invariant predictions and the spread-out properties of data are preserved. We demonstrate that the apparently simple auxiliary task provides powerful supervisions to enhance generalization. We conduct experiments on a CT dataset (4,758 samples) and an X-ray dataset (5,821 samples) assembled by open datasets and data collected in our hospital. Experimental results demonstrate that contrastive learning (as plugin module) brings solid accuracy improvement for deep learning models on both CT (5.49%-6.45%) and X-ray (0.96%-2.42%) without requiring additional annotations. Our codes are accessible online.
Keywords: Computed tomography, X-ray, COVID-19, Deep learning, Multi-task learning, Contrastive learning
1. Introduction
In December 2019, the coronavirus disease (COVID-19) broke out in Wuhan, China. The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was identified as the cause of COVID-19 and spread rapidly to other locations of China and the world. By December 1, 2020, there were more than 63,492,196 confirmed cases with 1469,003 deaths worldwide. The World Health Organization (WHO) has declared COVID-19 as a global pandemic. The prognosis of COVID-19 is poor. According to several studies, over 60% of patients died when the disease have developed to the severe/critical stage [1], [2]. The main causes of death include massive alveolar damage and progressive respiratory failure [3]. Moreover, the respiratory viruses SARS-CoV-2 spread amongst humans do transmit even in the absence of symptoms. Therefore, fast and accurate screening and diagnosis of COVID-19 is of great significance for planning early interventions, blocking the transmission path and formulating clinical schemes to improve the prognosis [4].
There are two main diagnostic methods for COVID-19. The first is nucleic detection implemented with real-time polymerase chain reaction (RT-PCR) test. RT-PCR has been widely used in clinical diagnosis, discharge assessment and recovery follow-up. However, the sensitivity of RT-PCR is low from swab samples [5], which may result in substantial false negative predictions [6]. The second approach to detect COVID-19 is chest medical imaging. Clinical studies have suggested that certain manifestations on computed tomography (CT) such as multiple small patches and ground glass shadow are associated with COVID-19 [7]. From the perspective of pathology, CT can provide detailed information to facilitate a quantitative assessment for pulmonary changes, which may have prognostic implications [8]. Despite its high sensitivity (97%) [5], CT is not suitable for large-scale screening due to the relatively high cost. Besides, CT has high radiation dose, which is harmful to human body [7]. Therefore, CT can be used for accurate clinical diagnosis, whereas not recommended in clinical applications where repetitive data-acquisition is required (e.g., recovery assessment and follow-up analysis). X-ray is another medical imaging to detect COVID-19. Considering that X-ray cannot provide 3D information like CT, radiologists generally use X-ray as a preliminary screening before CT diagnosis. However, X-ray has certain advantages. The low radiation dose brings less damage to human body. In addition, the relatively low cost makes it suitable for less developed countries and regions as an important method for COVID-19 diagnosis. At present, most researches have focus on CT diagnosis [5], [7], [8], whereas X-ray diagnosis has been less investigated [3].
Benefited from the strong representational learning ability of deep learning, artificial intelligence (AI) has demonstrated impressive capability in the automatic diagnosis of COVID-19 based on both CT [9], [10], [11], [12] and X-ray [13], [14]. AI has four advantages: (1) Diagnose quickly especially when the medical system is overloaded. (2) Reduce the burden of radiologists. (3) Assist undeveloped areas to realize accurate diagnosis. (4) Most importantly, as a new pandemic, current understandings on the sensitive and specific manifestations of COVID-19 lack systematic consensus. AI can automatically learn discriminative features in a data-driven manner, especially to distinguish COVID-19 from other pneumonia [4], [10], [13]. Despite the success of AI in both CT and X-ray diagnosis of COVID-19, the generalization of these models needs to be improved.
This paper proposes CMT-CNN, a novel multi-task framework for the improved generalization on unseen data. Different from typical multi-task models, CMT-CNN requires no additional supervisions from related tasks, but seeks for annotation-free performance improvement based on the self-supervised learning [15], [16], [17]. Our work is quite different from that of Doersch and Zisserman [18], which explored multiple self-supervised tasks in a multi-task framework, whereas no main task was explicitly appointed. Furthermore, different from recent advances in self-supervised learning who have focused on generating well-aggregated embedding [19], [20], [21], CMT-CNN pays more attention on fulfilling certain tasks. The main contributions of this paper are:
-
(1)
A novel CMT-CNN model that brings solid improvement in generalization without additional annotations. While completing specific tasks, the model seeks to evolve into an embedding function with fine spatial aggregating properties.
-
(2)
We present a series of effective augmentation methods for CMT-CNN based on distortion, painting and perspective transformations. They are not used as a data-preprocessing trick, but to enhance the representational learning at the model level. More importantly, these methods are highly related to the characteristics of CT/X-ray images, and thereby having good interpretability.
-
(3)
Experimental results on a large-scale CT dataset and an X-ray dataset both demonstrate that CMT-CNN has significant advantage over CNN for diagnosing COVID-19 from other pneumonia and normal controls.
2. Related works
We first review some representative works using AI to diagnosis COVID-19 (both CT and X-ray). Then, from the perspective of methodology, we present a literature review on some highly relevant concepts or works to our method.
Several studies have reported various results concerning deep learning in CT-based diagnosis. Li et al. [9] applied ResNet-50 as backbone and added a fully-connected (FC) layer with SoftMax activation to distinguish COVID-19 from community-acquired pneumonia. They considered 4356 chest CT instances, and the sensitivity was 90% with a specificity of 96% (AUC=0.96). However, the control group in the training set was randomly selected in hospital records, making the model being able to distinguish COVID-19 from all other conditions, whereas not being able to accurately distinguish COVID-19 from other pneumonia. Since COVID-19 and other types of pneumonia share similar imaging manifestations [8], exploiting and evaluating AI models that distinguish them is necessary. Bai et al. [22] added two FCs on backbone EfficientNet B4 to distinguish COVID-19 from other pneumonia. They considered 512 COVID-19 instances and 665 non-COVID-19 instances, and achieved an AUC of 0.95. This work demonstrated that AI model had higher test accuracy (96% vs. 85%), sensitivity (95% vs. 79%), and specificity (96% vs. 88%) than radiologists. The sample size they used was relatively small comparing with relevant researches. Zhang et al. [4] developed an AI model to diagnose COVID-19 and differentiate it from other common pneumonia and normal controls. They considered 4154 patients and selected 3D ResNet-18 as backbone and added a FC layer activated by SoftMax to conduct three-category classification. They reported 92% accuracy, 95% sensitivity and 91% specificity for distinguishing COVID-19 from other two classes (common pneumonia and normal controls), and 92% accuracy for the three-category classification. Most recently, some studies have exploited more sophisticated methods to diagnose COVID-19. For example, Ouyang et al. [10] proposed a dual-sampling attention network to diagnose COVID-19 from other pneumonia, where segmented lesions are used to refine the attention localization. Wang et al. [11] proposed a weakly-supervised framework to improve the lesion localization for diagnosing COVID-19 from other pneumonia. Kang et al. [12] incorporated multiple radiomic features and handcrafted features into a multi-view learning framework to separate COVID-19 from other pneumonia. The model used the complementary information from multiple types of features and achieved an accuracy of 94% using 2522 CT samples. Others have discussed about the feasibility of CT in screening and early diagnosis [7], the multi-modal diagnosis [23] and the correlation of CT and RT-PCR results [5], [6].
Compared with CT, X-ray has been less studied in the automatic diagnosis of this pandemic. Cohen et al. [24] contributed a dataset containing 221 COVID-19 instances, 4007 instances with other pneumonia (MERS, SARS, ARDS, etc.) and 1583 normal controls. Apostolopoulos and Mpesiana [13] considered 1427 X-ray images consisted of 224 COVID-19 instances, 700 common pneumonia instances and 504 normal controls. They systematically compared VGG-19, MobileNet, Inception, Xception and Inception ResNet v2. Experimental results showed VGG-19 achieved the highest three-category accuracy (93%). This work shows the potential of a low-cost, rapid and automatic diagnosis of the disease. Oh et al. [25] proposed a patch-based CNN for COVID-19 diagnosis, where decisions are made based on the majority voting from multiple patches at random locations within lungs on X-rays. The patch-based methods allow for a relatively small number of training samples and trainable parameters.
The medical imaging datasets used in the above researches are much smaller than those in computer vision tasks (e.g., ImageNet). To enhance the generalization ability of the diagnostic models, we integrate contrastive learning into CNN, and hereby proposing a multi-task learning framework: CMT-CNN. In the following, we review some concepts or works that are highly relevant to our method.
Multi-task Learning (MTL). MTL improves the generalization by leveraging the domain-specific information contained in the training signals of related tasks [26], [27]. MTL typically involves a main task and an auxiliary task. The auxiliary task enables the model to learn representations that are shared or helpful for the main task. Substantial studies have presented both theoretical [28] and empirical [29], [30] evidences that machine learning models would generalize better by sharing representations between related tasks. The labeled data for an auxiliary task are available in a typical MTL scenario. When labeled data are unavailable, pseudo-labels can be defined and used. For example, Ganin and Lempitsky [31] used domain label as the pseudo-label and applied an adversarial training scheme to reduce the representational differences between domains. Finding an effective and pervasive approach to improve the model generalizations in the absence of explicitly-annotated auxiliary tasks is a challenging and interesting topic. We think the self-supervised learning which develops rapidly in the near past is a feasible solution.
Self-supervised Learning (SSL). SSL designs pretext tasks to synthesize pseudo labels and then formulates it as a prediction task to learn the representations [15]. For example, Gidaris et al. [17] proposed to augment 2D images by applying rotations, and then learn image features to recognize the rotations. They demonstrated that this task provided a powerful supervisory signal for semantic feature learning and significantly closed the gap between supervised learning and unsupervised learning. Doersch et al. [32] proposed to learn representations by predicting context information of local patches. Noroozi and Favaro [33] approached SSL by predicting the position of randomly rearranged local patches of images. Pathak et al. [34] used inpainting to learn representations. Chen et al. [35] proposed a SSL strategy based on context restoration to exploit unlabeled medical images. They conducted classification on 2D ultrasound images, localization on CT images and segmentation on magnetic resonance (MR) images. Experimental results showed the semantic features learned by context restoration improved the performance of machine learning models on these tasks.
From the perspective of whether manual annotations are needed, SSL belongs to the unsupervised learning. In terms of the learning principle, SSL belongs to the discriminative learning. Discriminative approaches learn representations through objective functions associated with pretext tasks, and the models are optimized in a supervised manner. SSL approaches rely on heuristics to design pretext tasks. In particular, we regard Instance Recognition (IR) as one type of SSL, whose pretext task is to identify each instance. The model can be parametric or non-parametric. The Exemplar CNN [36] is a parametric example. The instance discrimination method [37] is a non-parametric example.
Contrastive Learning. This approach learns representations by contrasting sample pairs. IR is typically implemented using contrastive learning. The Exemplar CNN [36] represented each instance as a vector and trained a network to recognize each instance. Sampling is an important issue in contrastive learning. Wu et al. [37] constructed a memory bank to store instance vectors, and several works have used in-batch samples instead of memory banks to conduct contrastive learning. Recently, Chen et al. proposed the SimCLR [16], which generated augmentation-invariant embedding for input images. SimCLR outperformed previous SSL and semi-supervised learning on ImageNet. With a linear classifier trained on the learned representations, SimCLR reached a matching top-1 accuracy (76.5%) to that of ResNet-50. He et al. [38] proposed the momentum contrast (MoCo) for visual representational learning. MoCo built a large and consistent dictionary to facilitate efficient contrastive learning. Experimental results show that the representations learned by MoCo outperform their supervised pre-training counterparts in detection and segmentation tasks on benchmark PASCAL VOC, COCO, etc. These works showed that contrastive learning can effectively bridge the gap between unsupervised learning and supervised learning. The models in contrastive learning usually involve more parameters than their supervised counterparts. For example, to achieve a comparable top-1 accuracy, the parameters of SimCLR are 16 times of ResNet-50. We refer to contrastive learning as a special case of SSL. A typical SSL method defines pretext tasks on a same instance, whereas contrastive learning defines tasks on instance pairs.
Recent studies have proved that SSL can improve the quality of few-shot learning [39], transfer learning [40] and semi-supervised learning [37]. However, the value of SSL in the MTL framework remains undiscovered. We argue that SSL is able to provide annotation-free supervisions to improve the generalization performance of supervised learning in a MTL scheme. MTL, in turn, provides clear task-specific orientations for SSL. The CMT-CNN proposed in this paper integrates SSL as a plugin module into MT-CNN, which can improve the diagnostic accuracy of COVID-19 when the data amount is not very large.
3. Methods
Our goal is to learn a parametric model from a set of labeled imageswhere denotes the sample amount of the dataset. We hope that the representations generated by are able to (1) facilitate the high-accuracy classification model denoted as, and (2) exhibit good embedding properties. The former can be achieved by supervised training to yield inductive bias. The latter improves the quality of the representations to enhance the generalization on unseen samples, especially when the data amount is small. Recent works have shown evidences that high-quality embedding can achieve good classification performance with limited labeled data [16], [41], or even in the absence of labeled data [16], [38]. We therefore propose the CMT-CNN, a MTL solution to achieve these goals simultaneously. In the following, we will introduce the flowchart of CMT-CNN in an intuitive manner, formulate the objective function, and then explain the optimization procedure.
3.1. Data preparation
We conduct CT experiment using 3D volumes, which involve substantial pixels (e.g.,). To avoid overfitting, they are uniformly sampled as volumes to feed the CMT-CNN. The X-ray images are originally stored as 3-channel images. We resize every X-ray image to as a greyscale image. For every 3D CT volume and every 2D X-ray image, we normalize the pixels to using min-max scaling, and then every pixel is subtracted by 0.5. Therefore, the pixel values are normalized to.
3.2. Contrastive multi-task convolutional neural network (CMT-CNN)
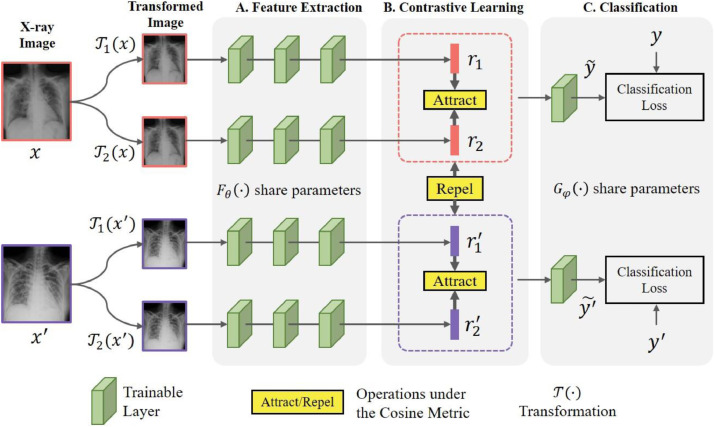
The flowchart of the proposed framework is summarized in Fig. 1 , which involves a main task: COVID-19 diagnosis, and an auxiliary task: Contrastive learning. The main task is implemented with the supervised training of neural networks, like relevant studies have done [4], [9], [10], [22]. The flexible SoftMax classification can be adopted to the binary classification (COVID-19 and other conditions), the ternary classification (COVID-19, other pneumonia and normal control) or any other tasks.
Fig. 1.
Flowchart of the proposed Contrastive Multi-task Convolutional Neural Network (CMT-CNN). The framework allows various choices of medical images. We opt for simplicity and adopt X-rays and two types of transformations for illustration. Images sampled in a training batch are transformed and fed into the feature extraction network (FEN) denoted as. On the top of FEN, contrastive learning is conducted to attract features belonging to a same instance closer, whereas features belonging to different instances are separated. The features are then fed into a FC layer activated by SoftMax denoted as for COVID-19 diagnosis. Note that the test images do not need to be transformed, and the original images are directly fed into the model.
The auxiliary task endows the model with the ability of making transformation-invariant predictions. Inspired by several studies in SSL [16], [21], [42], we adopt a series of data-augmentation methods as transformations. Then, contrastive learning is conducted based on the SoftMax embedding [21] to endow the model with fine spatial aggregation properties: (1) augmentations of a same instance are grouped together, and (2) augmentations of different instances are scattered across the embedding space.
3.2.1. Transformations applied to images
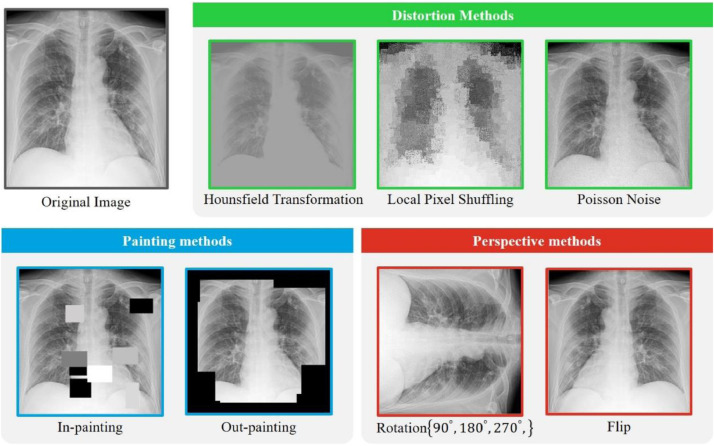
The transformations used in relevant literatures are quite different. Giving full consideration to the characteristics of CT/X-ray diagnosis of pneumonia, we define three categories of transformations, and new transformations can be easily added to our framework. Fig. 2 visualizes the applied transformations. We randomly pick out a COVID-19 X-ray for illustration.
-
(1)
Distortion methods. These methods change the value of pixels on the image. Because the pixel values at the micro level have changed, the model can pay more attentions to macro features such as shape and texture. We believe that this characteristic is beneficial to the diagnosis of COVID-19. We adopt three distortion methods. The first is Hounsfield transformation, which was recently proposed by Zhou et al. [42] to provide strong pixel-wise supervision. Restoring images distorted by the Hounsfield transformation can focus the model on learning lesion appearance in terms of shape and intensity distribution. We use the Bézier curve, a smooth and monotonous function to assign every pixel a value. The second is local pixel shuffling. For a given image, this method randomly samples some windows, and then shuffles the order of the pixels within the window. Apparently, the pixel values have not changed, but the relative positions between the pixels have changed. Zhou et al. [42] have demonstrated that to recover from local pixel shuffling, the model must memorize the global geometry, lesion spatial layout, local boundaries and texture, which encourage the model to learn the shapes and boundaries of disease-related lesions as well as the relative layout of different parts of them. To avoid changing the global content of the image, the window size should be smaller than the receptive field of the model. The third is Poisson noise. We propose the Poisson noise as a new training scheme for SSL. CT and X-ray images are based on X-ray. Physicists and radiologists have pointed out that the quantum noise of the X-ray imaging obeys the Poisson distribution [43] rather than the widely-used Gaussian noise in computer vision tasks [16]. Different from the Hounsfield transformation, which computes the values of every pixel based on its context pixels, the Poisson noise applies random noise to CT and X-ray images to improve the robustness to random noise, aside from the same benefits brought by Hounsfield transformation. The first row of Fig. 2 gives an example of how the distortion methods change an image.
-
(2)
Painting methods. These methods block part of the image randomly using random windows. As part of the image is lost, the model has to seek for complementary information in other pixels across the image. This characteristic will lead to context-awareness. Different from the context encoder [34] conducting in-painting at the central region of images, we adopt both in-painting and out-painting, which are suitable for SSL in medical image analysis. As for in-painting, the pixels inside the window are replaced by a constant, and the pixels outside the window are preserved. This characteristic allows the model to learn local continuities of lesions via interpolating [42]. As for out-painting, the pixels in the window are preserved, and the pixels outside the window are replaced by a constant. This characteristic compels the model to learn global geometry and lesion layout via extrapolating [42]. The in-painting and out-painting operations are complementary to each other. COVID-19 often manifests as multiple pathological changes on CT and X-ray images, and the painting operations allow the model to make context-aware predictions. We illustrate the effect of the painting methods at the second row of Fig. 2.
-
(3)
Perspective methods. These methods view the same image from different geometric perspectives, and then compel the model to recognize the semantic concept in them without difference. We adopt two perspective methods. The first is rotation, and the second is flip. Inspired by several recent studies in SSL [16], [17], we rotate the image according to an angle set . Different form the rotation prediction [17] which decodes the rotation at the output of the model via a classification task, we focus on making the rotated images with exact semantic information share similar representations. We argue that in order a model to be able to generate rotation-invariant representations it will require to understand the semantic concepts of objects (lesions) in the image, including the location, the shape, the appearance, and in particular, the type of the lesions. Based on similar motivations, we introduce the flip operation. The second row of Fig. 2 depicts how the perspective methods transform an image.
Fig. 2.
Illustrations of the transformations applied to X-ray images. The distortion methods, painting methods and perspective methods are highlighted in green, blue, and red, respectively. By making the representations of different transformations performed on a same image similar in a latent space, the model learns semantic features with transformation-invariant characteristics. The distortion methods change pixel values on a micro scale, making the model pay attention to macro features including the global geometry, lesion spatial layout, shape and texture. The painting methods lead to context-awareness by randomly sheltering part of the image, which encourages the model to learn the lesion spatial layout and local continuities. The perspective methods encourage the model to learn the location, shape and appearance information. These are important information for the diagnosis of COVID-19 and many other diseases using medical images. For CT, the transformations are applied to each slice. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2.2. Contrastive learning
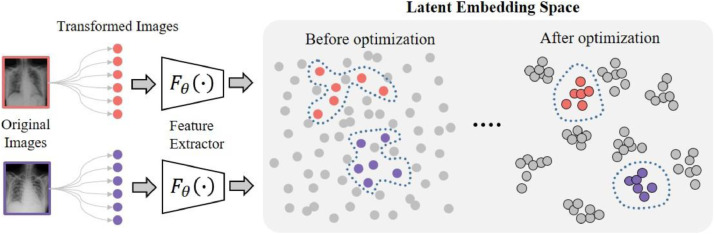
Contrastive learning is the auxiliary task of the CMT-CNN. Fig. 3 shows its effect in the representational learning. Numerous studies have demonstrated that well-optimized embedding through representational learning can provide high-quality representations for downstream tasks [15], [16], [38]. Motivated by this, we introduce contrastive learning as the auxiliary task to enhance the embedding quality so as to improve the model generalization. Note that the feature extractor can be seen as an embedding function, which is trained based on self-supervisions. The self-supervisions we use are the instance labels, i.e., the network can recognize every instance in a transformation-invariant manner and the distance between paired instances is encouraged.
Fig. 3.
Illustration of the effect of contrastive learning. We use different colors to distinguish different samples. Each image is transformed by a series of augmentation operations, and the feature extractor projects the transformed images to the latent embedding space. Contrastive learning is used to optimize the model for better spatial aggregating properties: representations belonging to a same image are concentrated together to formulate semantic clusters, whereas representations belonging to different images are scattered across the latent embedding space. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
For a clear description, we first introduce the algorithm using two images and two transformations (see Fig. 1). Let be a CT/X-ray image, where and are rows and columns of the image, respectively. The image is transformed by and, respectively. Then, and are fed into the feature extractor, which can be arbitrary CNN structures. After substantial layers of convolution and pooling, the augmented images are projected to a latent space, where vectors are defined as ( is the dimensionality of the latent space). We denote) as, and) as. Similarly, we obtain the representations of another image, i.e., and. We adopt the cosine distance to measure the distance between any two vectors in the latent space. Contrastive learning can be seen as an information retrieval task. Assuming that is fixed, we use as a query to search from a dictionary, where is the correct retrieval. Conversely, the correct retrieval for is.
Now we present the contrastive learning scheme within a training batch consisting of randomly-sampled images from the training set. We apply two transformations to the training batch, and data points are thus obtained. Let be a positive pair, i.e., they are representations of two views of a same image. When is fixed, the loss function can be written as
| (1) |
where is a temperature parameter controlling the concentration level of the sample distribution. Several studies have demonstrated that an appropriate can help learn from hard negatives [16], [21], [44]. The value of is generally small (e.g., 0.1). The indicator function is defined as
| (2) |
The loss function in Eq. (1) have been used in several works [37], and was termed the normalized temperature-scaled cross entropy (NT-Xent), which has shown significant advantage over NT-Logistic and Margin Triplet in contrastive learning [16]. The NT-Xent loss can guide the model to achieve good embedding results by learning transformation-invariant and spread-out features. Considering that both the logarithmic function and the exponential function are monotonically increasing, minimizing Eq. (1) requires maximizing and minimizing, . Note that is normalized. Maximizing means increasing the cosine similarity between and, and thereby drawing them closer in the latent space. This will result in transformation-invariance since is a positive pair. Meanwhile, minimizing means all the other vectors are separated from. Since the loss function will be computed across a training batch, the representations of different samples are separated from each other.
There are two notable issues about the NT-Xent. First, negative examples are not explicitly assigned, but rather, aside from the positive pair, the other examples are treated as negative examples. Second, both and will be computed across all positive pairs to yield a final loss for a training batch. Therefore, the loss function on a training batch is defined as
| (3) |
where is typically not equal with . We are seeking for parameter for according to
| (4) |
3.2.3. COVID-19 diagnosis
Diagnosing COVID-19 is the main task of CMT-CNN, which is formulated as a binary or ternary classification task. The latent representation of an image sample is, and is the predicted label. Our goal is to make similar to, which is the true label of the sample. In a training batch, we use the Kullback-Leibler (K-L) divergence to measure the distance between the true label distribution and the predicted label distribution:
| (5) |
where the first term is the (negative) entropy of , a constant in a training batch. The second term is the cross entropy loss, which we denote as
| (6) |
We are seeking for parameters and for and according to
| (7) |
3.3. Loss function and optimization
We write the overall loss function of CMT-CNN as
| (8) |
where balances the main task and the auxiliary task. In the optimization process, we use the following method to update the model parameters:
| (9) |
where is the learning rate. Note that the update procedure shown in (9) is applied on training batches with instances. Large batchsize tends to smooth the training curves, and existing studies have demonstrated large batchsize is beneficial to contrastive learning such as SimCLR [16]. In our training scheme, is applied to optimize both and, whereas is applied to optimize only.
3.4. Evaluation metrics
We regard the performance on the test data as the proxy to evaluate the generalization ability of CMT-CNN. For the binary classification, the performance is measured under four metrics: (1) Accuracy measuring by what percentage the test instances are correctly classified. It has been used in many works concerning COVID-19 diagnosis [4], [9], [13]. (2) Sensitivity measuring by what percentage the COVID-19 instances are correctly identified. (3) Specificity measuring by what percentage the non-COVID-19 instances are correctly excluded. (4) AUC depicting the area of the receiver operating characteristic curve (ROC). AUC comprehensively considers the sensitivity and the specificity. For the ternary classification, we use the accuracy as the metric, like several recent works have done [4], [13].
4. Experiments and results
We evaluate the method on a CT dataset [4] and an X-ray dataset [13] for both the binary and ternary classifications.
We use the CC—CCII1 dataset to validate the CMT-CNN. To our knowledge, it is currently the largest public dataset for CT-based COVID-19 diagnosis. There are totally 4356 CT scans, which encompass 1578 COVID-19 scans, 1614 common pneumonia scans, and 1164 normal control scans from 2778 peoples. Among these, COVID-19 diagnosis is made by confirmed RT-PCR positive. The common pneumonia includes viral pneumonia (adenoviral, influenza and parainfluenza pneumonia), bacterial pneumonia and mycoplasma pneumonia. All of the common pneumonias are diagnosed based on standard clinical, radiological and culture/molecular assay results. In addition to the CC—CCII dataset, our hospital has 402 CT scans from 108 diagnosed patients confirmed by RT-PCR test, which are added to the COVID-19 class.
The X-ray dataset contains three subsets of X-ray images of patients which are positive of COVID-19 or other viral and bacterial pneumonia plus X-ray images of normal controls. The COVID-19 instances are from three sources. The first is contributed by Cohen et al.,2 and the second is from a Kaggle competition.3 The third is from our hospital.4 There are 231 COVID-19 instances with confirmed RT-PCR diagnosis in total. The pneumonia (MERS, SARS, ARDS, etc.) and normal control instances are from one public resource, which can be accessed by searching for the ‘labeled optical coherence tomography (OCT) and chest X-ray images for classification’ on the Mendeley data website.5 There are 4007 instances for pneumonia, and 1583 instances for normal controls, respectively.
4.1. Model implementation details
MT-CNN embraces any kinds of CNNs without constriction. To demonstrate the advantages of contrastive learning in improving the generalization ability of the model, we explore the following three CNNs, which are the most commonly-used classical models, or the latest and most powerful model in computer vision tasks. Note that all these three CNNs have been applied to diagnose COVID-19 under single-task schemes [9], [13], [22].
-
(1)
VGG-19. This CNN won the localization and classification on ILSVRC 2014 with an architecture like the AlexNet. VGG-19 has 16 convolution layers using kernel with stride 1 for feature extraction followed by three FC layers to fulfill tasks. Note that five max-polling kernels are inserted in the sequential convolutions. VGG-19 has been used for analyzing X-ray images for COVID-19 diagnosis [13]. We use the output of the first FC layer (4096-D) as the latent representation of a given image.
-
(2)
ResNet-50. This CNN won the classification on ILSVRC 2015. Since then, it has been widely applied as backbone for computer vision tasks including medical image analysis. ResNet-50 introduces shortcut connections to combat the problem of gradient-vanishing in deep structures. ResNet-50 has been used for the automatic diagnosis of COVID-19 based on both X-ray [13] and CT [9]. For each CT/X-Ray image, we adopt the output of the last max-pooling layer (4096-D) as the latent representation.
-
(3)
EfficientNet. This CNN was proposed in 2019 and achieved state-of-the-art top-1 accuracy on ImageNet, and the overall performance surpassed those of the ResNet-101, NASNet-A, GPipe, etc. in both training speed and accuracy. EfficientNet B4 is adjusted in terms of model width, model depth and image resolution based on EfficientNet B0 obtained by network architecture search. EfficientNet B4 has been used in a recent research for the diagnosis of COVID-19 [22] and therefore we adopt this version. We use the output of the first FC layer (4096-D) as the latent representation of a given image.
The models are trained from scratch using PyTorch with Adam optimizer with a learning rate of 1e-3. Large batchsize (e.g., 2048, 4096) seems to be necessary in SSL for providing reliable gradients [45]. However, this will require a large-scale memory and high hardware requirements such as multiple TPU training. In our experiment, CMT-CNN considers supervisions from the disease labels, which allows for small batchsize for SSL during training. We find that the CMT-CNN works well with small batchsize, and assign the batchsize to 8 and 64 for CT and X-ray, respectively. We adopt the suggested setting in the literature [16] and set The optimal value of is determined by the validation accuracies. We search for with a step of 0.25, and finally determine . We use the NVIDIA TITAN X Pascal GPU for training and evaluation, and the training/test time for a single image is within 0.17/0.06 second, respectively.
4.2. Evaluation on the CT dataset
We use five-fold cross-validation to validate the CMT-CNN on the CT dataset. The results are summarized at Table 1 . To evaluate the three CNN models, we use the symbol ‘+’ to indicate statistical significant improvement ( evaluated by paired t-test) of EfficientNet with respect to VGG-19, and the symbol ‘*’ to indicate statistical significant improvement ( evaluated by paired t-test) of EfficientNet with respect to ResNet-50. For the binary classification, EfficientNet outperforms VGG-19 and ResNet-50 with significant advantages in accuracy and AUC. The highest AUC is 89.22% with an accuracy of 93.46%, a sensitivity of 90.57% and a specificity of 90.84%. For the ternary classification, EfficientNet outperforms VGG-19 with significant advantage, and the mean accuracy is higher than that of ResNet-50. The best accuracy is 91.45% in distinguishing COVID-19, common pneumonia and normal control. These results are comparable to the results achieved by Zhang et al. [4], where the best binary accuracy and ternary accuracy was both 92.49%. Their model also use the 3D CNN to analysis 3D CT volumes. Although using 2D slices enables more training samples, 3D models are more straight-forward in CT analysis. However, Zhang et al. [4] used a two-stage scheme: the pulmonary parenchyma segmentation and the classification on the cropped pulmonary area. As a comparison, the proposed CMT-CNN is a single-stage model working in an end-to-end manner.
Table 1.
The mean classification performance (%) of CMT-CNN on CT.
| CNN Model | Binary Cls |
Ternary Cls | |||
|---|---|---|---|---|---|
| Accuracy | Sensitivity | Specificity | AUC | Accuracy | |
| VGG-19 | 86.45 | 83.22 | 81.22 | 82.76 | 84.56 |
| ResNet-50 | 91.66 | 90.44 | 89.16 | 84.18 | 89.75 |
| EfficientNet | 93.46+* | 90.57+ | 90.84+ | 89.22+* | 91.45+* |
4.3. Evaluation on the X-ray dataset
We use five-fold cross-validation to validate the CMT-CNN on the X-ray dataset. Table 2 summarizes the results. For both the binary and ternary classification across all the considered metrics, ResNet-50 outperforms VGG-19, and EfficientNet outperforms both VGG-19 and ResNet-50 in terms of the mean value consistently.
Table 2.
The mean classification performance (%) of CMT-CNN on X-ray.
| CNN Model | Binary Cls |
Ternary Cls | |||
|---|---|---|---|---|---|
| Accuracy | Sensitivity | Specificity | AUC | Accuracy | |
| VGG-19 | 93.42 | 89.91 | 89.13 | 87.15 | 91.34 |
| ResNet-50 | 95.66 | 90.85 | 90.98 | 89.74 | 92.52 |
| EfficientNet | 97.23+* | 92.97+* | 91.91+ | 92.13+* | 93.49+ |
We use the symbol ‘+’ to indicate statistical significant improvement ( evaluated by paired t-test) of EfficientNet with respect to VGG-19, and the symbol ‘*’ to indicate statistical significant improvement ( evaluated by paired t-test) of EfficientNet with respect to ResNet-50. We find that EfficientNet has significant advantage over VGG-19 across tasks and metrics, and significantly better than ResNet-50 under most of the metrics except for specificity (for binary classification) and accuracy (for ternary classification). Generally, EfficientNet has better performance and fewer parameters. For the binary classification, we achieve an AUC of 92.13% with a sensitivity of 92.97% and a specificity of 91.91%. For the ternary classification, we achieved an accuracy of 93.49%, which is the same as the best report in the existing literature (93.48%) [13].
4.4. Ablation experiments
We conduct ablation experiments to evaluate the contribution of different parts of CMT-CNN's cost function to the results. First, we only use the typical supervised learning cost function to train the model. Then, we only perform SSL on the model based on contrastive learning. Note that when a change is applied to the algorithm, the remaining experimental settings and variables remain unchanged.
4.4.1. CMT-CNN with classification task
Five-fold cross-validation is used to validate the model when only classification loss is applied, and the results (both CT and X-ray) are summarized in Table 3 . For both the imaging modalities and both the binary and ternary classifications across all the considered metrics, EfficientNet outperforms ResNet-50 and VGG-19 in terms of mean value. Therefore, the EfficientNet is not only suitable for the multi-task scenario, but also shows advantage in the single-task scenario.
Table 3.
Ablation test results (%) of CMT-CNN with classification task.
| Binary Cls |
Ternary Cls | |||||
|---|---|---|---|---|---|---|
| Model | Accuracy | Sensitivity | Specificity | AUC | Accuracy | |
| CT | VGG-19- | 83.12 | 78.66 | 78.79 | 78.15 | 81.24 |
| ResNet-50- | 85.67 | 82.42 | 82.43 | 90.45 | 83.87 | |
| EfficientNet- | 87.21 | 84.12 | 85.21 | 84.87 | 85.96 | |
| X-ray | VGG-19- | 91.16 | 89.45 | 88.74 | 86.17 | 90.21 |
| ResNet-50- | 93.79 | 90.24 | 90.87 | 89.23 | 91.10 | |
| EfficientNet- | 94.81 | 92.16 | 91.01 | 91.47 | 92.53 | |
For all the results, the single-task CNN using classification loss only is inferior to the proposed CMT-CNN in terms of the mean value. We use the symbol ‘’ to indicate statistical significant decline ( evaluated by paired t-test) of the classification-only CNN with respect to the CMT-CNN. For CT, the best-performing EfficientNet encounters a significant decline on accuracy (6.25%), sensitivity (6.45%), specificity (5.63%) and AUC (3.45%) for binary classification, as well as a significant decline on accuracy (5.49%) for the ternary classification. For X-ray, the EfficientNet encounters a significant decline on accuracy (2.42%) for the binary classification. These observations indicate that CT is more sensitive to the auxiliary task.
4.4.2. CMT-CNN with contrastive learning task
Table 4 summarizes the results when only contrastive loss is applied under five-fold cross-validation. The model learns embedding through SSL in an unsupervised manner. We use the symbol ‘’ to indicate statistical significant decline ( evaluated by paired t-test) of the SSL-only CNN with respect to the CMT-CNN. For both CT and X-ray, both the binary and ternary classification tasks and all the metrics, the performance declines significantly. For CT, the binary-classification accuracy, sensitivity, specificity, AUC and ternary-classification accuracy of EfficientNet drop by 7.00%, 6.36%, 6.65%, 6.80% and 7.26%, respectively. For X-ray, the binary-classification accuracy, sensitivity, specificity, AUC and ternary-classification accuracy of EfficientNet drop by 5.51%, 4.20%, 1.74%, 2.16% and 3.02%, respectively. Despite of the significant differences, we argue that SSL is able to yield reasonable predictions even without disease labels. In particular, the accuracy achieved by SSL in the ternary classification of X-ray images reached 90.47%, demonstrating the potential of the unsupervised learning in learning discriminative representations.
Table 4.
Ablation test results (%) of CMT-CNN with contrastive learning task.
| Binary Cls |
Ternary Cls | |||||
|---|---|---|---|---|---|---|
| Model | Accuracy | Sensitivity | Specificity | AUC | Accuracy | |
| CT | VGG-19- | 82.53 | 76.55 | 76.34 | 77.73 | 80.56 |
| ResNet-50- | 84.76 | 81.43 | 79.67 | 79.54 | 80.25 | |
| EfficientNet- | 86.46 | 84.21 | 84.19 | 82.42 | 84.19 | |
| X-ray | VGG-19- | 87.24 | 84.31 | 85.88 | 83.24 | 88.24 |
| ResNet-50- | 89.88 | 87.32 | 86.76 | 86.33 | 89.31 | |
| EfficientNet- | 91.72 | 88.77 | 90.17 | 89.97 | 90.47 | |
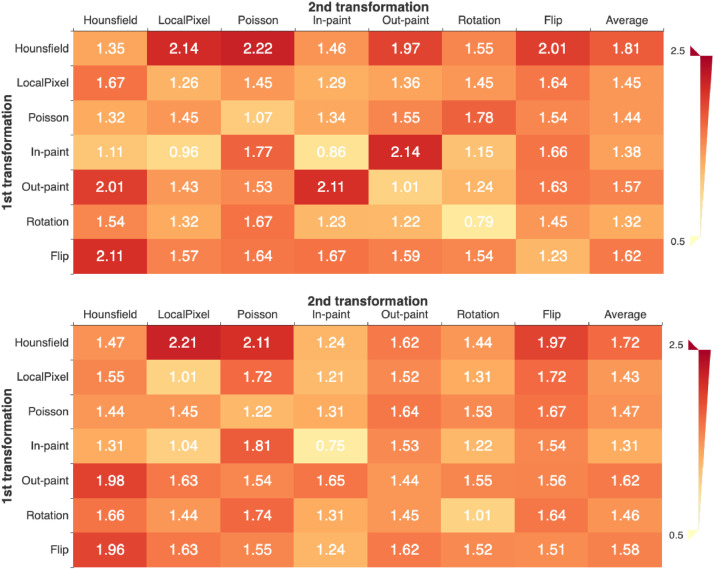
4.4.3. The influence of transformations on the results
To quantitatively measure the contribution of each transformation, we summarize the accuracy improvements of CMT-CNN with respect to the baseline CNN in Fig. 4 . We use the EfficientNet B4 for evaluation. The transformations are applied individually or in sequential pairs, which is similar to the protocol in SimCLR [16]. For each matrix, the last column shows the averaged accuracy over each row. According to the results, the top-3 effective transformations for both CT and X-ray are the same: Hounsfield transformation, out-painting and flip. Coincidentally, they belong to the distortion methods, painting methods and perspective methods, respectively. By changing substantial pixels, Hounsfield transformation can focus the model on the lesion shape and intensity distribution. Out-painting makes the model learn global geometry and lesion layout. Flip endows the model with the awareness of lesion location, especially when we consider that the two lung lobes have a certain symmetry. The above features are important features for diagnosing the pneumonia.
Fig. 4.
Accuracy improvement of CMT-CNN over the baseline under individual or composition of data transformations. The first matrix is for CT and the second matrix is for X-ray. For both the two matrices, the diagonal elements are for the single transformation, and the off-diagonal elements are for the composition of two sequentially-applied transformations. The last column shows the averaged accuracy improvement over each row.
5. Discussions
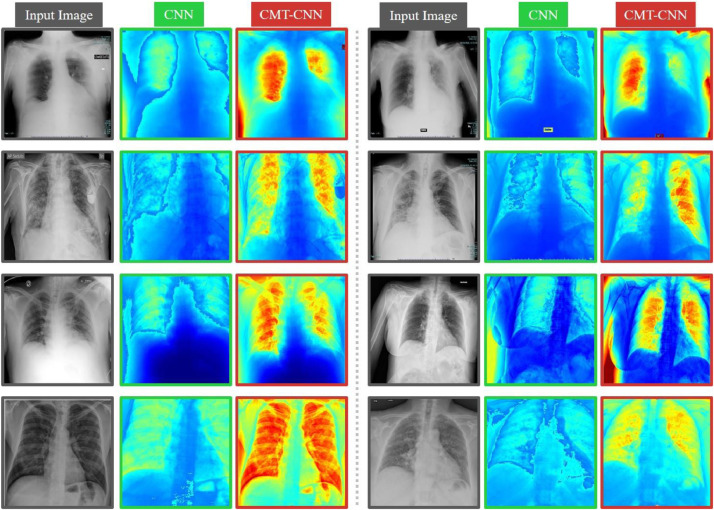
The acquisition of medical images is expensive, and the scales of well-annotated datasets are difficult to reach the scales of computer vision datasets. Specifically, in the face of emerging epidemics such as COVID-19, the number of samples is even more limited. This inspired us whether we can improve the generalization ability of the model on unseen samples under the condition of relatively limited training samples through SSL in addition to the typical supervised learning. SSL is used as an auxiliary task to encourage well-aggregated embedding characteristics based on contrastive learning. We have demonstrated theoretically that the auxiliary task can bring improvement to the automatic diagnosis of COVID-19. Empirical results have validated the assumption, and the accuracies we achieved are comparable to the best records using the same CT and X-ray datasets. In Fig. 5 , we visualize the attention maps of the typical CNN and the CMT-CNN based on the EfficientNet B4 model for further explanation. A Channel Average Pooling placed after the last feature map (with a size of 16 16 2048) generates the feature map with a size of 16 16 1 and two successive up-sampling operators (deconvolution with a scale of 2) generate the new feature map with a size of 64 64 1. To train these two up-sampling operators, we utilize two down-sampling operators (mean pooling with a scale of 2) to reduce the size of generated feature maps (from 64 64 1 to 16 16 1) and embed it into the original feature map with the element-wise product operation for all the channels. After obtaining the output of two upsampling operations, we utilize bilinear interpolation to resize it to 512 512 1, and a Sigmoid operation is used to reflect the focused areas in images. Then we embed the attention map into the original input images via the element-wise product operation to show where our model focused. We randomly select eight X-ray images for illustration. The CNN is trained on the typical classification loss, and the CMT-CNN is trained on the multi-task loss. There are two interesting phenomena. First, CMT-CNN can better distinguish between lung parenchyma and non-pulmonary tissues. Second, within the lung parenchyma, CMT-CNN can better focus on the lesions. Therefore, the auxiliary task designed to improve the embedding quality makes the model pay attention to lesions on the images. Upon these, the diagnostic performance on unseen data is enhanced. Similar phenomenon has been observed and reported by Gidaris et al. [17], where rotation-based SSL can help the model focus on the object. It is worth noting that the attention maps generated by deeper representations show better lesion-concentrating effects than the early layers do. Since the deeper layers of CNN learn semantic information, SSL improves the semantic learning ability remarkably.
Fig. 5.
Attention maps generated by the CMT-CNN and the baseline CNN based on the EfficientNet. We show eight X-ray images of COVID-19 patients for illustration. To restore the attention maps to the resolution of the input images, we use a decoder with two upsampling layers after the latent embedding to generate the attention maps. To train the decoder, we encoder the attention map and fuse it into the original feature map via an element-wise sum operator and then to feed them into the later layers. This method can locate the areas that the network pays attention to. For each X-ray image, we show the attention maps of the CNN using classification loss only (green rectangle) and the CMT-CNN model with classification loss together with contrastive loss (red rectangle). Compared with a typical CNN, the CMT-CNN can better focus on the lesion. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
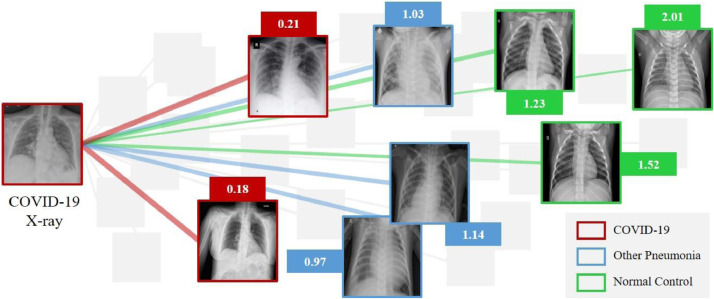
To further illustrate the semantic learning ability of SSL, Fig. 6 shows an example of the contrastive losses between X-rays with different semantic labels. For a given COVID-19 X-ray, its contrastive losses with respect to other COVID-19 X-rays are small (e.g., 0.2), its contrastive losses with respect to other pneumonia are larger (e.g., 1.03), and its contrastive losses with respect to normal controls are the largest (e.g., 1.52). These proves that the contrastive loss enables discrimination to semantic labels. Fig. 6 is only for an intuitive illustration. Considering the purpose of this paper, we did not evaluate the semantic classification performance by adding a simple classifier upon the representations. More in-depth analysis can be found elsewhere [16], [21].
Fig. 6.
Contrastive losses computed between a given COVID-19 X-ray and other X-rays from the dataset. We split the dataset into a training set and a test set. An EfficientNet is trained using contrastive loss only. Then, we randomly pick up a COVID-19 X-ray from the test set and contrast it with every X-ray in the test set. We use different colors to represent different semantic labels. The lines represent the connections between X-ray pairs. The smaller the contrastive loss, the bolder the line and the higher the similarity between the pair. SSL implemented with contrastive loss shows excellent ability to distinguish semantic labels.
Among the considered CNN structures, EfficientNet significantly outperforms the commonly-used VGG-19 and ResNet-50 for both the binary and ternary classifications for both CT and X-ray diagnosis of COVID-19. It is notable that the parameter amount of the EfficientNet B4 is 19 million, fewer than that of the VGG-19 (143 million) and the ResNet-50 (24 million). EfficientNet leverages the model depth, model width and input resolution to yield optimized model architectures. We have shown that this model is a feasible backbone for our task.
The data used in different studies are different, the experimental setups are different, and the evaluation protocols are also different. According to the literatures [4], [10], [12], [13], [25], the binary and ternary classification accuracies on CT are mainly between 90% and 96%, and the accuracies on X-ray are between 89% and 98%. Table 5 shows the comparison between CMT-CNN and the state-of-the-arts. In CT diagnosis, the metrics obtained by CMT-CNN are basically the same as those of Zhang et al. [4], except that the sensitivity has decreased by 4%. One reason is that they segmented the lung parenchyma to make the model focus more on the lesion. As a comparison, segmentation is not a precondition in CMT-CNN, and we implement the diagnosis in an end-to-end manner. Under an end-to-end framework, Ouyang et al. [10] generated attention maps to help the model focus on the lesions, and exploited a dual-sampling algorithm to reduce the model bias. Although being superior to CNN, their model is inferior to the CMT-CNN by large margins. One reason is that the training data (2186 scans) are less than what we used (3806 scans in each fold). The structured latent multi-view representation learning proposed by Kang et al. [12] shows advantage on sensitivity by 4% over CMT-CNN by considering 93 radiomic features and 96 handcrafted features, where substantial feature engineering and annotations are needed. It is noteworthy that Ouyang et al. [10] and Kang et al. [12] only considered the binary classification, whereas the ternary classification results were not presented.
Table 5.
Performance Comparison of CMT-CNN with State-of-the-arts.
| Data | Method | Binary Cls |
Ternary Cls | |||
|---|---|---|---|---|---|---|
| Accuracy | Sensitivity | Specificity | AUC | Accuracy | ||
| CT | Zhang et al. [4] | 92.49 | 94.93 | 91.13 | NP | 92.49 |
| Ouyang et al. [10] | 87.50 | 86.90 | 90.10 | NP | NP | |
| Kang et al. [12] | 93.90 | 94.60 | 91.70 | NP | NP | |
| CMT-CNN | 93.46 | 90.57 | 90.84 | 89.22 | 91.45 | |
| X-ray | Oh et al. [25] | 88.90 | NP | NP | NP | NP |
| Apostolopoulos et al. [13] | 96.78 | 98.66 | 96.46 | NP | 94.72 | |
| CMT-CNN | 97.23 | 92.97 | 91.91 | 92.13 | 93.49 | |
Results shown in mean value (%). NP: Not provided.
In X-ray diagnosis, Oh et al. [25] proposed a patch-based CNN to confront the shortage of annotated data in the binary classification. The accuracy (88.9%) is much lower than that of CMT-CNN (97.23%) due to the lack of data. Apostolopoulos et al. [13] conducted both the binary and ternary classification on a relatively small dataset (1427 images) using CNNs pre-trained on the ImageNet with superior sensitivity (98.66%) and specificity (96.46%), although the accuracies are basically the same as those of CMT-CNN.
Ablation results show that the gap between supervised learning and unsupervised learning has largely been bridged. For CT (EfficientNet), the gaps between supervised learning and unsupervised learning are 0.75% (binary accuracy), 2.39% (AUC) and 1.77% (ternary accuracy). For X-ray, the gaps are 3.09% (binary accuracy), 1.50% (AUC) and 2.06% (ternary accuracy). These phenomena imply SSL (unsupervised learning) has a bright future in (1) unsupervised semantic feature learning and (2) assisting supervised learning in a multi-task framework.
6. Conclusion
We have proposed the CMT-CNN, an effective multi-task learning framework for the automatic diagnosis of COVID-19. We have validated that the contrastive learning module can be easily integrated into existing CNN models without constrains to bring solid improvement to the performance. The module is based on self-supervisions and requires no additional human annotations. As a core element of self-supervised learning, we for the first time categorize the transformations into distortion methods, painting methods and perspective methods. These transformations have good interpretability in the medical image analysis. Among all the transformations, Hounsfield transformation, out-painting and flip have been identified to be the top-3 effective transformations in both the CT and the X-ray analysis. Experimental results on a CT dataset and an X-ray dataset show that CMT-CNN significantly outperforms CNNs in diagnosing COVID-19, and the results are comparable to the state-of-the-arts. CMT-CNN shows good generalization ability, which can help doctors make more objective diagnostic decisions. This study has two insufficiencies. First, we validate the effectiveness of the CMT-CNN on the classification task, whereas not on the localization and segmentation tasks. We believe that CMT-CNN can also improve the performance in localization and segmentation tasks since the contrastive loss can effectively focus the model on the lesions. With the completion of datasets with box-level and pixel-level annotations, we will evaluate our method in localization (under both the normal setting and weakly-supervised setting) and segmentation tasks. Second, like existing studies, we put together pneumonias such as MERS, SARS and ARDS collectively as ‘other pneumonia’ without further discrimination. One reason is that there are few samples for each type of pneumonia and the samples are unevenly distributed. Developing effective algorithms to solve the long-tail dataset is a valuable research direction. The main methods include, but are not limited to, more sophisticated cost-sensitive learning and sampling strategies for medical images.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported in part by Zhejiang Provincial Natural Science Foundation of China (LQ20F030013), Research Foundation of HwaMei Hospital, University of Chinese Academy of Sciences, China (2020HMZD22), Ningbo Public Service Technology Foundation, China (202002N3181), and Medical Scientific Research Foundation of Zhejiang Province, China (2021431314).
Biographies
Jinpeng Li received the Ph. D. degree (2019) in Pattern Recognition and Intelligent System from National Laboratory of Pattern Recognition, Institute of Automation, Chinese Academy of Sciences. His research interests include machine learning, deep learning, transfer learning algorithms and their applications in brain-computer interfaces and medical image analysis.
Gangming Zhao received the M. E. degree (2019) in Pattern Recognition and Intelligent System from Institute of Automation, Chinese Academy of Sciences. His research interest mainly include deep learning and its applications in computer vision tasks.
Yaling Tao received the M. S. degree (2016) in Immunology from Institute of Zoology, Chinese Academy of Sciences. Her research interests mainly include immunology, oncology, and clinical medical imaging.
Penghua Zhai received the M. E. degree (2019) in Automatic Control from Shenyang Institute of Automation, Chinese Academy of Sciences. His research interests mainly include machine learning, deep learning and their applications in medical image analysis.
Hao Chen received the M. E. degree (2019) in the University of Southampton. His research interests are machine learning, deep learning and their applications in medical image analysis.
Huiguang He received the Ph. D. degree in Pattern Recognition and Intelligent System with Institute of Automation, Chinese Academy of Sciences (CASIA). His research interests include pattern recognition, medical image processing, and brain-computer interfaces. He is now a full professor with CASIA.
Ting Cai received the M. M. degree (2012) in Clinical Medicine from Nankai University, China and Flinders University, Australia. He is now president of HwaMei Hospital, University of Chinese Academy of Sciences, and director of Key Laboratory of Diagnosis and Treatment of Digestive System Tumors of Zhejiang Province.
Footnotes
References
- 1.Guan W., Ni Z., Hu Y., Liang W., Ou C., He J., Liu L., Shan H., Lei C., Hui D.S.C., Du B., Li L., Zeng G., Yuen K.-.Y., Chen R., Tang C., Wang T., Chen P., Xiang J., Li S., Wang J., Liang Z., Peng Y., Wei L., Liu Y., Hu Y., Peng P., Wang J., Liu J., Chen Z., Li G., Zheng Z., Qiu S., Luo J., Ye C., Zhu S., Zhong N. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/nejmoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L., Tai Y., Bai C., Gao T., Song J., Xia P., Dong J., Zhao J., Wang F.S. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie X., Muruato A., Lokugamage K.G., Narayanan K., Zhang X., Zou J., Liu J., Schindewolf C., Bopp N.E., Aguilar P.V., Plante K.S., Weaver S.C., Makino S., LeDuc J.W., Menachery V.D., Shi P.Y. An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe. 2020;27:841–848. doi: 10.1016/j.chom.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.T. Ai, Z. Yang, H. Hou, C. Zhan, C. Chen, W. Lv, Q. Tao, Z. Sun, L. Xia, Correlation of Chest CT and RT-PCR Testing for Coronavirus Disease 2019 (COVID-19) in China: A Report of 1014 Cases. Radiology. 2020 Aug;296(2):E32-E40. doi: 10.1148/radiol.2020200642. Epub 2020 Feb 26. PMID: 32101510; PMCID: PMC7233399. [DOI] [PMC free article] [PubMed]
- 6.Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical Coronavirus Disease 2019 (COVID-19) pneumonia: relationship to negative RT-PCR testing. Radiology. 2020;296:E41–E45. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang Y., Cheng W., Zhao N., Qu H., Tian J. CT screening for early diagnosis of SARS-CoV-2 infection. Lancet Infect. Dis. 2020;20:1010–1011. doi: 10.1016/S1473-3099(20)30241-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi H., Han X., Jiang N., Cao Y., Alwalid O., Gu J., Fan Y., Zheng C. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect. Dis. 2020;20:425–434. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., Bai J., Lu Y., Fang Z., Song Q., Cao K., Liu D., Wang G., Xu Q., Fang X., Zhang S., Xia J., Xia J. Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: evaluation of the diagnostic accuracy. Radiology. 2020;296:E65–E71. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ouyang X., Huo J., Xia L., Shan F., Liu J., Mo Z., Yan F., Ding Z., Yang Q., Song B., Shi F., Yuan H., Wei Y., Cao X., Gao Y., Wu D., Wang Q., Shen D. Dual-sampling attention network for diagnosis of COVID-19 from community acquired pneumonia. IEEE Trans. Med. Imaging. 2020;39:2595–2605. doi: 10.1109/TMI.2020.2995508. [DOI] [PubMed] [Google Scholar]
- 11.Wang X., Deng X., Fu Q., Zhou Q., Feng J., Ma H., Liu W., Zheng C. A weakly-supervised framework for COVID-19 classification and lesion localization from chest CT. IEEE Trans. Med. Imaging. 2020;39:2615–2625. doi: 10.1109/TMI.2020.2995965. [DOI] [PubMed] [Google Scholar]
- 12.Kang H., Xia L., Yan F., Wan Z., Shi F., Yuan H., Jiang H., Wu D., Sui H., Zhang C., Shen D. Diagnosis of coronavirus disease 2019 (COVID-19) with structured latent multi-view representation learning. IEEE Trans. Med. Imaging. 2020;39:2606–2614. doi: 10.1109/TMI.2020.2992546. [DOI] [PubMed] [Google Scholar]
- 13.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020;43:635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shorfuzzaman M., Hossain M.S. MetaCOVID: a Siamese neural network framework with contrastive loss for n-shot diagnosis of COVID-19 patients. Pattern Recognit. 2020 doi: 10.1016/j.patcog.2020.107700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jing L., Tian Y. Self-supervised visual feature learning with deep neural networks: a survey. ArXiv. 2019:1–24. doi: 10.1109/tpami.2020.2992393. [DOI] [PubMed] [Google Scholar]
- 16.Chen T., Kornblith S., Norouzi M., Hinton G. A simple framework for contrastive learning of visual representations. ArXiv. 2020 [Google Scholar]
- 17.Gidaris S., Singh P., Komodakis N. Unsupervised representation learning by predicting image rotations. ArXiv. 2018:1–16. [Google Scholar]
- 18.Doersch C., Zisserman A. Proc. IEEE Int. Conf. Comput. Vis. 2017-Octob. 2017. Multi-task self-supervised visual learning; pp. 2070–2079. [DOI] [Google Scholar]
- 19.Zhuang C., Zhai A., Yamins D. Proc. IEEE Int. Conf. Comput. Vis. 2019-Octob. 2019. Local aggregation for unsupervised learning of visual embeddings; pp. 6001–6011. [DOI] [Google Scholar]
- 20.Huang J., Dong Q., Gong S., Zhu X. 36th Int. Conf. Mach. Learn. ICML 2019. 2019-June. 2019. Unsupervised deep learning by neighbourhood discovery; pp. 5090–5099. [Google Scholar]
- 21.Ye M., Zhang X., Yuen P.C., Chang S.F. Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit. 2019-June. 2019. Unsupervised embedding learning via invariant and spreading instance feature; pp. 6203–6212. [DOI] [Google Scholar]
- 22.Xiong Z., Wang R., Bai H.X., Halsey K., Mei J., Li Y.H., Atalay M.K., Jiang X.L., Fu F.X., Thi L.T., Huang R.Y., Liao W.H., Pan I., Choi J.W., Zeng Q.H., Hsieh B., CuiWang D., Sebro R., Hu P.F., Chang K., Shi L.B., Qi Z.Y. Artificial intelligence augmentation of radiologist performance in distinguishing COVID-19 from pneumonia of other origin at chest CT. Radiology. 2020;296:E156–E165. doi: 10.1148/radiol.2020201491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mei X., Lee H.C., yue Diao K., Huang M., Lin B., Liu C., Xie Z., Ma Y., Robson P.M., Chung M., Bernheim A., Mani V., Calcagno C., Li K., Li S., Shan H., Lv J., Zhao T., Xia J., Long Q., Steinberger S., Jacobi A., Deyer T., Luksza M., Liu F., Little B.P., Fayad Z.A., Yang Y. Artificial intelligence–enabled rapid diagnosis of patients with COVID-19. Nat. Med. 2020;26:1224–1228. doi: 10.1038/s41591-020-0931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paul Cohen J., Morrison P., Dao L. COVID-19 image data collection. ArXiv. 2020 http://arxiv.org/abs/2003.11597 [Google Scholar]
- 25.Oh Y., Park S., Ye J.C. Deep learning COVID-19 features on CXR using limited training data sets. IEEE Trans. Med. Imaging. 2020;39:2688–2700. doi: 10.1109/TMI.2020.2993291. [DOI] [PubMed] [Google Scholar]
- 26.Ruder S. An overview of multi-task learning in deep neural networks∗. ArXiv. 2017 [Google Scholar]
- 27.Adıyeke E., Baydoğan M.G. The benefits of target relations: a comparison of multitask extensions and classifier chains. Pattern Recognit. 2020;107 doi: 10.1016/j.patcog.2020.107507. [DOI] [Google Scholar]
- 28.Crammer K., Mansour Y. Learning multiple tasks using shared hypotheses. Adv. Neural Inf. Process. Syst. 2012;2:1475–1483. [Google Scholar]
- 29.Liu X., Gao J., He X., Deng L., Duh K., Wang Y.Y. NAACL HLT 2015 - 2015 Conf. North Am. Chapter Assoc. Comput. Linguist. Hum. Lang. Technol. Proc. Conf. 2015. Representation learning using multi-task deep neural networks for semantic classification and information retrieval; pp. 912–921. [DOI] [Google Scholar]
- 30.Ji Y., Sun S. Multitask multiclass support vector machines: model and experiments. Pattern Recognit. 2013;46:914–924. doi: 10.1016/j.patcog.2012.08.010. [DOI] [Google Scholar]
- 31.Ganin Y., Lempitsky V. Unsupervised domain adaptation by backpropagation. 32nd Int. Conf. Mach. Learn. ICML 2015. 2015;2:1180–1189. [Google Scholar]
- 32.Doersch C., Gupta A., Efros A.A. Proc. IEEE Int. Conf. Comput. Vis. 2015 Inter. 2015. Unsupervised visual representation learning by context prediction; pp. 1422–1430. [DOI] [Google Scholar]
- 33.Noroozi M., Favaro P. Lect. Notes Comput. Sci. (Including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics) 2016. Unsupervised learning of visual representations by solving jigsaw puzzles; pp. 69–84. [DOI] [Google Scholar]
- 34.Pathak D., Krahenbuhl P., Donahue J., Darrell T., Efros A.A. Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit. 2016-Decem. 2016. Context Encoders: feature Learning by Inpainting; pp. 2536–2544. [DOI] [Google Scholar]
- 35.Chen L., Bentley P., Mori K., Misawa K., Fujiwara M., Rueckert D. Self-supervised learning for medical image analysis using image context restoration. Med. Image Anal. 2019;58 doi: 10.1016/j.media.2019.101539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dosovitskiy A., Fischer P., Springenberg J.T., Riedmiller M., Brox T. Discriminative unsupervised feature learning with exemplar convolutional neural networks. IEEE Trans. Pattern Anal. Mach. Intell. 2016;38:1734–1747. doi: 10.1109/TPAMI.2015.2496141. [DOI] [PubMed] [Google Scholar]
- 37.Wu Z., Xiong Y., Yu S.X., Lin D. Unsupervised feature learning via non-parametric instance discrimination. Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit. 2018:3733–3742. doi: 10.1109/CVPR.2018.00393. [DOI] [Google Scholar]
- 38.He K., Fan H., Wu Y., Xie S., Girshick R. Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit. 2020. Momentum contrast for unsupervised visual representation learning; pp. 9726–9735. [DOI] [Google Scholar]
- 39.Gidaris S., Bursuc A., Komodakis N., Perez P.P., Cord M. Proc. IEEE Int. Conf. Comput. Vis. 2019-Octob. 2019. Boosting few-shot visual learning with self-supervision; pp. 8058–8067. [DOI] [Google Scholar]
- 40.Xu J., Xiao L., Lopez A.M. Self-supervised domain adaptation for computer vision tasks. IEEE Access. 2019;7:156694–156706. doi: 10.1109/ACCESS.2019.2949697. [DOI] [Google Scholar]
- 41.Tian Y., Wang Y., Krishnan D., Tenenbaum J.B., Isola P. Rethinking few-shot image classification: a good embedding is all you need? ArXiv. 2020 [Google Scholar]
- 42.Zhou Z., Sodha V., Siddiquee M.R., Feng R., Tajbakhsh N., Gotway M.B., Liang J. Models Genesis with the whole Supplementary Materials Models Genesis: generic Autodidactic Models for 3D Medical Image Analysis. Miccai. 2019:1–27. doi: 10.1007/978-3-030-32251-9_42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma L., Moisan L., Yu J., Zeng T. A dictionary learning approach for Poisson image Deblurring. IEEE Trans. Med. Imaging. 2013;32:1277–1289. doi: 10.1109/TMI.2013.2255883. [DOI] [PubMed] [Google Scholar]
- 44.Chen T., Kornblith S., Swersky K., Norouzi M., Hinton G. Big self-supervised models are strong semi-supervised learners. ArXiv. 2020:1–18. [Google Scholar]
- 45.Simonyan K., Zisserman A. 3rd Int. Conf. Learn. Represent. ICLR 2015 - Conf. Track Proc. 2015. Very deep convolutional networks for large-scale image recognition; pp. 1–14. [Google Scholar]